Abstract
Ribulose-1,5-bisphosphate carboxylase/oxygenase (Rubisco) is the major catalyst in the conversion of carbon dioxide into organic compounds in photosynthetic organisms. However, its activity is impaired by binding of inhibitory sugars such as xylulose-1,5-bisphosphate (XuBP), which must be detached from the active sites by Rubisco activase. Here, we show that loss of two phosphatases in Arabidopsis thaliana has detrimental effects on plant growth and photosynthesis and that this effect could be reversed by introducing the XuBP phosphatase from Rhodobacter sphaeroides. Biochemical analyses revealed that the plant enzymes specifically dephosphorylate XuBP, thus allowing xylulose-5-phosphate to enter the Calvin-Benson-Bassham cycle. Our findings demonstrate the physiological importance of an ancient metabolite damage-repair system in degradation of by-products of Rubisco, and will impact efforts to optimize carbon fixation in photosynthetic organisms.
Similar content being viewed by others
Introduction
The catalytic conversion of atmospheric CO2 into carbohydrates in photosynthetic organisms is largely mediated by a single enzyme – ribulose-1,5-bisphosphate carboxylase/oxygenase (Rubisco). It is estimated that Rubisco is the most abundant protein on Earth1 and fixes about 105 petagrams (10.511 tons) of CO2 annually2. Carboxylation of ribulose-1,5-bisphosphate (RuBP) is the first step in the Calvin-Benson-Bassham (CBB) cycle, which requires the activation of Rubisco through carbamylation of a conserved lysine residue in the active site by a non-substrate CO2 molecule3. The carbamylated lysine is then stabilized by the subsequent binding of a magnesium ion, which enables the efficient electrophilic attack of RuBP by the substrate CO2 molecule. Despite its central role in the CBB cycle, Rubisco has a low turnover rate of 3-10 CO2 molecules per sec4 and its complex reaction mechanism is prone to error. One of its by-products – 2-phosphoglycolate5 – effectively inhibits several enzymes of primary carbon metabolism in photosynthetic organisms, including triosephosphate isomerase6, phosphofructokinase7 and sedoheptulose 1,7-bisphosphate phosphatase8. In addition, 2-phosphoglycolate is recycled in a metabolic process called photorespiration which requires ATP and multiple enzymes, and involves no less than four subcellular compartments9. Another limitation for efficient carboxylation is that protonation and oxygenation of the RuBP enediolate intermediate gives rise to several isomeric pentulose bisphosphates [2,3-pentodiulose-1,5-bisphosphate (PDBP), 3-ketoarabinitol-1,5-bisphosphate and xylulose-1,5-bisphosphate (XuBP)]10, which are also produced during in-vitro studies with isolated Rubisco complexes. In such assays, the effects of these compounds become manifest in the progressive inactivation of Rubisco, which is referred to as ‘fallover’11. RuBP itself can also act as an inhibitory component when it binds to the uncarbamylated form of the enzyme12. Moreover, some plant species produce 2-carboxy-D-arabinitol 1-phosphate (CA1P), which accumulates in the dark or under low light13 and binds only to carbamylated Rubisco14. In contrast to other sugar phosphate inhibitors, CA1P is derived from fructose-1,6-bisphosphate (FuBP) and is synthesized via a pathway that is comprised of five reactions15. All of these inhibitors are structurally similar to transitory forms of RuBP that arise during catalysis and all bind to the enzyme’s active site, thus locking Rubisco into the closed, catalytically impaired complex.
The release of such inhibitors is in turn achieved by the action of the AAA+ (ATPase Associated with diverse cellular Activities) Rubisco activase (Rca), which facilitates conformational changes that convert Rubisco into a catalytically competent complex once again16,17,18. Although Rca enables the release of inhibitors, they must then be rapidly degraded to prevent them from rebinding to Rubisco. For instance, the inhibitor CA1P, which accumulates in the dark, is converted into the non-inhibitory component 2-carboxy-D-arabinitol (CA) by the phosphatase CA1Pase19.
Recently, another neutralising phosphatase has been identified, which is specific for the inhibitory sugar XuBP20. XuBP is generated by misprotonation of the 2,3-enediol derivative of RuBP21 and binds to both the decarbamylated and the carbamylated form of Rubisco22. In plants, rates of synthesis of XuBP are much higher than those of other sugar inhibitors23 and increase with temperature24,25. XuBP is a competitive inhibitor of RuBP and is also a poor substrate for the carboxylation reaction catalyzed by Rubisco26. XuBP phosphatases (CbbY) are conserved in plants and algae, as well as in many autotrophic bacteria, in which CbbY is encoded in the cbb operon (the Calvin-Benson-Bassham cycle operon). A combination of biochemical and structural analyses has revealed that CbbY proteins from Rhodobacter sphaeroides (RsCbbY) and its conserved counterpart in Arabidopsis thaliana (AtCbbYA) are highly specific XuBP phosphatases and transform XuBP into the harmless CBB cycle intermediate xylulose 5-phosphate (Xu5P), which can be recycled back into RuBP20.
In this study, we demonstrate that an ancient metabolite-repair system is required to maintain photosynthesis in plants. In addition, we describe a second metabolite damage-repair enzyme in Arabidopsis chloroplasts, which is able to recycle XuBP. XuBP dephosphorylation activity turns out to be critical for efficient carbon fixation. As shown here, its loss impairs photosynthesis and plant growth. Moreover, we demonstrate that the metabolite-damage repair system found in purple bacteria is compatible with that of land plants, as the introduction of the XuBP phosphatase from Rhodobacter sphaeroides complemented Arabidopsis plants that lacked their own XuBP phosphatases.
Results
The haloacid dehalogenase-like hydrolase AtCbbYB shares sequence similarity with AtCbbYA
In a screen for novel photosynthesis-related proteins in Arabidopsis, we focused on candidates that are (i) shared by photosynthetic eukaryotes from the green lineage and (ii) display a photosynthesis-specific mRNA expression profile. To this end, functional profiling analysis27 of Arabidopsis genes28 that were candidates from the GreenCut2 collection29 was carried out. One of the candidates that was co-expressed with genes (Supplementary Data 1) which exhibited a significant degree of enrichment (adjusted p-value of 4.3 × 10−12) for the functional category ‘photosynthesis’ (gene ontology term GO:0015979) was AT3G48420 (Supplementary Data 2), which is referred to as Arabidopsis CBBYA or AtCBBYA in the following. AtCbbYA belongs to the large class of haloacid dehalogenase-like hydrolases (HAD)30 and shares similarity with the protein product of AT4G39970.1, which is designated as AtCbbYB. A phylogenetic analysis revealed that homologues of AtCbbYB are found in green algae and land plants (Supplementary Fig. 1a, b, Supplementary Table 1). The 3D model of AtCbbYB (Fig. 1a) predicted by AlphaFold31 has a bipartite structure comprised of a core and a cap domain, which is also found in RsCbbY and AtCbbYA20, as well as in other members of the HAD family. Sequence alignment of AtCbbYA, AtCbbYB and RsCbbY (Fig. 1b) revealed that most of the catalytic (7 out of 8) and several signature residues (7 out of 13) defined by analyses of the crystal structure of RsCbbY bound to XuBP20 are conserved in AtCbbYB.
a Structures of CbbY from Rhodobacter sphaeroides (RsCbbY, 4UAT) and CbbYA from Arabidopsis thaliana CbbYA (AtCbbYA, 4UAV) were recently resolved20, and the structure of CbbYB from Arabidopsis thaliana CbbYB (AtCbbYB) was predicted by AlphaFold31. RsCbbY is shown in a complex with XuBP and Mg2+, both of which are highlighted in spherical shapes. Note that the predicted transit peptide (1-63 AA) of AtCbbYB was excluded from the 3D model. b Sequence alignment of At3g48420.1 (AtCbbYA), At4g39970.1 (AtCbbYB) and RsCbbY. Conservation is depicted on a color scale from red (0%) to blue (100%). The positions of catalytic and signature residues conserved in AtCbbYA and RsCbbY20 are depicted in black stars and triangles, respectively. The cap domain is indicated by a turquoise bar above the sequences.
Loss-of-function mutations in AtCbbYA and AtCbbYB impair plant growth
Arabidopsis T-DNA insertion lines for AtCBBYA (atcbbya, Salk Institute line SALK_025204) and AtCBBYB (atcbbyb, John Innes Centre line SM_3.15346) were identified, and the double mutant line atcbbyab was generated by crossing the two single mutants (Fig. 2a). Northern analyses indicated that the AtCBBYA mRNA was truncated in both the atcbbya and atcbbyab lines (Fig. 2b), whereas the AtCBBYB transcript was undetectable in atcbbyb and atcbbyab (Fig. 2c). To analyze the impact of the gene disruptions at the protein level (Fig. 2d), antibodies were raised against recombinantly expressed and purified AtCbbYA and AtCbbYB. Immunodetection assays showed that atcbbya and the double mutant atcbbyab failed to accumulate AtCbbYA, whereas AtCbbYA was still present in atcbbyb (Fig. 2d). Conversely, AtCbbYB could not be detected in either atcbbyb or the double mutant, but accumulated in atcbbya. No obvious growth phenotype was observed for atcbbya or atcbbyb, but growth of the double mutant atcbbyab was clearly reduced relative to wild-type plants under all conditions tested (Fig. 2e, f). We confirmed that the growth phenotype observed in atcbbyab resulted from the knockout of both genes by complementing the atcbbyab mutation with either the AtCBBYA or AtCBBYB gene fused to the eGFP-encoding reporter gene and placed under the control of the Cauliflower Mosaic Virus (CaMV) 35S promoter (Supplementary Fig. 2a–c). Transformed plants bearing eGFP fusion constructs were also employed to determine the subcellular localization of AtCbbYA and AtCbbYB (Supplementary Fig. 3). Both were targeted to the chloroplast, as demonstrated by fluorescence microscopy of isolated protoplasts (Supplementary Fig. 3a). Furthermore, stromal localization was confirmed by fractionation experiments carried out on wild-type plants, followed by immunodetection of AtCbbYA or AtCbbYB (Supplementary Fig. 3b). The fact that plant growth was visibly perturbed only when both proteins were absent indicates that AtCbbYA and AtCbbYB are functionally equivalent with respect to metabolite damage-repair of Rubisco during photosynthesis.
a Exon-intron structures of the AtCBBYA (AT3G48420.1) and AtCBBYB (AT4G39970.1) genes and sites of insertions in the respective T-DNA lines. Left and right T-DNA borders are labelled by “LB” and “RB”, respectively. Exons are shown as white rectangles, UTRs as black rectangles. Positions of T-DNA integrations are highlighted. b, c Northern analyses of AtCBBYA (b) and AtCBBYB (c) transcripts. Total RNA of Col-0, atcbbya, atcbbyb and atcbbyab was extracted, size-fractionated on denaturing agarose gels, transferred onto nylon membranes and examined by hybridization of radiolabeled AtCBBYA- and AtCBBYB-specific probes. As loading controls, abundant rRNAs were visualized by methylene blue staining (M.B.). Northern analyses were carried out once. d Immunodetection of AtCbbYA and AtCbbYB by Western analyses. A Coomassie-stained gel (C.B.B.) bearing size-fractionated proteins extracted from total leaves is shown as loading control. Immunodetections were repeated twice with similar results. e Col-0, atcbbya, atcbbyb and atcbbyab plants grown under a 12/12 h day/night cycle (80–100 µmol photons m−2 s−1) and photographed 26 days after germination. f Growth measurements. Rosette fresh weight was determined from plants grown under the indicated light regimes [8/16 h short day (80–100 µmol photons m−2 s−1), 12/12 h (80–100 µmol photons m−2 s−1), 16/8 h long day (80–100 µmol photons m−2 s−1), and under high light levels (~400 µmol photons m−2 s−1 on a 12/12 h day/night cycle) in the presence of fertilizer]. Ten plants per genotype (9 atcbbyb plants under long day, and 9 atcbbya plants under high light conditions) were examined. Centre lines show the medians, and boxes indicate the 25th and 75th percentiles. Whiskers denote 1.5x the interquartile range. Data points are plotted as circles and outliers are represented by dots. For statistical analyses, the non-parametric Kruskal–Wallis test was performed, followed by pairwise Dunn’s tests. The p-values were adjusted on an experiment level using the Benjamini–Hochberg method. P-values for comparison of each line with Col-0 are indicated and p ≤ 0.05 are marked in bold.
AtCbbYA and AtCbbYB are required for efficient photosynthetic electron transport and CO2 assimilation
The detrimental effects of simultaneous loss of AtCbbYA and AtCbbYB on plant growth prompted us to explore the influence of the two proteins on the light reactions of photosynthesis in more detail (Fig. 3). The maximum quantum yield of PSII (Fv/Fm) as determined in the dark-adapted state was slightly lower (Fig. 3a), whereas the effective quantum yield [Y(II)] at 200 µmol photons m−2 s−1 was reduced to about 50% of wild-type levels in atcbbyab (Fig. 3b). The reduction in PSII quantum efficiency in atcbbyab was shown to be attributable to a significant increase in the yield of regulated, non-photochemical quenching [Y(NPQ)] (Fig. 3c), while the level of non-regulated energy dissipation [Y(NO)] was unaltered (Fig. 3d). We also identified an increase in PSII excitation pressure (1-qP), indicating a higher reduction state of the plastoquinone pool in the double mutant relative to the wild type (Fig. 3e). A pronounced decrease in the quantum yield [Y(I)] of PSI was detected only in the double mutant (Fig. 3f). This effect could be ascribed to a sharp rise in the donor-side limitation [Y(ND)] of PSI (Fig. 3g), whereas the acceptor side limitation [Y(NA)] in the double mutant was slightly lower than in the wild type (Fig. 3h). As expected on the basis of their normal growth phenotype (Fig. 2f), the single mutants behaved like the wild type with respect to all photosynthetic parameters (Fig. 3). Moreover, the aberrant Y(II) and Y(NPQ) phenotypes of atcbbyab were corrected in the complementation lines P35S:AtCbbYA-eGFP and P35S:AtCbbYB-eGFP, as was confirmed by chlorophyll a fluorescence video imaging analyses (Supplementary Fig. 2).
Chlorophyll a fluorescence and P700 parameters of WT (Col-0), atcbbya, atcbbyb and atcbbyab plants grown under long-day conditions at 100 µmol photons m−2s−1. Plants were dark-adapted for 20 min and single leaves were exposed to 200 µmol photons m−2s−1 for 10 min. At the end of the actinic light treatment, a saturating pulse was applied and photosynthetic parameters were recorded. a Maximum quantum yield of PSII [Fv/Fm]. b Effective quantum yield of PSII [Y(II)]. c Regulated energy dissipation in PSII [Y(NPQ)], d Non-regulated energy dissipation in PSII [Y(NO)], e Reduction state of plastoquinone or excitation pressure [1-qP]. f Quantum yield of PSI [Y(I)]. g Donor-side limitation of PSI [Y(ND)]. h Acceptor-side limitation of PSI [Y(NA)]. Horizontal lines represent the medians, and boxes indicate the 25th and 75th percentiles. Whiskers denote 1.5x the interquartile range, outliers are represented by dots. Data points (n = 5) are plotted as open circles. For statistical analyses, the non-parametric Kruskal–Wallis test was performed, followed by pairwise Dunn’s tests. The p-values were adjusted on an experiment level using the Benjamini-Hochberg method. P-values for comparison of each line with Col-0 are indicated and p ≤ 0.05 are marked in bold.
We further investigated the pigment composition of atcbby mutants by high-performance liquid chromatography (Supplementary Table 2). Levels of chlorophylls a and b, as well as total carotenoids were unaltered in all atcbby mutants. However, the pool of violaxanthin, antheraxanthin and zeaxanthin was clearly increased in atcbbyab (72.2 ± 9.3 pmol mg−1 FW) with respect to the wild type (40.2 ± 4.6 pmol mg−1 FW), atcbbya (45.2 ± 5.9 pmol mg−1 FW) and atcbbyb (43.4 ± 6.3 pmol mg−1 FW). Indeed, antheraxanthin levels in atcbbyab (12.4 ± 1.3 pmol mg−1 FW) were about eight-fold higher than in the wild type (1.6 ± 0.5 pmol mg−1 FW), while zeaxanthin reached detectable levels only in the double mutant (8.4 ± 0.9 pmol mg−1 FW).
To quantify the impact of the loss of AtCbbYA and/or AtCbbYB function on CBB cycle activity, we performed gas-exchange measurements while simultaneously monitoring Chl a fluorescence (Fig. 4). First, we determined the response of carbon assimilation rates to photon flux (Fig. 4a) and included in our analysis the Rubisco small subunit (RbcS) knockdown line rbcs1a3b-1 as a control for reduced CBB cycle activity32. CO2 assimilation rates at saturating light intensities of 1200 µmol photons m−2 s−1 (Fig. 4a) were lower in atcbbyab (141 ± 23 µmol CO2 g−1 FW h−1) than in wild type (302 ± 49 µmol CO2 g−1 FW h−1), but not as severely impaired as in the control line rbcs1a3b-1 (79 ± 11 µmol CO2 g−1 FW h−1). As expected, PSII electron transport rates displayed identical trends (Fig. 4b), since the quantum yield of CO2 assimilation and the photochemical yield of PSII have been shown to correlate over a wide range of light intensities33. We also confirmed the high non-photochemical quenching phenotype of atcbbyab (Fig. 3c) and identified an even more pronounced Y(NPQ) phenotype for the RbcS knockdown line rbcs1a3b-1, which was particularly evident at light intensities of 300 and 600 µmol photons m−2 s−1 (Fig. 4c). Increasing the CO2 concentration under saturating light conditions (1200 µmol photons m−2 s−1) could not restore atcbbyab and rbcs1a3b-1 assimilation rates to wild-type levels (Fig. 4d). Analyses of gas-exchange kinetics and Chl a fluorescence in the single mutants atcbbya and atcbbyb did not reveal any obvious differences from the wild type (Supplementary Fig. 4a–c). Furthermore, the absence of either AtCbbYA or AtCbbYB or both had no effects on the level of either Rubisco or Rubisco activase, as demonstrated by immunodetection assays (Supplementary Fig. 4d). Based on these results, we deduced that both AtCbbYA and AtCbbYB are required for the maintenance of efficient photosynthetic electron transport and carbon dioxide assimilation. In contrast to rbcs1a3b-1, in which decreased CO2 assimilation rates were correlated with reduced Rubisco content32, the data for atcbbyab indicated that lower rates of carbon fixation could be due to inefficient degradation of inhibitors of Rubisco in the absence of both AtCbbYA and AtCbbYB.
CO2 assimilation and chlorophyll a fluorescence of wild type, atcbbyab and the Rubisco knockdown mutant rbcs1a3b-1 were studied using the gas-exchange and fluorescence system GFS-3000 (Walz®, Effeltrich, Germany). a CO2 assimilation rates were determined at various light intensities (0, 50, 100, 300, 600 and 1200 µmol photons m−2 s−1) while the ambient CO2 concentration in the measuring chamber was kept constant (46 Pa). b, c Electron transport rates through PSII [ETR(II)] and non-photochemical quenching [Y(NPQ)] parameters were recorded simultaneously with CO2 assimilation rates (a). d CO2 assimilation rates plotted as a function of increasing intercellular CO2 mole fraction [pCi] were determined at a saturating light intensity of 1200 µmol photons m−2 s−1. Three individual plants per genotype were analyzed. Averages (crosses) and standard deviations are provided.
AtCbbYA and AtCbbYB dephosphorylate XuBP
Since AtCbbYA had been shown to dephosphorylate XuBP20, we investigated the substrate specificity and kinetic behaviour of AtCbbYB. Fusions of AtCbbYA and AtCbbYB to the maltose-binding protein (MBP) were heterologously expressed in E. coli and subsequently purified under native conditions with the aid of an amylose matrix (Supplementary Fig. 5). Both fusion proteins released one molecule of phosphate per molecule of XuBP and RuBP, but were unable to dephosphorylate either xylulose-5-phosphate (Xu5P) or ribulose-5-phosphate (Ru5P) (Fig. 5a). In contrast, calf intestinal alkaline phosphatase (CIP), with its broad substrate specificity, liberated two phosphates per molecule of XuBP or RuBP, and was also able to dephosphorylate both Xu5P and Ru5P. These results indicated that MBP-AtCbbYB specifically cleaves the phosphate at position 1 in XuBP and RuBP, as has previously been demonstrated for AtCbbYA20. Kinetic analyses with XuBP as substrate revealed that the Michaelis-Menten constant (KM) for MBP-AtCbbYB (0.182 ± 0.03 mM) was ~4.5-fold higher than for MBP-AtCbbYA (0.04 ± 0.001 mM) (Fig. 5b, Table 1). Moreover, the turnover number (kcat) for MBP-AtCbbYB (0.54 ± 0.05 s−1) was ~4 times lower than that of MBP-AtCbbYA (2.13 ± 0.04 s−1). Analyses with RuBP as substrate revealed similar KM values for MBP-AtCbbYA (4.31 ± 0.11 mM) and MBP-AtCbbYB (4.84 ± 0.36 mM) (Fig. 5b, Table 1). Turnover numbers for RuBP relative to XuBP were ~50- and ~35-fold lower for MBP-AtCbbYA (0.043 ± 0.009 s−1) and MBP-AtCbbYB (0.015 ± 0.002 s−1), respectively, indicating that both phosphatases act selectively on XuBP. Consequently, MBP-AtCbbYA and MBP-AtCbbYB exhibited ~5000 and ~1000 higher catalytic efficiency (kcat/KM), respectively, with XuBP than RuBP.
a Substrate specificity of MBP-AtCbbYA and MBP-AtCbbYB. Calf intestinal phosphatase (CIP) served as a control. Phosphate release from the substrates XuBP, RuBP, Xu5P, and Ru5P was quantified using colorimetric malachite-green assays. Horizontal lines represent the medians, and boxes indicate the 25th and 75th percentiles. Whiskers denote 1.5x the interquartile range. Numbers of replicates are indicated. b Michaelis-Menten plots for MBP-AtCbbYA and MBP-AtCbbYB with XuBP and RuBP as substrates. Three measurements were conducted in each experiment. Note that a plot of CbbY kinetics with RuBP as substrate and a fitted scaling of the y-axis is provided in Supplementary Fig. 5b.
Overall, our biochemical analyses of MBP-AtCbbYA reproduced previously published data based on untagged AtCbbYA20 and demonstrated that Arabidopsis contains a second phosphatase that has the ability to degrade the Rubisco inhibitor XuBP to Xu5P. Both phosphatases displayed physiologically relevant high selectivity for XuBP, thus preventing dephosphorylation of the isomeric Rubisco substrate RuBP and subsequent impairment of the carboxylation reaction in the CBB cycle.
The XuBP phosphatase from Rhodobacter sphaeroides can substitute for AtCbbYA and AtCbbYB
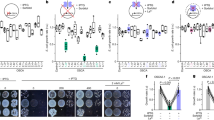
Since CbbY homologues have been identified in plants and algae, as well as in many autotrophic bacteria (Supplementary Fig. 1a), we investigated the functional compatibility of the XuBP phosphatase found in Rhodobacter sphaeroides (RsCbbY) with those of Arabidopsis. To this end, the codon usage of the RscbbY gene was optimized for efficient expression in Arabidopsis, and fused to the sequence coding for the predicted plastid transit peptide of AtCbbYA. Moreover, the construct was placed under the control of the cauliflower mosaic virus 35S promoter and fused to the eGFP-encoding reporter gene. After introduction of the overexpressor construct into the atcbbyab background, two independent lines (P35S:TP-RsCbbY-eGFP #1 and #2) were selected. The presence of the fusion was verified by immunodetection assays (Fig. 6a), and localization of TP-RsCbbY-eGFP to the chloroplasts was confirmed by fluorescence microscopy of isolated protoplasts (Fig. 6b). Remarkably, P35S:TP-RsCbbY-eGFP plants complemented the mutant photosynthetic phenotype, as indicated by the restoration of wild-type-like Y(II) and Y(NPQ) parameters during light induction experiments at 185 µmol photons m−2 s−1 (Fig. 6c).
a Immunodetection of TP-RsCbbY-eGFP using an eGFP-specific antibody. The absence of AtCbbYA and AtCbbYB in the two P35S:TP-RsCbbY-eGFP lines was confirmed using antibodies generated in this study. The PVDF membrane stained with Coomassie brilliant blue G-250 (C.B.B.) served as a loading control. Immunodetections were repeated twice with similar results. b Localization of TP-RsCbbY-eGFP to chloroplasts was verified by fluorescence microscopy. The integrity of the protoplasts was assessed by bright-field microscopy and chloroplasts were detected by chlorophyll fluorescence (Auto). Fluorescence signals derived from TP-RsCbbY-eGFP (eGFP) were merged with chloroplast-derived chlorophyll fluorescence signals (Merged). c Photosynthetic activity in Col-0, atcbbyab and two independent P35S:TP-RsCbbY-eGFP overexpressor lines (atcbbyab background) was quantified with an Imaging-PAM system (Walz®, Effeltrich, Germany) in a light induction experiment (185 µmol photons m−2 s−1). Effective quantum yields of PSII [Y(II)] and levels of non-photochemical quenching [Y(NPQ)] are displayed on a false-color scale. Data were obtained from 5 leaves of 5 plants (n = 25) per genotype. Horizontal lines represent the medians, and boxes indicate the 25th and 75th percentiles. Whiskers denote 1.5x the interquartile range, outliers are represented by dots. Data points are plotted as circles. For statistical analyses, the non-parametric Kruskal–Wallis test was used, followed by pairwise Dunn’s tests. The p-values were adjusted on an experiment level using the Benjamini–Hochberg method. P-values for comparison of complemented lines with atcbbyab are indicated and p ≤ 0.05 are marked in bold.
Discussion
Metabolite damage is a widespread phenomenon that occurs in all living organisms and has been studied in more detail in several metabolic contexts, such as glycolysis34, amino-acid synthesis35 and damage to phosphometabolites36. In particular, plants that have to cope with rapidly changing and extreme environmental conditions are expected to be more susceptible to metabolite damage, and it is estimated that large numbers of uncharacterized genes are associated with the repair of metabolite damage37. Although the inhibitory effect of XuBP on Rubisco activity has long been known38, the metabolite-repair mechanism has been elucidated at the biochemical level only recently20. In our study, we reveal now its physiological importance in plant photosynthesis which has remained unrecognized until now due to redundant functions of the two XuBP phosphatases. Taken together, the identification of a second XuBP phosphatase in Arabidopsis (Fig. 1) and the pronounced phenotype of the atcbbyab mutant underline the significance of functional cooperation between Rubisco activase and sugar phosphatases for the maintenance of normal growth yields (Fig. 2e, f) and photosynthesis (Figs. 3, 4). Both findings demonstrate in planta that the release of XuBP from Rubisco, mediated by Rubisco activase, is insufficient to keep photosynthesis running efficiently, unless the CbbYA/CbbYB degradation system is available to prevent the inhibitor from rebinding (Fig. 7).
The Rubisco inhibitor XuBP is efficiently converted into xylulose-5-phosphate by functional cooperation between Rubisco activase (Rca) and the two XuBP phosphatases CbbYA and CbbYB in the wild type (WT, left panel). The absence of CbbY phosphatases in the atcbbyab double mutant results in Rubisco inhibition, downregulation of CBB cycle activity and the onset of photosynthetic control (right panel) including enhanced non-photochemical quenching [Y(NPQ)] and increased donor-side limitation of PSI [Y(ND)]. Xylulose-5-phosphate (Xu5P), 3-phosphoglycerate (3PGA), glyceraldehyde 3-phosphate (G3P), Ribulose 1,5-bisphosphate (RuBP).
Interestingly, atcbbyab plants that were grown under high levels of light (~400 µmol photons m−2 s−1) were much smaller than the wild type (Fig. 2f), This may be due to the increased accumulation of XuBP that occurs under conditions in which carbon assimilation rates are enhanced. However, photosynthesis was already affected in atcbbyab plants that were exposed to moderate light intensities (~100 µmol photons m−2 s−1; Fig. 3) and this could be ascribed to the reduced activity of the CBB cycle (Fig. 4), and the ensuing limited availability of both ADP and the final electron acceptor NADP+ for light reactions (Fig. 7). As a consequence, the trans-thylakoid proton gradient was enhanced in atcbbyab, and ΔpH-dependent quenching mechanisms were triggered, which then led to increased xanthophyll cycle activity (Supplementary Table 2) and regulated heat dissipation in the PSII antenna (Fig. 3c). In addition, increased luminal acidification downregulated plastoquinol oxidation in the Q cycle of the Cyt b6f complex in the double mutant (Fig. 3e). This mechanism was previously referred to as ‘photosynthetic control’39,40, since it prevents excessive photosynthetic electron transport to PSI. In fact, PSI photochemistry was donor-side-limited in atcbbyab (Fig. 3g) and over-reduction at the acceptor side of PSI was alleviated (Fig. 3h). Accordingly, the photosynthetic apparatus was efficiently protected from photodamage by downregulation of linear electron transport in atcbbyab, as shown by the fact that maximal quantum yields (Fv/Fm) of PSII remain close to wild-type levels (Fig. 3a).
Although both AtCbbYA and AtCbbYB can dephosphorylate XuBP to Xu5P (Fig. 5) and substitute for each other in complementation experiments (Supplemental Fig. 2), the question remains as to why two different XuBP phosphatases evolved in the green lineage (Supplemental Fig. 1). One possible explanation is that AtCbbYB can dephosphorylate sugar phosphates other than XuBP that inhibit Rubisco activity. Two observations support this idea: (i) several CbbY signature and catalytic residues are specific to AtCbbYA and not found in AtCbbYB (Fig. 1b) and (ii) its catalytic efficiency on XuBP is lower than that of AtCbbYA (Table 1). A comparable case may be advanced for CA1Pase, which dephosphorylates both CA1P and PDBP19. This catalytic promiscuity was revealed upon the addition of recombinant CA1Pase to in-vitro assays, which prevented PDBP-dependent fallover of Rubisco activity.
Despite the reduction in carbon assimilation rates, the atcbbyab phenotype did not result in lethality (Fig. 2). This suggests the existence of an additional process responsible for XuBP removal in the absence of AtCbbYA and AtCbbYB. One possible explanation is that Rubisco carboxylation of XuBP26 is sufficient to maintain its residual enzymatic activity required for photoautotrophic growth. Alternatively, an unidentified phosphatase or enzyme could exist that recycles XuBP. In future studies, the simultaneous determination of Rubisco and Rubisco inhibitor concentrations in the chloroplasts of the different mutant lines will provide a deeper understanding of CbbY substrate specificity and the extent of Rubisco inactivation.
The restoration of wild-type-like photosynthesis observed upon targeting of the bacterial XuBP phosphatase RsCbbY to chloroplasts revealed that CbbY enzymes represent an ancient metabolite-repair system (Fig. 6). Compared to only a minor reduction in specific Rubisco activity of 10% in the R. sphaeroides strain without RsCbbY and the phosphoglycolate phosphatase RsCbbZ41, assimilation rates dropped considerably to about 50% of wild-type levels in atcbbyab (Fig. 4). One explanation for this discrepancy is that purple bacteria and plants exhibit different kinetics in Rubisco misfiring and repair enzyme reactions. Another possibility is that purple bacteria have alternative, unknown mechanisms to remove both XuBP and 2-phosphoglycolate. In addition, XuBP and other inhibitory sugar phosphates derived from Rubisco’s side reactions might be part of as yet unknown, plant-specific regulatory networks42 that orchestrate the downregulation of photosynthetic electron transport and CBB cycle activity.
In conclusion, the discovery of the physiological significance of instances of ‘misfiring’ by Rubisco also has implications for efforts to increase CO2 fixation efficiency in photosynthetic organisms. Since XuBP is inherently produced by Rubisco and binds directly to its active site, Rubisco encapsulation strategies, such as engineering a pyrenoid-based CO2-concentrating mechanism43,44 or cyanobacterial carboxysomes45 in crops, need to be re-evaluated in terms of efficient release and degradation of Rubisco inhibitors in synthetic organellar subcompartments.
Methods
Growth conditions and plant material
Plants were grown in potting soil (A210, Stender AG, Schermbeck, Germany) exposed to a light regime of 80–120 μmol photons m−2 s−1 (12/12 h, light/dark) under temperature-controlled conditions (22 °C in the light phase, 18 °C in the dark phase). Plants used for crossing and seed propagation were kept under greenhouse conditions on a 14/10 h light/dark cycle, and an additional light source (HQI Powerstar 400 W/D, Osram) was used to provide supplemental illumination, resulting in a daytime light intensity of 180 µmol photons m−2 s−1. Wuxal Super fertilizer (8% nitrogen, 8% phosphorus, and 6% potassium; MANNA Wilhelm Haug GmbH & Co. KG, Ammerbuch-Pfäffingen, Germany) was used according to the manufacturer’s instructions. High light treatment was carried out at a light intensity of 300–400 μmol photons m−2 s−1 (12/12 h, light/dark) under a high-pressure mercury-vapor lamp (Powerstar, HQI-T 1000 W/D, Osram, Germany).
The single mutants atcbbya (SALK_025204, insertion in AT3G48420) and atcbbyb (SM_3.15346, insertion in AT4G39970) were obtained from the SALK46 and SLAT (John Innes Centre) collections47, respectively. The double mutant atcbbyab was generated by crossing the single mutants and homozygous plants were screened by genotyping plants from the segregating T2 generation. DNA was extracted as described previously48 and combinations of gene- and T-DNA-specific primers were employed in PCR assays (see Supplementary Table 3 for primer information). Generation and characterization of the Rubisco knockdown line rbcs1a3b-1 was described previously32.
The AtCbbYA-eGFP and AtCbbYB-eGFP constructs used in overexpression experiments were assembled as follows. Coding regions (gene models AT3G48420.1 and AT4G39970.1 according to TAIR) were cloned into the binary Gateway destination vector pB7FWG249, thus placing them under the control of the 35S promoter. For cross-species complementation, the codon usage of the RscbbY gene (GenBank: U67781.1) was optimized for efficient expression in Arabidopsis, synthesized as a gBlock® (IDT®) and fused to the sequence coding for the predicted plastid transit peptide (TP) of AtCbbYA (1-74 aa). The fusion was subsequently cloned into the Gateway vector pB7FWG2, which placed TP-RsCbbY under the control of the CaMV 35S promoter and fused it with the eGFP-encoding reporter gene. All constructs were transformed into Agrobacterium tumefaciens strain GV3101, and then into atcbbyab plants by the floral-dip method50. Positive transformants were selected by restored plant growth, the presence of the GFP reporter system, and immunodetection of the fusion proteins. For each transformation, two independent, homozygous lines were isolated from the T3 generation and subjected to biochemical and physiological analyses.
Chlorophyll a fluorescence, P700 and gas exchange measurements
Chlorophyll a fluorescence and P700 parameters were determined with the Dual-PAM system (Walz, Effeltrich, Germany). The measurements shown in Fig. 3 were carried out on non-fertilized plants grown under long-day conditions (16/8 h, light/dark) for 4 weeks. Plants were dark-incubated for 20 min and then subjected to a light induction curve protocol at a light intensity of 196 µmol photons m−2 s−1 for 10 min. The maximum quantum yield of PSII [Fv/Fm = (Fm – Fo) / Fm)] was determined from fluorescence parameters (Fo and Fm), which were derived from the first saturating light pulse after dark incubation. Then, saturating light pulses were applied every 30 s and photosynthetic parameters were calculated by the Dual-PAM software based on equations previously described51,52. Photosynthetic parameters of complemented lines were determined with an Imaging-PAM system (Walz, Effeltrich, Germany). Plants grown without fertilizer on a 12/12 h light/dark cycle (80–120 μmol photons m−2 s−1) for 4-5 weeks were dark-incubated for 20 min. Saturating light pulses were applied every 20 s over 10 min in a light induction experiment (185 µmol photons m−2 s−1) to determine fluorescence parameters in the light (Fm’ and F). The effective quantum yield of PSII [Y(II)] and the yield of non-photochemical quenching [Y(NPQ)] were calculated by the equations Y(II) = (Fm’ - F)/Fm’ and Y(NPQ) = F/Fm’ - F/Fm, respectively52.
Rates of gas exchange were determined simultaneously with measurements of chlorophyll a fluorescence parameters using a GFS-3000 system (Walz, Effeltrich, Germany) equipped with an Arabidopsis Chamber (3010-A, Walz, Effeltrich, Germany). Col-0, atcbbya and atcbbyb were analyzed after 4 weeks of growth on a 12/12 h light/dark cycle (80–120 μmol photons m−2 s−1). The genotypes atcbbyab and rbcs1a3b-1 were grown for 2 and 3 weeks longer, respectively, allowing them to reach developmental stages comparable to Col-0, atcbbya and atcbbyb plants. First, assimilation rates and chlorophyll fluorescence parameters were recorded at six different light intensities (0, 50, 100, 300, 600, and 1200 µmol photons m−2 s−1) at a constant CO2 concentration (46 Pa). Subsequent measurements were then carried out at CO2 pressures of 5, 20, 91 and 122 Pa. CO2 assimilation rates were expressed relative to rosette leaf fresh weight (µmol CO2 h−1 g−1 FW), which was determined for each plant after the gas-exchange measurement. The intercellular CO2 mole fraction pCi was determined and calculated according to Caemmerer and Farquhar53.
Pigment analyses
Pigments of rosette leaves were isolated from four-week-old plants grown on a 12/12 h light/dark cycle (80–120 μmol photons m−2 s−1). Leaves were harvested 3 h after the shift from the dark into the light and immediately homogenized in liquid nitrogen. Pigments were extracted with 99.5% acetone and separated from leaf debris by centrifugation (16,000 g) for 20 min at 4 °C. Pigments were separated by reverse-phase HPLC using a LiChrospher 100 RP-18 column with a particle size of 5 µm (Merck, Darmstadt, Germany)54. Solvent A consisted of a mixture of acetonitrile, methanol, and 0.1 M Tris/NaOH (pH 8.0), in a ratio of 87:10:3. Solvent B contained methanol and hexane in a ratio of 4:1. The gradient from solvent A to solvent B was developed from 9 to 12.5 min at a flow rate of 2 mL/min and eluted pigments were detected at 440 nm. The concentration of the pigments was calculated from the integrated peak area using conversion factors determined by calibration with pure pigments.
Northern analyses
Total RNA was isolated with the aid of the TRIzol reagent (Invitrogen) from Col-0, atcbbya, atcbbyb and atcbbyab leaves that had been frozen and homogenized in liquid nitrogen. Northern analyses were carried out according to standard protocols55. Samples equivalent to 10 µg of total RNA were size-fractionated by electrophoresis in formaldehyde-containing agarose gels (1.2%). RNA was transferred overnight onto nylon membranes (Hybond-N+, Amersham Bioscience) by capillary transfer and subsequently fixed by UV radiation (Stratalinker® UV Crosslinker 1800). Ribosomal RNAs on nylon membranes were quantified by staining with methylene blue solution (0.02% [w/v] methylene blue, 0.3 M sodium acetate pH 5.5) to control for equal loading. Amplicons derived from cDNA were labeled with radioactive [α-32P]dCTP (Hartmann Analytic, Braunschweig, Germany) and subsequently used as probes for the detection of AT3G48420 and AT4G39970 transcripts in hybridization experiments (see Supplementary Table 3 for primer information). Signals were recorded with the Typhoon Phosphor Imager System (GE Healthcare).
Immunoblot analyses, protein purification, and antibody generation
Rosette leaves were harvested, frozen in liquid nitrogen, and homogenized in loading buffer (100 mM Tris-HCl, pH 6.8, 50 mM DTT, 8% [w/v] SDS, 24% [w/v] glycerol, and 0.02% [w/v] bromophenol blue). Proteins were denatured for 5 min at 70 °C. After centrifugation (5 min, 12,000 g, room temperature), solubilized proteins in the supernatant were fractionated on Tricine-SDS-PAGE (10% gels)56. Loading was adjusted to leaf fresh weight. Protein transfer to PVDF membranes (Immobilon-P, Millipore) was conducted using a semidry blotting system (Bio-Rad) following the supplier’s instructions. PVDF membranes were stained with a Coomassie brilliant blue G-250 solution56 to control for efficient transfer and sample loading. PVDF membranes were subsequently blocked with TBST buffer (10 mM Tris-HCl, pH 8, 150 mM NaCl, and 0.1% [v/v] Tween 20) supplemented with 3% (w/v) skim milk powder. Then, membranes were incubated overnight at 4 °C in blocking buffer containing primary antibodies. After washing three times with TBST buffer, membranes were treated with secondary antibodies in TBST supplemented with 3% (w/v) skim milk powder. After three more washes with TBST, signals were detected by enhanced chemiluminescence (Pierce, Thermo Scientific) using an enhanced chemiluminescence reader system (Fusion FX7, PeqLab).
For protein purification, AtCbbYA and AtCbbYB were heterologously expressed in BL21 (DE3) E. coli cells. Sequences encoding AtCbbYA66-319aa and AtCbbYB47-316aa were cloned into the expression vectors pET151 D-TOPO (ThermoFisher) and pET101 D-TOPO (ThermoFisher), respectively, resulting in 6xHis N-terminally tagged AtCbbYA66-319aa and 6xHis C-terminally tagged AtCbbYB47-316aa fusions, which were subsequently purified under native conditions using nickel-nitrilotriacetic acid agarose (Qiagen) following the manufacturer’s instructions. For enzyme assays and antibody purification, AtCbbYA66-319aa and AtCbbYB47-316aa were fused to the maltose-binding protein (MBP). To this end, coding sequences were cloned into the pMal-c5x vector (New England Biolabs, NEB, Ipswich, MA, USA) and transformed into BL21 (DE3) E. coli cells. After heterologous expression, MBP-AtCbbYA and MBP-AtCbbYB were purified using an amylose resin (New England Biolabs) in accordance with the manufacturer’s instructions.
For antibody production, 6xHis-AtCbbYA66-319aa and AtCbbYB47-316aa-6xHis were injected into rabbits. The 6xHis-AtCbbYA66-319aa antibody was generated in Roberto Barbato’s laboratory. AtCbbYB47-316aa-6xHis antibodies were generated by Pineda (Berlin, Germany) and subsequently purified by affinity chromatography with immobilized MBP-AtCbbYB. Detection of the RsCbbY-eGFP fusion was performed with a commercially available GFP-specific antibody (A-6455, ThermoFisher Scientific) at a dilution of 1:7500. Rubisco activase (AS10 700, dilution of 1:5000) and the large subunit of Rubisco RbcL (AS03 037, dilution of 1:5000) were immunodetected with antibodies obtained from Agrisera. The secondary antibody (Sigma-Aldrich A9169, Goat Anti-Rabbit, IgG Antibody, HRP-conjugate) was used in a 1:30,000 dilution.
Enzyme kinetics
Xylulose-1,5-bisphosphate (XuBP) was synthesized by enzyme-catalyzed aldol addition of dihydroxyacetone phosphate to glycolaldehyde phosphate20. The purity of XuBP-Li2 was estimated by MBP-CbbYA-mediated phosphate release. Ribulose-1,5-bisphosphate (RuBP), xylulose-5-phosphate (Xu5P) and ribulose-1,5 phosphate (Ru5P) were purchased from Merck (Darmstadt, Germany). MBP-AtCbbYA and MBP-AtCbbYB were isolated as described above and were employed for enzyme activity assays, because they were of higher purity than the non-fused proteins. Calf intestinal phosphatase (CIP) was obtained from New England Biolabs and served as a positive control. Enzyme activity was assessed by determining the release of inorganic phosphate from sugar phosphates using the colorimetric malachite green assay57. Malachite Green was purchased from Merck. Assays were carried out in a phosphate-free buffer (MOPS-KOH pH 7.5, 100 mM KCl, 10 mM MgCl2) in the presence of the CbbY cofactor Mg2+ at 25 °C20.
Subcellular localization analyses
Chloroplasts were isolated on a two-step Percoll gradient58. Leaves (20 g) from 4-week-old and dark-adapted wildtype plants were homogenized in a solution containing 450 mM sorbitol, 20 mM Tricine-KOH (pH of 8.4), 10 mM EDTA, 10 mM NaHCO3, and 0.1% (w/v) bovine serum albumin (BSA). The homogenate was filtered using two layers of Miracloth (Calbiochem) and then centrifuged (500 g) for 6 min at 4 °C. The sedimented chloroplasts were suspended in a solution containing 800 µL of 0.3 M sorbitol, 20 mM Tricine-KOH (pH of 8.4), 2.5 mM EDTA, and 5 mM MgCl2, placed on top of a two-step Percoll gradient (40–80% [v/v]) and centrifuged (6500 g) for 20 min at 4 °C. Intact chloroplasts from the interface were collected and ruptured in a buffer containing 20 mM HEPES (pH 7.5, adjusted with KOH), and 10 mM EDTA. After 30 min on ice, insoluble proteins were separated from the soluble fraction by centrifugation at 35,000 g for 30 min at 4 °C. Mitochondria were isolated as described previously59. Successful fractionation was verified by immunodetection of the following marker proteins: cytochrome oxidase subunit II (CoxII) for mitochondria (Agrisera AS04 053 A), LHCII type III chlorophyll a/b-binding protein (Lhcb3) for the insoluble chloroplast fraction (Agrisera, AS01 002), and Csp41b for the soluble chloroplast fraction (the Csp41b antibody was kindly provided by David Stern).
For protoplast isolation, leaves from 3- to 5-week-old plants (P35S:AtCbbYA-eGFP, P35S:AtCbbYB-eGFP and P35S:RsCbbY-eGFP) were cut into strips, vacuum-infiltrated with enzyme solution (2.6 mg/mL macerozyme, 10 mg/mL cellulase, 10 mM MES pH 5.7, 20 mM KCl, 0.5 M mannitol, 10 mMCaCl2, and 1 mg/mL BSA) and incubated for 3-4 h in the dark at room temperature on a shaker (50 rpm). Protoplasts were then filtered through a nylon membrane (72 μm pore size), pelleted by centrifugation (50 × g, 10 min), resuspended in 8.5 mL of a saccharose-containing buffer (10 mM MES pH 5.7, 20 mM MgCl2, and 120 mg/mL saccharose) and overlaid with 2 mL of a mannitol-containing buffer (10 mM MES pH 5.7, 10 mM MgCl2, 10 mM MgSO4, and 0.5 M mannitol). Protoplasts found at the interphase after centrifugation (50 × g, 10 min) were washed once and then resuspended in the overlay buffer. Chlorophyll autofluorescence and eGFP signals were detected using an Axio Imager fluorescence microscope (Zeiss).
Bioinformatic and statistical analyses
Co-expression studies of AtCBBYA (AT3G48420) were done with ATTED28 and functional profiling of co-expressed genes was analyzed by g:Profiler27. Transit peptides were predicted by TargetP60. Prediction of AtCbbYB structure was conducted by AlphaFold31. Protein sequence alignments and the phylogenetic tree were constructed with the CLC workbench software (version 20). The evolutionary history of AtCbbYA, AtCbbYB and RsCbbY was inferred from sequence comparisons using the maximum-likelihood method61. The tree was drawn to scale, with branch lengths measured in the number of substitutions per site according to the Whelan and Goldman method62. The phylogenetic tree involved 24 amino-acid sequences (sequence identifiers are found in Supplementary Table 1) and a bootstrap analysis with 1000 replicates.
Boxplots were drawn with the web-tool BoxPlotR63. Statistical analyses were performed in R v3.5.2 (https://www.r-project.org/). Non-parametric Kruskal-Wallis tests were used, which were followed by pairwise Dunn’s tests employing the R package dunn.test. P-values were adjusted on an experiment level using the Benjamini–Hochberg method.
Accession numbers
AtCbbYA (At3g48420), AtCbbYB (At4g39970), RsCbbY (UniProt ID P95649).
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Data availability
The authors declare that all data presented in this study are available in the figures and the accompanying Supplementary Information file. The source data underlying Fig. 2 b–f; Fig. 3; Fig. 4; Fig. 5; Fig. 6a, c; Supplementary Fig. 2b, c; Supplementary Fig. 3b; Supplementary Fig. 4; Supplementary Fig. 5; and Supplementary Table 2, as well as detailed corresponding statistics, are provided as a Source Data file. Other data that support the study are available from the corresponding author. Source data are provided with this paper.
References
Bar-On, Y. M. & Milo, R. The global mass and average rate of rubisco. P. Natl. Acad. Sci. USA. 116, 4738–4743 (2019).
Field, C. B., Behrenfeld, M. J., Randerson, J. T. & Falkowski, P. Primary production of the biosphere: integrating terrestrial and oceanic components. Science (N. Y., NY) 281, 237–240 (1998).
Lorimer, G. H. & Miziorko, H. M. Carbamate Formation of the e-Amino Group of a Lysyl Residue as the Basis for the Activation of Ribulosebisphosphate Carboxylase by CO2 and Mg2. Biochem.-Us 19, 5321–5328 (1980).
Hartman, F. C. & Harpel, M. R. Structure, Function, Assembly of Carboxylase/Oxygenase. Annu Rev. Biochem. 63, 197–234 (1994).
Tolbert, N. E. The C2 oxidative photosynthetic carbon cycle. Annu. Rev. Plant Physiol. Plant Mol. Biol. 48, 1–25 (1997).
Anderson, L. E. Chloroplast and cytoplasmic enzymes. II. Pea leaf triose phosphate isomerases. Biochim Biophys. Acta. 235, 237–244 (1971).
Kelly, G. J. & Latzko, E. Chloroplast Phosphofructokinase: II. Partial Purification, Kinetic and Regulatory Properties. Plant Physiol. 60, 295–299 (1977).
Flugel, F. et al. The Photorespiratory Metabolite 2-Phosphoglycolate Regulates Photosynthesis and Starch Accumulation in Arabidopsis. Plant Cell. 29, 2537–2551 (2017).
Hagemann, M. et al. Evolution of photorespiration from cyanobacteria to land plants, considering protein phylogenies and acquisition of carbon concentrating mechanisms. J. Exp. Bot. 67, 2963–2976 (2016).
Parry, M. A. et al. Rubisco regulation: A role for inhibitors. J. Exp. Bot. 59, 1569–1580 (2008).
Edmondson, D. L., Badger, M. R. & Andrews, T. J. A Kinetic Characterization of Slow Inactivation of Ribulosebisphosphate Carboxylase during Catalysis. Plant Physiol. 93, 1376–1382 (1990).
Jordan, D. B. & Chollet, R. Inhibition of ribulose bisphosphate carboxylase by substrate ribulose 1,5-bisphosphate. J. Biol. Chem. 258, 13752–13758 (1983).
Moore, B. & Seemann, J. R. Evidence That 2-Carboxyarabinitol 1-Phosphate Binds to Ribulose-1,5-Bisphosphate Carboxylase in Vivo. Plant Physiol. 105, 731–737 (1994).
Gutteridge, S. et al. A nocturnal inhibitor of carboxylation in leaves. Nature 324, 274–276 (1986).
Andralojc, P. J., Keys, A. J., Kossmann, J. & Parry, M. A. J. Elucidating the biosynthesis of 2-carboxyarabinitol 1-phosphate through reduced expression of chloroplastic fructose 1,6-bisphosphate phosphatase and radiotracer studies with 14CO2. P Natl Acad. Sci. USA. 99, 4742–4747 (2002).
Portis, A. R. Jr. Rubisco activase-Rubisco’s catalytic chaperone. Photosynthesis Res. 75, 11–27 (2003).
Portis, A. R., Li, C., Wang, D. & Salvucci, M. E. Regulation of Rubisco activase and its interaction with Rubisco. J. Exp. Bot. 59, 1597–1604 (2008).
Hauser, T., Popilka, L., Hartl, F. U. & Hayer-Hartl, M. Role of auxiliary proteins in Rubisco biogenesis and function. Nat. plants 1, 1–11 (2015).
Andralojc Paul, J. et al. 2-Carboxy-D-arabinitol 1-phosphate (CA1P) phosphatase: evidence for a wider role in plant Rubisco regulation. Biochemical J. 442, 733–742 (2012).
Bracher, A., Sharma, A., Starling-Windhof, A., Hartl, F. U. & Hayer-Hartl, M. Degradation of potent Rubisco inhibitor by selective sugar phosphatase. Nat. Plants 1, 14002 (2015).
Edmondson, D. L., Kane, H. J. & Andrews, T. J. Substrate isomerization inhibits ribulosebisphospate carboxylase-oxygenase during catalysis. Febs Lett. 260, 62–66 (1990).
Zhu, G. & Jensen, R. G. Xylulose 1,5-Bisphosphate Synthesized by Ribulose 1,5-Bisphosphate Carboxylase/Oxygenase during Catalysis Binds to Decarbamylated Enzyme. Plant Physiol. 97, 1348–1353 (1991).
Pearce, F. G. Catalytic by-product formation and ligand binding by ribulose bisphosphate carboxylases from different phylogenies. Biochem. J. 399, 525–534 (2006).
Salvucci, M. E. & Crafts-Brandner, S. J. Mechanism for deactivation of Rubisco under moderate heat stress. Physiologia Plant. 122, 513–519 (2004).
Schrader, S. M., Kane, H. J., Sharkey, T. D. & Von Caemmerer, S. High temperature enhances inhibitor production but reduces fallover in tobacco Rubisco. Funct. Plant Biol. 33, 921–929 (2006).
Yokota, A. Carboxylation and detoxification of xylulose bisphosphate by spinach ribulose bisphosphate carboxylase/oxygenase. Plant Cell Physiol. 32, 755–762 (1991).
Raudvere, U. et al. g:Profiler: a web server for functional enrichment analysis and conversions of gene lists (2019 update). Nucl. Acids Res. 47, W191–W198 (2019).
Obayashi, T., Aoki, Y., Tadaka, S., Kagaya, Y. & Kinoshita, K. ATTED-II in 2018: A Plant Coexpression Database Based on Investigation of the Statistical Property of the Mutual Rank Index. Plant Cell Physiol. 59, e3 (2018).
Karpowicz, S. J. & Prochnik, S. E. Grossman aR, Merchant SS. The GreenCut2 Resource, a Phylogenomically Derived Inventory of Proteins Specific to the Plant Lineage. J. Biol. Chem. 286, 21427–21439 (2011).
Burroughs, A. M., Allen, K. N., Dunaway-Mariano, D. & Aravind, L. Evolutionary Genomics of the HAD Superfamily: Understanding the Structural Adaptations and Catalytic Diversity in a Superfamily of Phosphoesterases and Allied Enzymes. J. Mol. Biol. 361, 1003–1034 (2006).
Varadi, M. et al. AlphaFold Protein Structure Database: massively expanding the structural coverage of protein-sequence space with high-accuracy models. Nucl. Acids Res. 50, D439–D444 (2022).
Izumi, M., Tsunoda, H., Suzuki, Y., Makino, A. & Ishida, H. RBCS1A and RBCS3B, two major members within the Arabidopsis RBCS multigene family, function to yield sufficient Rubisco content for leaf photosynthetic capacity. J. Exp. Bot. 63, 2159–2170 (2012).
Genty, B., Briantais, J.-M. & Baker, N. R. The relationship between the quantum yield of photosynthetic electron transport and quenching of chlorophyll fluorescence. Biochimica et. Biophysica Acta. (BBA) - Gen. Subj. 990, 87–92 (1989).
Bommer, G. T., Van Schaftingen, E. & Veiga-da -Cunha, M. Metabolite Repair Enzymes Control Metabolic Damage in Glycolysis. Trends Biochem Sci. 45, 228–243 (2020).
Linster, C. L., Van Schaftingen, E. & Hanson, A. D. Metabolite damage and its repair or pre-emption. Nat. Chem. Biol. 9, 72–80 (2013).
Huang, L. et al. A family of metal-dependent phosphatases implicated in metabolite damage-control. Nat. Chem. Biol. 12, 621–627 (2016).
Hanson, A. D., Henry, C. S., Fiehn, O. & de Crécy-Lagard, V. Metabolite Damage and Metabolite Damage Control in Plants. Annu. Rev. Plant Biol. 67, 131–152 (2016).
McCurry, S. D. & Tolbert, N. E. Inhibition of ribulose-1,5-bisphosphate carboxylase/oxygenase by xylulose 1,5-bisphosphate. J. Biol. Chem. 252, 8344–8346 (1977).
Tikhonov, A. N., Khomutov, G. B., Ruuge, E. K. & Blumenfeld, L. A. Electron-Transport Control in Chloroplasts - Effects of Photosynthetic Control Monitored by the Intrathylakoid Ph. Biochim Biophys. Acta. 637, 321–333 (1981).
Joliot, P. & Johnson, G. N. Regulation of cyclic and linear electron flow in higher plants. P. Natl Acad. Sci. USA. 108, 13317–13322 (2011).
Gibson, J. L. & Tabita, F. R. Analysis of the cbbXYZ operon in Rhodobacter sphaeroides. J. Bacteriol. 179, 663–669 (1997).
Orr, D. J., Robijns, A. K. J., Baker, C. R., Niyogi, K. K. & Carmo-Silva, E. Dynamics of Rubisco regulation by sugar phosphate derivatives and their phosphatases. J. Exp. Bot. 74, 581–590 (2023).
Fei, C., Wilson, A. T., Mangan, N. M., Wingreen, N. S. & Jonikas, M. C. Modelling the pyrenoid-based CO(2)-concentrating mechanism provides insights into its operating principles and a roadmap for its engineering into crops. Nat. Plants 8, 583–595 (2022).
Adler, L. et al. New horizons for building pyrenoid-based CO2-concentrating mechanisms in plants to improve yields. Plant Physiol. 190, 1609–1627 (2022).
Long, B. M. et al. Carboxysome encapsulation of the CO(2)-fixing enzyme Rubisco in tobacco chloroplasts. Nat. Commun. 9, 3570 (2018).
Alonso, J. M. et al. Genome-wide insertional mutagenesis of Arabidopsis thaliana. Science (N. Y., NY) 301, 653–657 (2003).
Tissier, A. F. et al. Multiple independent defective suppressor-mutator transposon insertions in Arabidopsis: a tool for functional genomics. Plant Cell 11, 1841–1852 (1999).
Ihnatowicz, A. et al. Mutants for photosystem I subunit D of Arabidopsis thaliana: effects on photosynthesis, photosystem I stability and expression of nuclear genes for chloroplast functions. Plant J.: Cell Mol. Biol. 37, 839–852 (2004).
Karimi, M., Inzé, D. & Depicker, A. GATEWAY vectors for Agrobacterium-mediated plant transformation. Trends Plant Sci. 7, 193–195 (2002).
Clough, S. J. Bent aF. Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J.: Cell Mol. Biol. 16, 735–743 (1998).
Klughammer, C. & Schreiber, U. Saturation Pulse method for assessment of energy conversion in PS I. PAM Appl. Notes 1, 4 (2008).
Klughammer, C. & Schreiber, U. Complementary PS II quantum yields calculated from simple fluorescence parameters measured by PAM fluorometry and the Saturation Pulse method. PAM Appl. Notes 1, 9 (2008).
von Caemmerer, S. & Farquhar, G. D. Some relationships between the biochemistry of photosynthesis and the gas exchange of leaves. Planta 153, 376–387 (1981).
Farber, A., Young, A. J., Ruban, A. V., Horton, P. & Jahns, P. Dynamics of Xanthophyll-Cycle Activity in Different Antenna Subcomplexes in the Photosynthetic Membranes of Higher Plants (The Relationship between Zeaxanthin Conversion and Nonphotochemical Fluorescence Quenching). Plant Physiol. 115, 1609–1618 (1997).
Sambrook J., Russell D. W. Molecular Cloning: A Laboratory Manual, 3th edn, Cold Spring Harbor Laboratory, New York. Cold Spring, Harbor Laboratory, New York, 3th edn, (2001).
Schägger, H. Tricine-SDS-PAGE. Nat. Protoc. 1, 16–22 (2006).
Geladopoulos, T. P., Sotiroudis, T. G. & Evangelopoulos, aE. A malachite green colorimetric assay for protein phosphatase activity. Anal. Biochem. 192, 112–116 (1991).
Kunst, L. Preparation of physiologically active chloroplasts from Arabidopsis. Methods Mol. Biol. (Clifton, NJ) 82, 43–48 (1998).
Escobar, M. A., Geisler, D. A. & Rasmusson, A. G. Reorganization of the alternative pathways of the Arabidopsis respiratory chain by nitrogen supply: opposing effects of ammonium and nitrate. Plant J. 45, 775–788 (2006).
Armenteros, J. J. A. et al. Detecting sequence signals in targeting peptides using deep learning. Life Sci. Alliance 2, e201900429 (2019).
Le, S. Q. & Gascuel, O. An improved general amino acid replacement matrix. Mol. Biol. Evolution 25, 1307–1320 (2008).
Whelan, S. & Goldman, N. A general empirical model of protein evolution derived from multiple protein families using a maximum-likelihood approach. Mol. Biol. Evol. 18, 691–699 (2001).
Spitzer, M., Wildenhain, J., Rappsilber, J. & Tyers, M. BoxPlotR: a web tool for generation of box plots. Nat. Methods 11, 121–122 (2014).
Acknowledgements
We thank Paul Hardy for critical reading of the manuscript, the German Research Foundation (DFG, grant TRR175 to D.L., grant RU 1945/2−1 and RU 1945/4-1 to T.R.) for financial support, Silke Ruberg and Lea Rosenhammer for technical support, Hiroyuki Ishida for rbcs1a3b-1 seeds, Ute Armbruster for atcbbya (SALK_025204) seeds, and David Stern for providing the CSP41b antibody.
Funding
Open Access funding enabled and organized by Projekt DEAL.
Author information
Authors and Affiliations
Contributions
T.R. and D.L. designed experiments. Experiments in Arabidopsis were performed by T.R., N.K., B.R., V.P., and S.B., A.S. and M.P. synthesized XuBP and carried out enzyme kinetics studies. T.N. designed and performed gas exchange experiments. P.J. analyzed pigment composition by HPLC and R.B. generated the AtCbbYA antibody. T.R. was responsible for conceptualization and management of the entire study and wrote the paper with contributions from all authors.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Nature Communications thanks Elizabete Carmo-Silva and Oliver Martin Mueller-Cajar for their contribution to the peer review of this work. A peer review file is available.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Source data
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Leister, D., Sharma, A., Kerber, N. et al. An ancient metabolite damage-repair system sustains photosynthesis in plants. Nat Commun 14, 3023 (2023). https://doi.org/10.1038/s41467-023-38804-y
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41467-023-38804-y
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.