Abstract
Cyclophilins, or immunophilins, are proteins found in many organisms including bacteria, plants and humans. Most of them display peptidyl-prolyl cis-trans isomerase activity, and play roles as chaperones or in signal transduction. Here, we show that cyclophilin anaCyp40 from the cyanobacterium Anabaena sp. PCC 7120 is enzymatically active, and seems to be involved in general stress responses and in assembly of photosynthetic complexes. The protein is associated with the thylakoid membrane and interacts with phycobilisome and photosystem components. Knockdown of anacyp40 leads to growth defects under high-salt and high-light conditions, and reduced energy transfer from phycobilisomes to photosystems. Elucidation of the anaCyp40 crystal structure at 1.2-Å resolution reveals an N-terminal helical domain with similarity to PsbQ components of plant photosystem II, and a C-terminal cyclophilin domain with a substrate-binding site. The anaCyp40 structure is distinct from that of other multi-domain cyclophilins (such as Arabidopsis thaliana Cyp38), and presents features that are absent in single-domain cyclophilins.
Similar content being viewed by others
Introduction
Cyclophilins constitute a sub-family of immunophilins, next to FK-506-binding proteins (FKBPs) and parvulin-like proteins1,2. They can be found in all subcellular organelles and almost all forms of life, ranging from archaea to animals and plants1,2,3. Immunophilins were originally recognized as receptors for immunosuppressive drugs like the cyclic undecapeptide cyclosporin A, FK506, the macrocyclic lactones rapamycin and juglones, which hold only clinical relevance4. Despite of their low sequence and structural similarity, most of them possess peptidyl-prolyl cis-trans isomerase activity that catalyzes the rotation of X-Pro peptide bonds from cis to trans1,2,3,4. This is a rate-limiting step in protein folding and of prime importance, as 90% of all proteins comprise trans prolyl imide bonds5,6. Cyclophilins have been categorized into (i) single catalytic cyclophilin domain containing proteins and (ii) multidomain cyclophilins, which contain additional functional domains like WD40, RRM, TPR, PsbQ like domains, Zinc Finger domains and others2.
Immunophilins have diverse functions in protein folding, trafficking, maturation and scaffolding, in RNA processing, in spliceosome and in RISC assembly, stabilization and signaling of receptor complexes and in cell cycle regulation7,8,9,10,11,12. In plants, they further function in assembly and maintenance of photosystems, and stress responses in general13,14,15,16. In contrast, cyanobacterial cyclophilins remained largely unexplored. Cyanobacteria comprise a group of photoautotrophic prokaryotes and represent the ancestors of modern plastids, thereby they were of great impact in development of eukaryotic algae and plants17. Furthermore, diazotrophic cyanobacteria are dominant nitrogen fixers in oceans and rice fields18. Proteomic studies uncovered an increase of cyclophilin protein abundance in the diazotrophic Anabaena sp. PCC 7120 (abbreviated as Anabaena sp. hereafter) under salt, herbicide and UV-B stress19,20,21,22. This suggests an impact of cyanobacterial cyclophilins on stress response.
Cyanobacterial cyclophilins might function in photosynthetic regulation in addition to stress management. In spinach, a 40 kDa cyclophilin-type peptidyl-prolyl cis-trans isomerase (PPIase) was found to reside in the thylakoid lumen and was annotated as thylakoid lumen PPIase of 40 kDa (TLP40). The protein contains a PPIase domain and a phosphatase binding module, and negatively regulates the thylakoid protein phosphatase13,14,15. The ortholog in Arabidopsis thaliana, atCyp38 (cyclophilin of 38 kDa) shares 82% sequence identity to the spinach protein and participates in early biogenesis of photosynthetic protein complexes14,23. It is assumed that atCyp38 assists in D1 and possibly in CP43 folding during assembly of the oxygen evolving complex (OEC)14. However, the direct targets and the mode of action remain unclear. The presence of a similar cyclophilin family protein (Alr5059) in Anabaena sp. is remarkable, as the regulation of OEC assembly is different in cyanobacteria, because the D1 turnover is not dependent on D1 phosphorylation state24.
Apart from minor differences in subunit composition, the PSII-core is conserved from cyanobacteria to plants. The PSII-complex normally functions as a dimer25,26, whereas functional monomeric complexes have also been isolated. Those are considered as intermediates from assembly or the damage–repair cycle27. PSII assembly and turnover requires molecular chaperones and other auxiliary proteins which are transiently involved in the biogenesis, maintenance and stabilization of the dimers of PSII. However, the functions of many of these proteins remain poorly understood28,29.
PSI-complexes of cyanobacteria and plants are remarkably similar as well25, whereas differences exist in the occurrence of extrinsic protein subunits: (i) subunits PsaG and PsaH are present exclusively in plants, whereas PsaX and PsaM are found in cyanobacteria only, and (ii) subunit PsaL is structurally different in plants and cyanobacteria25. These differences cause alterations in supramolecular organization of the PSI complex between cyanobacteria and plants. The cyanobacterial PSI complex can be isolated as a monomer, trimer (most species) or tetramer30. In eukaryotes (algae and higher plants), PSI exists as monomer which can be part of a supercomplex with the light-harvesting Chl complex of PSI (LHCI)26. PsaM and PsaL are considered responsible for trimer formation in cyanobacteria30, whereas the PsaG subunit found in plants PSI provides an anchoring point for Lhca subunits, leading to the formation of a PSI-LHC supercomplex31. Moreover, the small subunits PsaF, PsaJ, PsaK, and PsaX located at the distal side of the PSI complexes may be involved in the interaction of PSI with its external antenna systems, such as IsiA or phycobilisomes32.
Although the PS-core shows high conservation from cyanobacteria to plants, antenna complexes are diverse. In cyanobacteria and red algae, phycobilisomes (PBS) are peripheral water-soluble antenna complexes that harvest light energy and transfer it to the photosynthetic centres33. PBS constitute supercomplexes, which consist of the chromophore-bearing biliproteins phycocyanin (PC), allophycocyanin (APC) and phycoerythrin (PE). Those are connected by non chromophorylated linker (L) proteins. L-proteins also stabilize and regulate PBS quaternary structure, and by that optimize the energy transfer33. PBS transfer energy to PSII and to PSI in a megacomplex in cyanobacteria34, or to PSI as discussed in alternative models35. The coexistence of alternative complexes is seen as indication for high flexibility of the PBS-PS complex formation. In contrary to the cyanobacterial system, the higher plant light-harvesting complex of PSII (LHCII) (genes: lhcb1-3) constitutes about one-third of the total chloroplast protein content. LHCII trimers bind half of the total chlorophyll located in the thylakoid membrane, whereas the LHCI antenna (genes: lhca1-4) associate entirely with PSI36.
Taking recourse to above, we report the function of a multidomain cyclophilin (47% identical to atCyp40) from Anabaena sp. through generation of Anabaena sp. single recombinant mutants, the in vivo analysis of the localization and the interactome, as well as by determination of the crystal structure of the protein.
Results
The gene encoded by alr5059 is a cyclophilin family member
The 40 kDa protein encoded by alr5059 from Anabaena sp. is most similar to bacterial cyclophilins or plant thylakoid lumen PPLases (TLP40, atCyp38), therefore it is annotated as anaCyp40. The computed phylogenetic relation shows a global conservation of anaCyp40 in cyanobacteria, algae chlorophytes, bryophytes and plantae (Fig. 1a, Supplementary Table 1). Sequence analysis of anaCyp40 shows the presence of a C-terminal cyclophilin domain and an N-terminal conserved domain of unknown function (Fig. 1b). The utilized transmembrane domain prediction programs (Supplementary Table 2) consistently predict a transmembrane domain (TM) between amino acids 13 and 30. A putative signal sequence (Sec/SPI) of about 36 amino acids length was predicted at the N-terminal region including the TM domain, though with a low likelihood of 0.51 (SignalP)37. Worth mentioning, the predicted cleavage site (Signal P) stands in contrast to the N-terminal peptide obtained by mass spectrometry, predicting a polypeptide starting at amino acid 1538. The cyanobacterial and plant proteins differ in their signal sequence, the region preceding the transmembrane domain and in the region annotated as domain of unknown function (DUF3084)39.
a Shown is the phylogenetic analysis of the sequences of the plant and cyanobacterial cyclophilin family as described in Materials and Methods (Supplementary Table 1). b The predicted domains of anaCyp40 are shown as bar diagram. The conserved domains were extracted from NCBI database (CDD)91 and additional regions for membrane anchoring or targeting predicted as described (Supplementary Table 2). TM: transmembrane region, DUF: domain of unknown function. c Purified anaCyp40ΔTM-His (5 nM, blue) or BSA (5 nM, red) were incubated with 40 μM N-succinyl-ala-ala-pro-phe-p-nitroanilidine and the catalytic reaction monitored by the increase of absorption at 390 nm. Source data are provided as a Source data file. The values were normalized to the baseline and to the maximum. A representative experiment is shown. The average of the determine rate constant (n > 5 repetitions of the experiment) is presented and the standard deviation is shown as error bar. d E. coli BL21 (DE3) transformed with pET21a (upper panel) or pET21a-anaCyp40 (lower panel) were spotted on agar plates or plates containing the indicated concentrations of arsenic (upper left) or sodium chloride (lower right). Bacteria on normal plates were exposed to 50 °C or to UV-B (2.9 mWm−2 nm−1) for the indicated time. Representative images of growth after 24 h are shown.
To test the catalytic activity of anaCyp40, a recombinant version without the N-terminal transmembrane helix (aa 1–35) but with C-terminal His tag was expressed in E. coli. The purified protein was used to hydrolyze N-succinyl-ala-ala-pro-phe-p-nitroanilidine in order to determine a putative peptidyl prolyl cis-trans isomerase activity40. For anaCyp40 an activity with a rate of 0.07 ± 0.01 sec−1 was observed, while a background rate of 0.02 ± 0.01 s−1 was determined for bovine serum albumin (Fig. 1c; Supplementary Fig. 1; BSA). The measured rate constant of purified anaCyp40 is comparable to the activity reported for homologs from other organism (Supplementary Table 3).
Next, the protective capacity of anaCyp40 against specific stresses was probed, to further confirm the assignment as cyclophilin. The recombinant protein was expressed in E. coli, which was subsequently exposed to arsenic, salt, UV-B or heat stress. At high doses of the stressors, E. coli transformed with the empty plasmid showed a clear growth defect, while the bacteria transformed with the protein coding plasmid were resistant to elevated stress levels (Fig. 1d). This result is consistent with the functional assignment of anaCyp40 as cyclophilin.
A functional anaCyp40 is required for cyanobacterial salinity stress response
The anaCyp40-coding gene is expressed under normal growth conditions of Anabaena sp., while the mRNA abundance is enhanced in response to abiotic stress. qRT-PCR analysis on isolated mRNA showed that the exposure of Anabaena sp. to arsenic, to desiccation by incubation at 30 °C for 10 h, to heat stress at 45 °C, to enhanced salinity by addition of 100 mM sodium chloride or to UV-B stress by application of 2.9 mW m−2 nm−1 for 30 min enhances the transcript abundance of anacyp40 by 2.5–4.5 fold compared to normal growth conditions (Fig. 2a). Hence, anacyp40 transcription is regulated in response to various abiotic stresses, and appears to be required for cellular homeostasis of Anabaena sp. This is consistent with the observed ability of anaCyp40 to enhance the stress resistance of E. coli (Fig. 1).
a Wild-type Anabaena sp. was grown in BG11 and cells were treated as indicated followed by RNA isolation and qRT-PCR analysis with specific oligonucleotides for anaCyp40 (Supplementary Table 4). The mean of the ratio to the mRNA abundance in non-treated cultures is shown and error bars indicate the standard deviation (n = 4). The statistical significance determined by ANOVA (Duncans) of the difference to non-treated cultures is indicated. b Two independently generated AFS-I-anaCyp40 strains and wild-type cells were spotted at two indicated concentrations at BG11 supplemented with indicated concentrations of NaCl. The plates were imaged after 8 days of growth. c Wild-type (blue) and AFS-I-anaCyp40 (orange) were grown in BG11 liquid medium (left) with additional 100 mM NaCl (right). The culture density was imaged after 8 days (top) and the average values and the standard deviation for the growth of three independent cultures normalized to the OD at the start of the analysis are shown (bottom). Source data are provided as a Source data file.
An insertion mutant created by single recombination and annotated as AFS-I-anacyp40 (Anabaena sp. mutant created in Frankfurt by Schleiff group with plasmid insertion)41 was generated to explore the function of anaCyp40 in cyanobacterial stress response (Supplementary Table 4). The insertional mutant was segregated, documenting that anaCyp40 is not essential under normal growth conditions (Supplementary Fig. 2). It was reported recently that an antisense RNA to alr5059 is expressed in the absence of fixed nitrogen, which might be involved in heterocyst differentiation42. However, the strategy of mutant generation does not inhibit the induction of the antisense RNA in the absence of fixed nitrogen (Supplementary Fig. 2). Thus, the phenotype of the mutant is dedicated to the loss of anaCyp40 function.
In total, three independent strains of AFS-I-anacyp40 were isolated and analyzed. All three strains had the same phenotype as exemplified for the strain (1) and (2) (Fig. 2b). Subsequently, the results for strain (1) are shown as representative if not otherwise noted. Consistent with the expression profile of anacyp40, AFS-I-anacyp40 showed an enhanced sensitivity to salt stress (Fig. 2b). The growth of the mutant strain on solid BG11 medium under standard light conditions was comparable to wild-type Anabaena sp., but was inhibited on solid BG11 medium containing 75 mM NaCl. Similarly, the mutant strain does not show any growth distinction to wild type in liquid BG11 medium (Fig. 2c, left), but is reduced in the presence of 100 mM NaCl (Fig. 2c, right). Worth mentioning, the growth of wild-type Anabaena sp. is reduced in the presence of 100 mM NaCl as well when compared to the growth of this bacterium in normal media.
AnaCyp40 interacts with different proteins and complexes
The importance of anaCyp40 in stress response (Figs. 1, 2) prompted the analysis of the substrate spectrum of the cyanobacterial cyclophilin. For isolation of cyclophilin-substrate complexes a strain coding for the protein bearing a C-terminal Strep-tag was created (AFS-anacyp40-strep; Supplementary Fig. 4). The AFS-I-anacyp40-strep strain exhibited a phenotype comparable to the wild type in response to enhanced salinity (Supplementary Fig. 5), indicating that the function of anaCyp40 is not impaired by the Strep-tag. The complex formation of anaCyp40 was probed by mass spectrometry of proteins after affinity tag purification of the Strep-tagged protein from cell lysate (Supplementary Fig. 4). In total, 65 proteins have been detected with a p-value smaller than 0.05 (Table 1, Supplementary Data 2).
Many, but not all proteins of the ribosomes and the RNA polymerase were precipitated with anaCyp40 as bait. This could suggest that anaCyp40 interacts with substrates during translation. Notably, the cell lysate was not treated with RNAse or DNAse, which would disrupt RNA-polymerase - ribosome complexes43. In favor of this interpretation is the discovery of proteins from small and large ribosomal subunits, the former interacting with the RNA polymerase and the latter representing the proteins at the ribosomal exit tunnel. Interestingly, the PPIase-like domain containing trigger factor anchors to the ribosome through interaction between its tail domain and L23, being in vicinity of L29 and L1944. In here, L23 and L19 were identified among the ribosomal proteins. Alternatively, ribosomal and polymerase proteins could also be substrates of anaCyp40, because all of them contain at least one proline (Table 1). However, in the latter case, a broader range of precipitated ribosomal proteins would have been expected45.
A second group represented by 29 proteins presumably involves substrates of anaCyp40, as these could not be assigned to a specific functional complex (Table 1). All of these proteins contain a high number of prolines, and especially proteins like ClP, Ssb3 or CbbS have a higher proline content than the average in the theoretical proteome of Anabaena sp. Thus, an assignment as substrates would be consistent with the proposed function of anaCyp40.
A third category unifies proteins involved in photosynthesis, with the highest abundance found for PBS components (Table 1). Some of these genes belong to the highest expressed genes in Anabaena sp. wild-type cells46, which could argue for a classification as substrate. However, this does not hold true for all genes coding for the identified PBS proteins and in turn, not all PBS proteins encoded by highly expressed genes, or containing a large number of prolines, were identified (Supplementary Table 5). This might suggest that anaCyp40 has an additional function related to photosynthetic performance.
AnaCyp40 is important for the photosynthetic capacity of Anabaena sp. PCC 7120
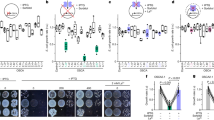
Consistent with a function of anaCyp40 in regulation of photosynthesis, the mutant strain was significantly reduced in its growth when cultivated at 120 µE illumination (Fig. 3a, b). Interestingly, the chlorophyll content, but not the phycocyanin or allophycocyanin content, was significantly reduced in the mutant compared to wild type under normal growth conditions (Fig. 3c–e). The chlorophyll, phycocyanin or allophycocyanin content was generally reduced in wild type and AFS-I-anacyp40 under high-light conditions. Here, the difference of the chlorophyll content between the wild-type and mutant strain was not significant, while the phycocyanin or allophycocyanin content was significantly lower in the mutant.
a, b Wild-type (blue) and AFS-I-anaCyp40 (orange) were grown in BG11 liquid medium at 120 µE illumination (right). The culture density was imaged after 8 days (a) and the average values for the growth of three independent cultures normalized to the OD at the start of the analysis are shown (b). c The chlorophyll content in wild-type (blue) and AFS-I-anaCyp40 (orange) cells grown under 40 µE or 120 µE (transparent color) was analyzed and normalized to cell density for three independent experiments. d, e The PC (d) and the APC (e) content in wild-type (blue) and AFS-I-anaCyp40 (orange) cells grown under 40 µE or 120 µE (transparent color) was analyzed and normalized to cell density. The statistical significance was analyzed and p < 0.001 is indicated. f The oxygen evolution rate was determined for wild-type (green) and AFS-I-anaCyp40 (orange) grown in BG11 liquid medium at 40 (dark color) or 120 µE illumination (transparent color). The average values and the standard deviation are shown. Stars indicate statistical significance with p < 0.001. g The photosynthetic parameters were determined by PAM. The Φ(II) for wild-type and mutant culture grown at 40 or 120 µE are shown. The statistical significance of the change was analyzed for same illumination condition and p < 0.001 is indicated. Error bars in b–g indicate standard deviation and the bare represents the mean. In b–g the statistical significance was analyzed by ANOVA (Duncans) and the star indicates a p < 0.001. h, i The 77 K fluorescence emission spectra of Anabaena sp. wild-type (green) or the insertion mutant of anaCyp40 (I-anaCyp40, orange) grown at normal (solid line) or high-light (dashed line) were recorded by excitation with 590 nm (f) or 440 nm (g). Normalization was done at 800 nm, and the spectra are the average of at least three independent biological replicates. The color and line coding shown in (f). Source data are provided as a Source data file.
The analysis of the oxygen evolution revealed a comparable efficiency of wild type and AFS-I-anacyp40 when grown under normal light conditions in BG11 (Fig. 3f). Cultivation under high-light conditions resulted in an enhanced maximal rate of net oxygen production at light saturation (Pm) at 1000 µE m−1 s−1 in wild type, but a reduced oxygen production in the mutant (Fig. 3f, Supplementary Table 6). Moreover, the compensation irradiance at which oxygen production and oxygen consumption are balanced (Ec), that can be estimated from Pm and the measurement at 100 µE m−1 s−1, was enhanced in AFS-I-anacyp40 when grown under high light (Fig. 3f, Supplementary Table 6). The maximal effective PSII quantum yield (Φ(II)) probed by pulse-amplitude modulation (PAM-)measurements was comparable between wild-type Anabaena sp. and AFS-I-anacyp40 grown under normal light (Fig. 3g, left). In contrast, a significant reduction of the maximal effective PSII quantum yield was observed for the mutant when compared to wild-type after growth at high-light (Fig. 3g, right).
Subsequently the 77 K fluorescence spectra was recorded. Excitation of the PBS at 590 nm (Fig. 3h) resulted in a large phycocyanin (~650 nm) and allophycocyanin peak (~665 nm), as well as in a somewhat smaller peak at ~685 nm likely representing the fluorescence of ApcE (~680 nm) and PSII (~685–695 nm)47,48,49,50. The shoulder at ~720–740 nm is related to the fluorescence of PSI. Under normal light conditions, a difference between mutant and wild-type strain was not observed (Fig. 3h, solid lines). When grown under high light, especially the allophycocyanin and the PSII fluorescence was reduced in the mutant, while this fluorescence was only moderately affected in wild type (Fig. 3h, dashed lines). Excitation of the chlorophyll at 440 nm yielded highest fluorescence of PSI (~720–740 nm) under normal light conditions, and a comparable fluorescence of PSI and PSII (~685–695 nm) at high light (Fig. 3i, dashed lines). In addition, a basal fluorescence of the PBS is observed as well (~650 nm/~665 nm). Remarkably, the mutant strain grown under high light showed an enhanced phycocyanin peak when compared to the wild type.
The apparent energy-transfer from PBS to PSI in relation to the energy transfer from PBS to PSII deduced from the ration FPSI/FPSII after PBS excitation51 does not change in the mutant or through high-light treatment (Supplementary Table 6). The ratio of the activity of the two photosystems, which was calculated based on chlorophyll excitation changed in response to high-light, but again, not through mutagenesis of gene coding for anaCyp40.
The dimerization of photosystem I is affected in the anaCyp40 mutant
The influence of the mutation on the assembly of complexes involved in photosynthesis was determined by BN-PAGE analysis, using isolated membranes of Anabaena sp. wild type and AFS-I-anacyp40 grown under normal or high-light conditions. Thylakoid membranes were solubilized by addition of 1% n-Dodecyl β-D-maltoside. At normal growth conditions, the abundance of the PBS, the cytochrome b6f complex and PSII was not affected in the mutant, while the PSII dimer was not detected in AFS-I-anacyp40 (Fig. 4a; Supplementary Fig. 3). In contrast, the abundance of the monomeric PSI is enhanced, while the level of the dimeric state of PSI is reduced. Remarkably, the abundance of the tetrameric state is only moderately affected (Fig. 4a, PSITE). When grown at high light, the wild type shows the typical reduction of the dimeric PSI. Moreover, the PSII dimer is reduced under strong light as well. In the mutant, PBS and monomeric PSI were enriched, while the dimeric and tetrameric state of PSI was reduced when compared to wild type. Hence, anaCyp40 influences PSI dimerization, and one explanation could be that the functions of PsaL might have been lost, which is discussed to be important for dimerization of PSI52,53.
a Membranes of wild-type (green) and AFS-I-anaCyp40 (orange) grown in BG11 liquid medium at 40 or 120 µE illumination were solubilized and subjected to BN-PAGE. The chlorophyll staining (left) and the Coomassie Blue staining (right) are shown. On the left migration of complexes and between the two images the migration of the molecular weight standards is show. The Coomassie Blue staining was quantified by ImageJ and the intensity of the individual complexes was normalized to the intensity in wild-type grown at 40 µE light intensity. b Phycobilisomes were isolated from cultures grown as in (a) and subjected to a 10–50% sucrose gradient. A representative profile is shown. c Equal amounts of phycobilisomes (5 µg protein) isolated from the strains as in (a) were subjected to SD-PAGE as indicated. The migration of the molecular weight is shown on the right and the proteins are assigned54. d The strain AFS-anaCyp40-strep was grown in BG11, cells harvested and solubilized in low concentrated buffer and subjected on top of a 10–50% sucrose gradient. The fractions were subjected to SDS-PAGE followed by Western blotting (DB71-staining shown) and incubation with indicated antibodies. PBS and Thylakoid membrane fractions are indicated. e Cell lysate (CL) and phycobilisomes (PBS) were isolated in buffer with high concentration of phosphate from AFS-anaCyp40-strep and subjected to SDS-PAGE followed by Western blotting (DB71 staining shown) and incubation with indicated antibodies.
AnaCyp40 localizes to thylakoids and physically interacts with the phycobilisomes
The changes of PBS association with membranes in the mutant strain (Fig. 4a) led to the analysis of the cellular PBS content and composition. The PBS were isolated and fractionated on a 10–50% sucrose gradient (Fig. 4b). While an alteration between normal and high-light conditions was observed, the sub-complex profile was not affected in the mutant strain. When isolated phycobilisomes were subjected to SDS-PAGE, comparable profiles annotated according to previous reports54 were observed as well (Fig. 4c). Only an increase of ApcE was detected in AFS-I-anacyp40.
To further probe for an interaction of anaCyp40 with phycobilisomes, AFS-anacyp40-strep (Supplementary Figs. 4, 5) cells were lysed and the cell lysate was fractionated by sucrose gradient centrifugation. A small fraction of anaCyp40 migrated with the thylakoid membrane fraction, identified by the protein profile and the immunodetection of PsbA (Fig. 4d; Supplementary Fig. 6). Most of the protein migrated at a lower density fraction that contained PBS as judged from the protein profile (Fig. 4d) and the color of the fraction (not shown). To distinguish between an interaction with PBS and a co-migration of anaCyp40 in this fraction, PBS total lysate was isolated from AFS-anacyp40-strep, and probed with antibodies against the strep tagged protein or the periplasmic localized anaTic2255. While both proteins were detected in the cell lysate (Fig. 4e, CL; Supplementary Fig. 7), the Strep-tagged anaCyp40 protein was enriched in the PBS fraction (Fig. 4e, PBS).
The association with the phycobilisomes prompted the analysis of the intracellular localization of anaCyp40 with near-molecular spatial resolution. We used single-molecule super-resolution microscopy in combination with DNA-labeled antibodies (DNA-points accumulation for imaging in nanoscale topography, DNA-PAINT)56 to determine the nanoscale structural organization of anaCyp40 in cells. The localization of the Strep-tagged protein was visualized in AFS-anacyp40-strep together with endogenous fluorophores in the thylakoid membrane. The outer membrane and plasma membrane were stained with NileRed (Fig. 5a). The endogenous fluorophores of Anabaena sp. exhibited a photoswitching behavior and were sufficiently bright and photostable to perform single-molecule detection, which allowed tracing the thylakoid structure in cells. After photobleaching of the auto-fluorescence signal, a DNA imager strand was added to the imaging buffer in order to visualize AFS-anacyp40-strep in the same cells. The super-resolution images show the distribution of the anti-Strep signal inside the cell (Fig. 5a, overlay). The spatial resolution of the super-resolution images was determined by decorrelation analysis to be 24 nm (Supplementary Fig. 8)57,58. A co-localization analysis from intensity profiles (Fig. 5b) showed a spatial separation of the NileRed and the anti-Strep signal, but a co-occurrence of the thylakoid localized auto-fluorescence and the anti-Strep signal. The calculation of the Pearson correlation coefficient (PCC) of multiple regions indicates a co-localization of anaCyp40 with thylakoid membranes, but not with the membrane system surrounding the cell (Fig. 5c). This is consistent with the obtained phenotype, as well as with an association of anaCyp40 with phycobilisomes. The specificity of the antibody staining was confirmed in wild type Anabaena sp. (Supplementary Fig. 9).
a AFS-anaCyp40-strep was grown in BG11 and processed as described in “Methods”. Using three-color super-resolution microscopy, the nanoscale localization of anaCyp40 (left), the thylakoid membrane (second from left) and the plasma membrane (second from right) were visualized (overlay shown on the right, scale bar = 5 µm). b Enlarged section as indicated in (a) with two boxed regions (1, 2) from where intensity profiles were extracted (scale bar = 1 µm). c Intensity profiles for the two regions shown in (b) for anaCyp40 (cyan), the thylakoid membrane (red) and the plasma membrane (green) are shown. d The Pearson correlation coefficient (PCC) was calculated from intensity values for pixels and for anaCyp40 and thylakoid (PT), anaCyp40 and membrane (PM) as well as for the membrane and thylakoid (MT). Values were calculated in 10 individual regions and are shown as box plot (horizontal lines indicate median, box sizes represent the 25th and 75th percentile of all values, whiskers indicate the standard deviation). Source data are provided as a Source data file.
AnaCyp40 contains two functional domains
A more precise characterization of the enzymatic function of anaCyp40 at a molecular level requires high-resolution structural information. To achieve this, high-quality single crystals of anaCyp40 lacking the N-terminal trans-membrane helix (anaCyp40ΔTM) were grown by vapor diffusion in hanging drops39 and used to collect diffraction data to 1.2 Å (Supplementary Table 7). The structure was determined by molecular replacement with atCyp38 (pdb-id: 3rfy)23 as search model. The asymmetric unit contains one anaCyp40 molecule with two domains: a helical N-terminal domain and a C-terminal cyclophilin domain (Fig. 6a). Except of five N-terminal amino acids, all residues could be traced and are present in the final model. This includes the “LEVLFQ” sequence from the PreScission protease recognition site at the C-terminus that remained after the cleavage of the purification tag. The C-terminal region contributes to the crystal contacts between the individual molecules by interacting with the C-domain of the next molecule (Fig. 6b, Supplementary Figs. 10, 11). The N-terminal domain of anaCyp40 (residues 42–162) has a total of five helices, which are all arranged parallel to each other and connected by short loops (Fig. 6a). The first Helix (α1) is about half as long as the other four helices (α2–α5), which form a four-helix bundle. The topology of the four-helix bundle is very similar to the PsbQ proteins from photosystem II of higher plants (e.g., pdb-id: 1vyk, 2mwq, 3ls1; Supplementary Fig. 12)59,60,61 leading to the annotation of this domain as Q-domain (Fig. 6a). However, helix α1 has no counterpart in other known PsbQ structures (Supplementary Fig. 12).
a Ribbon presentation of anaCyp40 from two orientations related to each other by a 180° vertical rotation. The N-terminal four-helix bundle (Q-domain) is shown in blue and the cyclophilin domain (C-domain) is shown in green colors. The two domains are linked by a flexible loop. The interaction between the domains is mediated by the N-terminal helix (α1). The cyclophilin domain is formed by a distorted β-barrel with eight β-strands (β2–5 and β10–13). The β-barrel is flanked on one side by helix α6 and the remaining β-strands and on the other side by helix α7. The N- and C-termini are indicated. b The intermolecular interactions between two symmetry-related anaCyp40 molecules are shown on the left side. The interaction is mediated by the last seven C-terminal residues shown in purple. The boxed region is magnified on the right side and shows the interactions of the C-terminal peptide with the potential substrate binding site on the outer surface of the β-barrel. The metal at the site within close proximity to the C-terminus of the symmetry-related monomer was interpreted as heptagonal coordinated calcium ion (Ca2+) that obviously is part of the active site of anaCyp40.
Helix α1, located between the four-helix bundle and cyclophilin domain, is the main structural element that mediates the interaction of the Q- and C-domain. Helix α3 in the bundle is interrupted at about 2/3 of its length, and the shorter part is tilted by about 45° with respect to the rest of that helix and the bundle. The four-helix bundle is held together by interactions of hydrophobic residues that are pointing to the center, forming a hydrophobic core. On the surface ionic and hydrophilic interactions predominate, where the predominantly positively charged surface of helices α2 and α3 stands out (Supplementary Fig. 13). Together with the N-terminal area of helix α1, they form a fairly large positively charged area on that side, while more or less neutral to slightly negatively charged residues are present on the surface of helix α4 and α5 (Fig. 7). The topological arrangement of the four-helix bundle of anaCyp40 is also present in other unrelated proteins, as found by the structural analysis of the Q-domain using DALI62. In addition to the PsbQ proteins and among others, cytochrome C (pdb-id: 1CPR; 1QQ3)63,64, complement inhibitory domain of BBK32 (pdb-id: 6N1L)65, or even the transmembrane subunit of the plasma membrane CO2 channel LCI1 (pdb-id: 6BHP)66 have the same helix topology and therefore exhibit high structural similarity and low RMSDs (<2 Å) to the anaCyp40 Q-domain (Supplementary Fig. 12).
a The topological organization of structural elements in anaCyp40 is shown on the left and of atCyp38 (pdb-id: 3rfy) on the right. In anaCyp40 the Q-domain is shown in blue colors and the C domain in red colors. The loop regions interacting with helix α1 are shown as blue lines. β-strands β2- β5 and β10-β13 form the 8-stranded β-barrel. In atCyp38 the structural elements of Q-domain are shown in black and of the C-domain in gray. Strong discrepancies to the topology of anaCyp40 are indicated in red. b The structural comparison of anaCyp40 and atCyp38 from two perspectives shows a very similar organization of the Q domain whereas the similarities in the C-domain are restricted to the β-barrel and the helices surrounding the β-barrel. Compared to the unstructured long loops that connect the helices and β-strands of the β-barrel in atCyp38, the loops in anaCyp40 are shorter and have structural elements like β-hairpins and short β-sheets. Coloring of the structural elements in anaCyp40 is according to Fig. 6. The topological discrepancies are highlighted in red also in the structure of atCyp38.
The cyclophilin domain (C-domain, residues 163-368) is formed by 14 β-strands, of which 8 antiparallel β-strands form a distorted β-barrel that is flanked by an α-helix on each side (Fig. 6a). Helix α6 between β3 and β4 and helix α7 between β12 and β13 are connected to the β-barrel by short loops. Helix α6 is surrounded by the β-hairpins formed by β6/7 and β8/9, and by a β-sheet formed by the β-strands β1 and β14. In contrast to the short loops connecting the β-strands β2 to β3 and β4 to β5 in the β-barrel, the loops between the other β-strands are relatively long (8–20 amino acids). The Q- and C-domain in anaCyp40 are well separated from each other. In contrast, the Q- and C-domain in atCyp38 are intertwined with β-strands β10 and β11, which are topologically placed to the N-terminus of the protein (Figs. 7; 8a). Consequently, the C-domain topology of anaCyp40 does not agree with that of atCyp3823, but it is in perfect topological agreement with all other single-domain cyclophilins with known structures like hCypA67 and PfCyP1968 (Fig. 8).
a The C-domain of anaCyp40 is superimposed to single-domain cyclophilins identified by DALI62 with an RMSD < 2 Å. Three of the compared domains (1cwa:67 red; 4jjm:125 orange; 1qng:67 blue) have a bound cyclosporin A at the corresponding site of anaCyp40, where the C-terminal peptide from the symmetry-related molecule is bound. The other three remaining domains (1vbt:126 magenta; 4jjm:125 orange; 6i42:127 cyan) have at the same position peptides of different length. b Individual comparison of the substrate binding sites are highlighted.
AnaCyp40 crystallized in the absence of substrates or inhibitors. However, the C-terminal tail of the second molecule with the leftover residues from PreScission cleavage (amino acids 370LEVLFQ375) occupies the potential substrate binding pocket (Figs. 6b; 8, Supplementary Fig. 10) and probably mimics a substrate of anaCyp40. The binding of the peptide stretch is mediated by sidechain interactions of Glu226 and Gln231 from β5, Asp234 and Gln240 from the loop downstream to β5, and Gln306 from β11 with the backbone of the “substrate”. Also, hydrophobic interactions of Phe308 and Phe310 from β11 contribute to the binding. In close proximity to the C-terminus of the “substrate” we also identified a metal binding site, formed by the carboxyl group of Asp234 and backbone of oxygens of Asp234, Pro235, Gly237, Glu239. These residues surround the ion, together with three water molecules. The heptagonal coordination geometry of the metal ion and the electron density suggests a Ca2+ ion binding at this position (Fig. 6b), which remained bound to the protein during the purification steps, indicating a tight binding of that ion.
Discussion
AnaCyp40 belongs to the multidomain cyclophilin protein family (TLP40 like) with a predicted transmembrane helix at its N-terminus (aa 13-30), followed by a domain of unknown function (DUF, aa 54–148) and a C-terminal peptidyl-prolyl cis–trans isomerase domain, first identified in spinach (TLP40)13,14,15. These proteins are present in the green lineage ranging from cyanobacteria to algae and higher plants. In contrast, they do not exist, or at least have not been identified so far, in heterotrophic/chemoautotrophic bacteria, fungi or mammals, suggesting a specialized function for photosynthetic organisms (Fig. 1). TLP40 contains two important N-terminal segments (GLKALDSVERN158 and AGLAESKKDRG185) with similarity to the sequence of FKBP12, which is known to participate in recruiting the protein phosphatase calcineurin (~60% similarity)69. These are involved in the regulation of thylakoid protein phosphorylation by TLP4070. Interestingly, these sequences are either not present (motif 1, loop between helix α2 and α3; Fig. 6) or replaced in anaCyp40 (motif 2, loop between helix α3 and α4; TSVPEERQTQA114; ~10% similarity to the phosphatase binding sequences of FKBP12; Fig. 6), which precludes its possibility to regulate thylakoid protein dephosphorylation. In addition, a sequence alignment revealed differences between the cyanobacterial and the plant protein, particularly in the Q-domain39. These two observations might be the reason for an at least in parts different function of the cyanobacterial and the plant protein. Nevertheless, anaCyp40 is required for the regulation of the photosynthetic activity as well, especially under high light conditions (Fig. 3).
The N-terminal anaCyp40 Q-domain, and the homologous domains in atCyp3823 share structural similarity to several unrelated proteins. Exemplarily the oxygen evolving subunit of PSII (PsbQ)59,60,61, cytochrome C63,64, the complement inhibitory domain of BBK3265, or even the transmembrane subunit of the plasma membrane CO2 channel LCI166 have high structural similarity to the anaCyp40 Q-domain (Fig. 7), indicating a high versatility for this domain architecture. In contrast to PsbQ (pdb-id 1vyk)59 and cytochromes63,64, which have specifically bound Zn2+ ions (Fig. 7b), metal binding was not identified in the anaCyp40 Q-domain (Fig. 7a). However, the surface charge distribution with positively charged clusters that is present on one side of the Q-domain indicates that the function of this domain is likely to position the catalytic domain with respect to the membrane (Supplementary Fig. 13).
The cyclophilin domain of anaCyp40 is enzymatically active (Fig. 1), as shown previously for TLP4015. Consistent with this, the expression of the cyanobacterial gene in E. coli increases the resistance of the bacteria against various abiotic stresses. On the other hand, the loss of function of anaCyp40 in Anabaena sp. increases the stress sensitivity (Figs. 1 and 2). This phenotype can be explained by a function of anaCyp40 in maintaining the correct protein folding during stress in cooperation with chaperones, as reported for other cyclophilins1,2,3. Indeed, anaCyp40 interacts with proteins of the transcription coupled translation machinery, as well as with 29 proteins that contain a high percentage of prolines, which are likely substrates of anaCyp40 (Table 1).
The structure presented here is consistent with the observed enzymatic property of the anaCyp40 C-domain, which interacts with the flexible C-terminus of another molecule that represents an unfolded or not properly folded amino acid stretch. The substrate binding pocket is similar to other biochemically active single domain cyclophilins (Figs. 6–9) with one exception. In the reported structure for atCyp38, two β-strands, which correspond to the β-strands β10 and β11 in the anaCyp40 β-barrel, were placed topologically N-terminal to the atCyp38 Q-domain (Fig. 7). These served as indication for the inactivity of atCyp38, as this “β-sheet insertion” would prevent substrate binding to the active site. However, as atCyp38 residues 78-83, which are located in the N-terminal half of the first β-strand, originate from the thrombin cleavage site of the construct used for the crystallization, we assume an incorrect assignment and subsequent topological misinterpretation of the atCyp38 structure. In addition to the topological mismatch found in the N-terminal β-strands, direct structural comparison of anaCyp40 and atCyp38 shows more discrepancies. While in anaCyp40 the conserved canonical cyclophilin-like domain that comprises two α-helices (amino acids 201–214 and 337–344) flanks the distorted β-barrel with eight β-strands in a (β2-3)-α6-(β4,5,10-12)-α7-β13 architecture (Figs. 6, 7), the loops between the β-strand and α-helices of the atCyp38 C-domain are remarkably long. The analysis of the electron density and the available atCyp38 model deposited in the PDB (pdb-id: 3rfy)23 gives the explanation for this. For most of the residues in these long loops, no electron density is present. Moreover, a metal binding site where Ca2+ was bound is present in anaCyp40 in close proximity to the C-terminus of the “substrate”. This might represent a specific regulatory mode for substrate recognition. Metal binding close to the cyclophilin domain is reported so far only for the phloem cyclophilin BnCYP19-171.
a AnaCyp40ΔTM is placed on the membrane (green) in that way that the positively charged surface of the Q-domain interacts with the membrane lipids. In this orientation the N-terminus is close at the membrane, so that the trans membrane helix in the full-length anaCyp40 can be attached to the N-terminus of anaCyp40ΔTM. In this orientation also the substrate binding site is accessible for the substrate of anaCyp40. b The structure of the tetrameric PSI71, the dimeric PSII72, 73 and the PBS74 as well as the information on the positioning of the PBS with respect to PSII74 was used to create a model for the positioning on/in the thylakoid membrane. The different domains of the PBS (black letter) and the positioning of the proteins found to complex with anaCyp40 (purple letter) are indicated. The red circles indicate the maximal dimension of the soluble domain of anaCyp40 that would fit into the cavity in the PBS between PSII and PSI according to the first hypothetical functional model. Note: PBS-PSII-PSI megacomplex formation used here to illustrate a putative function of anaCyp40 is only one mode of complex organization at membranes as current results point toward a flexible arrangement. Hence, reader should keep in mind that this is only one possible ensemble.
In vivo multi-color super-resolution microscopy revealed the structural organization of AnaCyp40 directly in cells. We have developed a simple and robust procedure to visualize AnaCyp40 in respect to the thylakoid and cellular membrane with a spatial resolution of 24 nm. Imaging data showed that AnaCyp40 is localized in, or at least attached to the thylakoid membrane (Figs. 5, 9), where the protein interacts with components of the PBS and the photosystems (Fig. 4, Table 1). These results support the findings obtained in this study on the association of AnaCyp40 to PBS and the photosystems in a cellular context. This interaction appears to be specific and is not dependent on the expression level or a high proline content of the interaction partner (Table 1; Supplementary Table 4). Analyzing the position of the interaction partners in existing structures of tetrameric PSI52, dimeric PSII72,73 and phycobilisomes74,75,76 shows an enrichment of those between the two photosystems and near a cavity in the PBS between the core (A1/A2) and the Rb rods. Remarkably, the cavity has a dimension of about 6–7 nm.
One model based on the obtained results would place anaCyp40 as part of the assembled structure between PBS and the photosystems (Fig. 9; green circle). In line with such interpretation, PsaE exposes a domain toward the cytoplasm52 while PsbC is a transmembrane protein facing the outer rim of PSII72,74. The other two identified PSII proteins, PsbO and PsbP (CyanoP) directly interact with PsbC77,78. Both proteins are present in the thylakoid lumen and thus either they interact with anaCyp40 through its lumenal exposed N-terminus, or the three PSII proteins together with anaCyp40 form a subcomplex that remains stable during the solubilization of the thylakoid membrane. Worth mentioning, PsbP is discussed to be important for PSII assembly79,80,81. However, we did not observe a significant reduction of PSII monomer or dimer, only of the PSI dimer (Fig. 4). Thus, this model suggests that the membrane-anchored anaCyp40 is required for the arrangement or stabilization of the photosynthetic super complex involving PBS, PSI and PSII. Consistent with such hypothesis, AFS-I-anacyp40 is sensitive to high light, which was reflected by a reduced growth and a reduction of the oxygen evolution rate and the maximal effective PSII quantum yield in the mutant strains (Figs. 2, 3). Moreover, a lower fluorescence emission from APC and PSII after excitation with 580 nm light was observed in the mutants under high light, which suggests a disturbed PBS function. A similar loss of APC activity was observed for a mutant of Synechocystis sp. PCC 6803, which cannot assemble functional phycobilisomes82. Consequently, our results are indicative for a reduced energy transfer between phycobilisomes and the photosystems in general, as the transfer between PBS and PSI or PSII is comparable between wild type and the mutant strain. The phycocyanin fluorescence after excitation of chlorophyll was enhanced in the mutant compared to wild-type when grown under high light, this can be linked to the previously discussed assembly of PBS with IsiA83, which is induced under high light conditions84. Again, the increased association of PBS with IsiA in the mutant supports the hypothesis of a destabilized assembly of PBS with the photosystems. The absence of PSI complex dimers and the reduction of tetramers observed in the mutants (Fig. 4a), the latter is likely required for PSI-PBS connectivity82, had a considerable negative effect on energy transfer from the PBS.
An alternative model could be built on the cyclophilin activity of anaCyp40. The peptidyl-prolyl cis-trans isomerase activity of the membrane anchored protein could be important for the proper folding of the PBS and PS proteins, as prerequisite of the subsequent assembly. Remarkably, 9.3% of all amino acids of PsaL are prolines and although not detected in our experiment, this protein could be a prominent target. Its malfunction would lead to reorganization of the oligomeric state of the PSI52,53. This model would define a functional relevance of anaCyp40 comparable to that of HspA, which is involved in the salt stress response85 and the protection of the photosynthetic machinery86,87,88. Such functional model for anaCyp40 would still be consistent with the membrane localization observed, because thylakoid associated ribosomes have been observed which would link translation and protein folding in the vicinity of these membranes89. However, based on electron tomography, membrane associated ribosomes and the PBS-PS complexes do not co-localize89, which leaves the association of anaCyp40 with specific PBS and PS proteins unexplained.
Summarizing, the cyclophilin anaCyp40 from the cyanobacterium Anabaena sp. is an enzyme which might have a dual functionality. One the one hand, the structural composition in conjunction with the functional properties denote the protein as factor for the general stress response, due to its proposed activity in general protein folding. On the other hand, its association with the thylakoid membrane (Fig. 9a), with the photosystems and PBS (Fig. 9b), in combination with the physiological properties of the mutant strain mark anaCyp40 as an important factor for the regulation of cyanobacterial photosynthesis. The exact mode of this regulatory function can only be speculated on at stage, and further investigation is required to provide a detailed explanation. Moreover, its relation to the dimerization of PSI and PSII and thus, to PsaL, has to be further resolved. The discrepancy to the plant proteins is in accordance with the fact that cyanobacterial D1 is not phosphorylated24, while plants do no longer contain PBS82. Thus, the function of plant TLP40 has likely been adjusted during evolution. Moreover, our structural analysis places the TLP40 family into the family of typical cyclophilins. Hence, anaCyp40 likely provides a link between photosynthetic performance and stress response. Whether these two functions co-exist, or whether anaCyp40 switches between the proposed two functions during stress remains to be established.
Methods
Bioinformatics analysis
Anabaena sp. Cyp40 orthologs were identified from OMA orthology database90 using default settings. Putative functional domains were searched using the Conserved Domain Database (CDD)91. Multiple sequence alignments were computed using MUSCLE and phylogeny was inferred by using Maximum Likelihood method with default settings of MEGA692. Branches corresponding to partitions reproduced in less than 60% bootstrap replicates are collapsed. Initial tree(s) for the heuristic search were obtained by applying the Neighbor-Joining method to a matrix of pairwise distances estimated using a JTT model. A discrete Gamma distribution was used to model evolutionary rate differences among sites (5 categories (+G, parameter = 2.4832)). The analysis involved 72 amino acid sequences. All positions with less than 95% site coverage were eliminated. That is, fewer than 5% alignment gaps, missing data, and ambiguous bases were allowed at any position. There was a total of 153 positions in the final dataset. List of genes used for phylogenetic analysis is present in Supplementary Table 1. Prediction of signal peptide is described in the text and of transmembrane domains in Supplementary Table 2.
Stress tolerance assay utilizing E. coli
anacyp40 was cloned into pET21a followed by transformation of pET21a—anacyp40 and pET21a into E. coli strain BL21 (DE3). Primers used are listed in Supplementary Table 8. The impact of abiotic stresses on E. coli BL21(DE3) strains transformed with empty pET21a and pET21a—anacyp40 was determined by spot assay after subjecting E. coli BL21 cells to different treatments of arsenic (Na3AsO4), heat, salinity (NaCl) and UV-B. Prior to stress treatment, transformed cells were grown in LB medium with 100 μg/ml ampicillin at 37 °C up to OD600nm = 0.6, followed by IPTG addition (final concentration 0.3 mM) and growth for 5 h at 16 °C. Cultures were serially diluted (10−1, 10−2, and 10−3 fold) and 3 μl from each dilution was spotted on LB-ampicillin plates comprising indicated concentrations Na3AsO4 or NaCl. For heat (50 °C) and UV-B treatment (12.9 mW m−2 nm−1), induced E. coli cells were exposed for indicated times followed by spotting on LB ampicillin plates. All plates were incubated at 37 °C overnight and images were captured.
Enzymatic analysis of PPIase activity
PPIase activity was examined at 15 °C for indicated times in a coupled reaction with chymotrypsin40. The 1 ml mixture contained 40 μM N-succinyl-ala-ala-pro-phe p-nitroanilidine in 50 mM HEPES (pH 8.0), 150 mM NaCl, 0.05% Triton X-100 and 5 nM of protein. The reaction was started by the addition of chymotrypsin (300 μg/ml) at 15 °C and the change in absorbance at 390 nm was recorded, the number of replicas is indicated in the figure legend.
Cyanobacterial and bacterial strains and plasmids generation
All strains used are listed in Supplementary Table 4. For the generation of anacyp40 single-recombinant insertion mutants (AFS-I-anacyp40), an internal fragment (168–665 bp) of the coding region was amplified by PCR from Anabaena sp. genomic DNA (oligonucleotides: Supplementary Table 8) introducing BamH1 restriction sites. PCR product was cloned into pGEM-T vector (Promega), producing plasmid pGEMT-I-anacyp40 (bp168–665) and further subcloned in pCSV3 vector93 containing an Spr Smr gene cassette, resulting in plasmid pCSV3-I-anacyp40 (Supplementary Table 9). In addition, a plasmid with a gene coding for a translational fusion of anaCyp40 with the C-terminal Strep-tag (WSHPQFEK) was constructed. The gene and Strep-tag were amplified by PCR (oligonucleotides: Supplementary Table 8) and cloned into the EcoRI sites into pGEM-T-Easy vector (Promega), producing plasmid pGEMT-Easy-anacyp40-strep. The product was cloned in pCSV3 vector93 containing an Spr/Smr gene cassette, resulting in plasmid pCSV3-anacyp40-strep (Supplementary Table 9). The plasmid was transferred into wild-type Anabaena sp. through conjugation94 producing single recombination mutant AFS-anacyp40-strep. The fusion was confirmed by colony PCR.
Transcript analysis
RNA extraction using the RNASure mini kit (Nucleo-pore) was performed from 50 mL culture (OD750nm = 0.6) of Anabaena sp. grown in BG11 before and after 1 day of incubation in 100 mM NaCl, 40 mM Na3AsO4, exposure to 42 °C for 1 h, 30 °C for 10 h or 12.9 mWm−2 nm−1 UV-B for 30 min21,22,95,96,97. Total RNA (1 µg) was reverse transcribed in a 20 μl using the iScript cDNA synthesis kit (BioRad). Transcript analysis was performed using gene specific primer for anacyp40 and for reference 16S rRNA (Supplementary Table 8). For qRT-PCR using a CFX-96 (Bio-Rad), 15 ng of cDNA extracted from each sample was used in 20 μl including 10 pmol of each, forward and reverse primers and 1x Sso fast evagreen qPCR supermix (BioRad). Transcript levels were normalized to 16S transcript and calculated relative to 0 h using the 2−ΔΔCt method98 to evaluate the relative quantities of each amplified product. The threshold cycle (Ct) was automatically determined for each reaction by the system (default parameters). The specificity of the PCR was determined by melting curve analysis of the amplified products.
Anabaena sp. growth, spotting assay and chlorophyll content estimation
Anabaena sp. was grown in BG11 medium99 under photoautotrophic conditions buffered with 8 mM TES-NaOH, pH 7.5 and 17.6 mM NaNO3 as the nitrogen source at 30 ± 2 °C under constant illumination (70 μmol photon m−2 s−1, photosynthetically active radiation). Cultures of AFS-I-anacyp40 and AFS-anacyp40-strep contained 3 μg ml−1 streptomycin/3 μg ml−1 spectinomycin. Agar plates were prepared with 1% Bacto™ Agar (Otto Nord Wald, G).
For growth curve analysis, cells were grown in BG11 liquid medium, washed three times by centrifugation (5000 × g, RT, 3 min) and resuspended in BG11 adjusted to an OD750 of 0.05. All experiments were performed in triplicates in 50 ml culture volumes. Alternative additions ore treatments are described in the text. OD750 values were recorded once a day for 8 days. For growth analysis on solid medium, cells were spotted in triplicates at different OD750 (0.25, 1.0) on standard BG11 agar medium either in the presence or absence of the indicated amounts of NaCl concentrations. Chlorophyll a concentration was determined41.
77 K Fluorescence emission spectra
77 K fluorescence measurements were performed using an JASCO spectrofluorimeter (FP-8700 spectrofluorimeter, JASCO). Cuvettes were kept immersed in liquid nitrogen and transferred to the measuring cell of the fluorimeter one at a time. Chla excitation was performed at 440 nm and PBS excitation at 590 nm. Fluorescence emission was measured from 600 to 800 nm. Assignment of peaks was made according to literature47,48,49,50.
Chlorophyll fluorescence analysis and oxygen evolution measurements
Chlorophyll fluorescence analysis were acquired using a Closed FluorCam FC 800-C (Photon System Instruments, Brno, Czech Republic) with a CCD camera and four fixed LED panels. All images were captured using the FLUORCAM 7 software (Photon System Instruments). Chl a fluorescence images were taken using the imaging PAM fluorimeter100. In brief, cells in 2 ml of liquid culture were dark-adapted for 15 min and the minimum fluorescence level (Fo) was determined with a measuring light. After start of the measurement, a 0.8 s pulse of saturating light was applied (determination of Fm′dark) followed by exposure to actinic light for 300 s with saturating pulses in between (determination of Fm′). The actinic light was then turned off and saturating light was applied to monitor recovery kinetics of Fm′ in the absence of actinic light. Fluorescence parameters were determined by established protocols101.
Oxygen evolution was quantified in 2 mL culture samples (adjusted to 20 μg chlorophyll mL−1) in BG11 with 10 mM NaHCO3 using a Clark-type electrode and DW2 Oxygen Electrode Chamber (Hansatech) with constant stirring at 30 °C. Light was introduced to samples using an LS2/H tungsten-halogen 100 W light source. All measurements were performed in triplicate.
Thylakoid membrane preparation and BN-PAGE
Isolated thylakoid membranes102 equivalent to 1 mg Chl ml–1 was solubilized with 1% n-Dodecyl β-D-maltoside (β-DM) on ice for 15 min, followed by centrifugation at 9391 × g rpm for 10 min at 4 °C. The supernatant (equivalent to 7 μg of Chl) was subjected to BN-PAGE103 with a gradient of 5–13.5% (w/v) acrylamide in the separation gel. Complex assignments were performed according to existing data in the lab based on previous reports104.
Phycobilisome (PBS) Isolation
PBS was isolated according to established protocols105 with few modifications. 500 ml cells grown in BG11 were harvested, washed twice with 0.9 M potassium phosphate buffer (pH 7.0), and crushed in liquid nitrogen thrice. The cell extract was treated with 2% (vol/vol) Triton X-100 in 0.9 M phosphate buffer for 30 min and centrifuged at 20,000 × g for 20 min at 18 °C to separate the extract into the upper green Triton X-100 layer and the lower blue aqueous layer. The blue layer was loaded onto a 10–50% (wt/vol) linear sucrose density gradient with 0.9 M phosphate buffer and centrifuged at 130,000 × g for 16 h at 18 °C.
Affinity Purification of anaCyp40 Complexes
500 ml culture (OD ~ 3.0) of AFS-anacyp40-strep or wild-type Anabaena sp. was centrifuged at 8000 rpm for 10 min. Cell pellets were resuspended in 15 ml of ice-cold buffer (100 mM Tris, pH 8.0, 50 mM NaCl, 5 mM β-mercaptoethanol, 1 mM PMSF). Following three cycles of lysis in liquid nitrogen, 1 mg/ml lysozyme, 1% β-DM and Protease inhibitor cocktail was added. After incubation for 60 min at 4 °C on rotary shaker, the whole cell lysate was centrifuged at 3500 rpm for 2 min and biotin blocking buffer was added. This fraction was loaded onto MagStrep “type3” XT beads (IBA Life Sciences) preequilibrated with lysis buffer and left on rotary shaker for 60 min at 4 °C. The column was washed with 100 mM Tris, pH 8.0, 1 mM β-mercaptoethanol, 1 mM PMSF, 1 mM EDTA containing 50 mM NaCl (WB1), 100 mM NaCl (WB2) or 100 mM NaCl and 0.5% Nonidet P-40 (WB3). anaCyp40 protein complexes were eluted WB1 containing 50 mM D-biotin. Protein abundance was tracked using Strep-Tactin® HRP conjugate (IBA Life Sciences).
Tryptic in solution digestion and liquid chromatography electrospray ionization MS
Elutions form affinity purification were pooled and and 2X Lysis Buffer (5% DCA, 5 mM TCEP, 100 mM NH4HCO3) was added, mixed at ~1000 rpm in a thermo-mixer for 10 min at 90 °C. Subsequent alkylation was performed (15 mM IAA, 3 mM NH4HCO3, pH 8) and samples were kept in the dark for 30 min followed by dilution in dilution buffer up to final concentration of 0.5% DCA, 0.5 mM TCEP, 100 mM NH4HCO3. Trypsin was added to the samples (1:500 w/w) and incubated at 37 °C for overnight. Reaction was stopped by addition of 1.0% TFA. The samples were desalted using C18 column106. Tryptic peptides recovered from C18 column were fractionated using strong cation-exchange (SCX) StageTips107. In total, 4 SCX fractions were collected by eluting first 3 fractions in 75, 200, 400 NH4HCO3/30% ACN/0.5% TFA solutions followed by a final elution with 5% mM ammonium hydroxide/50% ACN. Fractions eluted were dried in a SpeedVac, resuspended in ddH2O and subjected to LC-MS analysis as described55.
The three experiments (experiment 1: one biological probe and 12 controls, experiment 2 and 3: three biological replicas and three controls) were analyzed to determine the LFQ value for each experiment. For further analysis, only proteins with identified peptides in all bait-dependent precipitations and an LFQ > 0 in at least 2 of the bait containing probes were considered. The ratio of log10(LFQ + 1)BAIT-log10(LFQ + 1)CONTROL was calculated and the p-value for the distribution of the ratios was determined for the hypothesis of a median ≤0 by one way t-test (Sigma Plot, SPSS).
Super-resolution imaging
Fixation of cells was performed according to existing protocols108. In brief, 20 µl of cyanobacterial culture (OD750 = 0.3) was added to poly-lysine coated surfaces and dried for 20 min at 55 °C followed by fixation in 70% ethanol and incubation for 30 min at −20 °C. The surfaces were air-dried for 20 min at room temperature and permeabilized by adding a drop of 0.05% Triton X-100 in PBS for 2 min at room temperature, and repeated it three times by removing the drop each time with a pipette. Afterward, incubated with a drop of 3% BSA (bovine serum albumin), 0.2% Triton X-100 in PBS for 1 h at 4 °C in a moisture chamber and removed the blocking solution. Cells were incubated with anti-strep primary antibody for 2 h at 4 °C in a moisture chamber and subsequently washed three times at room temperature. Afterward, incubated with oligolabelled secondary antibody for 1 h at 4 °C in a moisture chamber followed by three washing steps. The docking strand (DNA strand at the secondary antibody) was P1.
Super-resolution imaging was performed on a home-built microscope as described109. The software μManager110 was used to control the microscope hardware and the Andor Ixon Ultra EMCCD camera (DU-897U-CS0-#BV; Andor). Measurements were performed applying a highly inclined and laminated optical sheet111. First, samples were illuminated with a 647 nm excitation laser to detect the autofluorescence until all cellular fluorophores were bleached. Typically, 60 min of imaging data were recorded at a frame rate of 20 Hz (72000 frames). Second, 400 pM P1-Atto655 imager strand in PBS supplemented with 500 mM NaCl was added to the sample and excited at 647 nm using the same laser. 48,000 frames with a frame rate of 20 Hz were recorded to visualize the P1 labeled prolylisomerase. Third, 500 pM NileRed in PBS was added to the sample to visualize the outer and plasma membrane112, illuminated with a 568 nm excitation laser and 15,000 frames with a frame rate of 50 Hz were recorded. Laser densities range between 0.5 and 5 kW/cm2.
Super-resolution images were generated using rapidSTORM113 or Picasso114 and post-processed using Fiji100. Single-molecule localizations were fitted using free PSF fit parameters in rapidSTORM. For autofluorescence and NileRed, a global threshold of 132 photons was applied, and for Atto655, a global threshold of 1320 photons was used. Localizations were filtered by their full width at half maximum (220 nm < FWHM < 440 nm for the autofluorescence and Atto655, 280 nm < FWHM < 360 nm for NileRed) and PSF asymmetry (0.7 < FWHM(x)/FWHM(y) < 1.3). Brightness and contrast were adjusted for optimal visualization. The software Origin2019 (OriginLab Corporation) was used for calculations and graphical data representation. Parameter-free image resolution estimation based on decorrelation analysis was performed using the Fiji plugin ImageDecorrelationAnalysis57.
Cluster analysis was performed using the DBSCAN algorithm implemented in the LAMA software115. Localizations with equal or more than 10 neighbors (minpts = 10) within a 20 nm radius were assigned to a cluster.
Protein Purification for crystallization
The 10x-His fusion proteins (F7XC3H-anacyp40ΔTM(1-35)-HRV 3C-His10 construct) was overexpressed in E. coli BL21 (DE3). Cells were grown in Luria-Bertani medium to an OD600 of 0.5–0.7 at 37 °C. Expression of the recombinant protein was induced with 1.0 mM isopropylthio-β-galactoside at 25 °C for 16 h. Cells were harvested by centrifugation (4 °C; 6000 × g; 10 min). The bacterial pellet was suspended in ice-cold lysis buffer (20 mM Tris-HCl pH 7.0, 150 mM NaCl), lysed by French press (Thermo Scientific, Waltham, USA) at 1200 psi followed by centrifugation (30.000 × g, 4 °C, 30 min). The supernatant was loaded onto Ni-NTA affinity resin (Qiagen, Hilden, Germany), pre-equilibrated with lysis buffer and washed trice with lysis buffer containing 40 mM imidazole to remove contaminants. On-column cleavage of histidine tag was performed using PreScission protease116 in 50 mM Tris-HCl pH 7.0, 150 mM NaCl, 1 mM EDTA and 1 mM DTT at an enzyme: protein ratio of 1:50. The cleaved protein has additional seven C-terminal residues. The flow through was collected and cleared by size exclusion chromatography (Superdex 200, GE Healthcare, Solingen, Germany) pre-equilibrated in lysis buffer and subsequently concentrated using amicon ultra centrifugal filters (30 kDa molecular-weight cutoff filter, Merck, Darmstadt, G) up to ~15 mg/ml.
Crystallization, data collection, and structure determination
Single crystals of anaCyp40ΔTM were obtained after 7 days from drops consisting of 0.2 M ammonium citrate, 20% PEG 3350, and 10% Morpheus additive screen (PEG 3350 precipitant 25.00 % w/v)39. Crystals were immersed for 5 min at 293 K in cryoprotectant solution consisting of 20% Glycerol and 18% sucrose followed by flash-cooling in liquid nitrogen. A total of 7600 X-ray diffraction images were collected on the id29 beamline at the European Synchrotron Radiation Facility (ESRF, France) at 100 K in a nitrogen-gas stream and at a wavelength of 0.9795 Å. The data were indexed, integrated and scaled with the XDS program package117 and the structure was solved by molecular replacement118 using PHASER119 from the CCP4 program suite120 with the structure of atCyp38 (pdb-id 3RFY) as search model23. The high-resolution model was refined by iterative cycles of manual rebuilding using COOT121 and phenix.refine122 Figures were drawn using ChimeraX123. Electronic potentials for surface presentation were calculated using PDB2PQR124 and APBS.
Accession numbers
Sequence data from this article can be found in the UniProtKB under identifier Q8YM80. The coordinates and structure factors for anaCyp40ΔTM have been deposited at the Protein Data Bank under the entry code 7A73.
Reporting summary
Further information on research design is available in the Nature Research Reporting Summary linked to this article.
Data availability
The atomic coordinates for anaCyp40ΔTM have been deposited in the Protein Data Bank with the accession code 7A73. The source data underlying Figs. 1, 2, 3, 5 are provided as a Source data file. Other data that support the findings of this study are available upon request to the corresponding author. Source data are provided with this paper.
References
Andrzej, G. Peptidylprolyl cis/trans isomerases (immunophilins): biological diversity—targets—functions. Curr. Top. Med. Chem. 3, 1315–1347 (2003).
Schiene-Fischer, C. Multidomain peptidyl prolyl cis/trans isomerases. Biochim. Biophys. Acta 1850, 2005–2016 (2015).
Romano, P., Gray, J., Horton, P. & Luan, S. Plant immunophilins: functional versatility beyond protein maturation. N. Phytol. 166, 753–769 (2005).
Schreiber, S. L. Chemistry and biology of the immunophilins and their immunosuppressive ligands. Science 251, 283–287 (1991).
Stewart, D. E., Sarkar, A. & Wampler, J. E. Occurrence and role of cis peptide bonds in protein structures. J. Mol. Biol. 214, 253–260 (1990).
Schmidpeter, P. A. & Schmid, F. X. Prolyl isomerization and its catalysis in protein folding and protein function. J. Mol. Biol. 427, 1609–1631 (2015).
Ferreira, P. A., Nakayama, T. A., Pak, W. L. & Travis, G. H. Cyclophilin-related protein RanBP2 acts as chaperone for red/green opsin. Nature 383, 637–640 (1996).
Krzywicka, A. et al. KIN241: a gene involved in cell morphogenesis in paramecium tetraurelia reveals a novel protein family of cyclophilin-RNA interacting proteins (CRIPs) conserved from fission yeast to man. Mol. Microbiol. 42, 257–267 (2001).
Horowitz, D. S., Lee, E. J., Mabon, S. A. & Misteli, T. A cyclophilin functions in pre-mRNA splicing. EMBO J., 21, 470–480 (2002).
Iki, T., Yoshikawa, M., Meshi, T. & Ishikawa, M. Cyclophilin 40 facilitates HSP90-mediated RISC assembly in plants. EMBO J. 31, 267–278 (2012).
Brazin, K. N., Mallis, R. J., Fulton, D. B. & Andreotti, A. H. Regulation of the tyrosine kinase Itk by the peptidyl-prolyl isomerase cyclophilin A. Proc. Natl Acad. Sci. USA 99, 1899–1904 (2002).
Heitman, J., Movva, N. R. & Hall, M. N. Targets for cell cycle arrest by the immunosuppressant rapamycin in yeast. Science 25, 905–909 (1991).
Fu, A. et al. A chloroplast cyclophilin functions in the assembly and maintenance of photosystem II in Arabidopsis thaliana. Proc. Natl Acad. Sci. USA 104, 15947–15952 (2007).
Sirpio, S. et al. AtCYP40 ensures early biogenesis, correct assembly and sustenance of photosystem II. Plant J. 55, 639–651 (2008).
Fulgosi, H., Vener, A. V., Altschmied, L., Herrmann, R. G. & Andersson, B. A novel multi-functional chloroplast protein: identification of a 40 kDa immunophilin-like protein located in the thylakoid lumen. EMBO J. 17, 1577–1587 (1998).
Vasudevan, D. et al. Plant immunophilins: a review of their structure-function relationship. Biochim. Biophys. Acta 1850, 2145–2158 (2015).
Ku, C. et al. Endosymbiotic origin and differential loss of eukaryotic genes. Nature 524, 427–432 (2015).
Whitton, B. A. In The Ecology of cyanobacteria (Kluwer Academic Publishers, Dordrecht, 2000).
Hassidim, M. et al. A cyanobacterial gene encoding peptidyl-prolyl cis-trans isomerase. Plant Phys. 100, 1982–1986 (1992).
Agrawal, C. et al. Comparative proteomics reveals association of early accumulated proteins in conferring butachlor tolerance in three N2-fixing Anabaena spp. J. Proteom. 96, 271–290 (2014).
Rai, S., Agrawal, C., Shrivastava, A. K., Singh, P. K. & Rai, L. C. Comparative proteomics unveils cross species variations in Anabaena under salt stress. J. Proteom. 98, 254–270 (2014).
Shrivastava, A. K. et al. UV-B stress induced metabolic rearrangements explored with comparative proteomics in three Anabaena species. J. Proteom. 127, 122–133 (2015).
Vasudevan, D., Fu, A., Luan, S. & Swaminathan, K. Crystal structure of Arabidopsis cyclophilin38 reveals a previously uncharacterized immunophilin fold and a possible autoinhibitory mechanism. Plant Cell 24, 2666–2674 (2012).
Pursiheimo, S., Rintamäki, E., Baena-Gonzalez, E. & Aro, E.-M. Thylakoid protein phosphorylation in evolutionally divergent species with oxygenic photosynthesis. FEBS Lett. 423, 178–182 (1998).
Shen, J. R. The structure of photosystem II and the mechanism of water oxidation in photosynthesis. Annu. Rev. Plant. Biol. 66, 23–48 (2015).
Nelson, N. & Junge, W. Structure and energy transfer in photosystems of oxygenic photosynthesis. Annu. Rev. Biochem. 84, 659–683 (2015).
Hankamer, B., Morris, E. P. & Barber, J. Revealing the structure of the oxygen-evolving core dimer of photosystem II by cryoelectron crystallography. Nat. Struct. Biol. 6, 560–564 (1999).
Plöchinger, M., Schwenkert, S., von Sydow, L., Schröder, W. P. & Meurer, J. Functional update of the auxiliary proteins PsbW, PsbY, HCF136, PsbN, TerC and ALB3 in maintenance and assembly of PSII. Front. Plant Sci. 7, 423 (2016).
Heinz, S., Liauw, P., Nickelsen, J. & Nowaczyk, M. Analysis of photosystem II biogenesis in cyanobacteria. Biochim. Biophys. Acta 1857, 274–287 (2016).
Li, M. et al. Physiological and evolutionary implications of tetrameric photosystem I in cyanobacteria. Nat. Plants 5, 1309–1319 (2019).
Sener, M. K. et al. Comparison of the light-harvesting networks of plant and cyanobacterial photosystem I.” Biophys. J. 89, 1630–1642 (2005).
Grotjohann, I. & Fromme, P. Structure of cyanobacterial photosystem I. Photosynth. Res. 85, 51–72 (2005).
Watanabe, M. & Ikeuchi, M. Phycobilisome: architecture of a light-harvesting supercomplex. Photosynth Res. 116, 265–276 (2013).
Liu, H. et al. Phycobilisomes supply excitations to both photosystems in a megacomplex in cyanobacteria. Science 342, 1104–1107 (2013).
Watanabe, M. et al. Attachment of phycobilisomes in an antenna–photosystem I supercomplex of cyanobacteria. Proc. Nat. Acad. Sci. USA 111, 2512–2517 (2014).
Pan, X., Liu, Z., Li, M. & Chang, W. Architecture and function of plant light-harvesting complexes II. Curr. Opin. Struct. Biol. 23, 515–525 (2013).
Almagro Armenteros, J. J. et al. SignalP 5.0 improves signal peptide predictions using deep neural networks. Nat. Biotechnol. 37, 420–423 (2019).
Sazuka, T. Proteomic analysis of the cyanobacterium Anabaena sp. strain PCC 7120 with two-dimensional gel electrophoresis and amino-terminal sequencing. Photosynth. Res. 78, 279–291 (2003).
Yadav, S. et al. Purification and preliminary X-Ray crystallographic analysis of the peptidyl-prolyl cis-trans isomerase Alr5059 from Anabaena sp. PCC 7120. Crystallography Rep. 65, 1226–1230 (2020)
Fischer, G., Bag, H. & Mech, C. Determination of enzymatic catalysis for the cis-trans-isomerization of peptide binding in proline-containing peptides. Biomed. Biochim. Acta 43, 1101–1111 (1984).
Nicolaisen, K. et al. Alr0397 is an outer membrane transporter for the Siderophore Schizokinen in Anabaena sp. strain PCC 7120. J. Bacteriol. 190, 7500–7507 (2008).
Brenes-Álvarez, M. et al. Elements of the heterocyst-specific transcriptome unravelled by co-expression analysis in Nostoc sp. PCC 7120. Environ. Microbiol. 21, 2544–2558 (2019).
McGary, K. & Nudler, E. RNA polymerase and the ribosome: the close relationship. Curr. Opin. Microbiol. 16, 112–117 (2013).
Ferbitz, L. et al. Trigger factor in complex with the ribosome forms a molecular cradle for nascent proteins. Nature 431, 590–596 (2004).
Gudavicius, G. et al. The prolyl isomerase, FKBP25, interacts with RNA-engaged nucleolin and the pre-60S ribosomal subunit. RNA 20, 1014–1022 (2014).
Flaherty, B. L., Van Nieuwerburgh, F., Head, S. R. & Golden, J. W. Directional RNA deep sequencing sheds new light on the transcriptional response of Anabaena sp. strain PCC 7120 to combined-nitrogen deprivation. BMC Genomics. 12, 332 (2011).
Su, X., Fraenkel, P. G. & Bogorad, L. Excitation energy transfer from phycocyanin to chlorophyll in an apcA-defective mutant of Synechocystis sp. PCC 6803. J. Biol. Chem. 267, 22944–22950 (1992).
Shen, G., Boussiba, S. & Vermaas, W. F. Synechocystis sp PCC 6803 strains lacking photosystem I and phycobilisome function. Plant Cell. 5, 1853–1863 (1993).
Masuda, T. et al. Diel regulation of photosynthetic activity in the oceanic unicellula diazotrophic cyanobacterium Crocosphaera watsonii WH8501. Environ. Microbiol. 20, 546–560 (2018).
Calzadilla, P. I. et al. The cytochrome b6f complex is not involved in Cyanobacterial state transitions. Plant Cell. 31, 911–931 (2019).
Kondo, K., Ochiai, Y., Katayama, M. & Ikeuchi, M. The membrane-associated CpcG2-phycobilisome in Synechocystis: a new photosystem I antenna. Plant Physiol. 144, 1200–1210 (2007).
Kato, K. et al. Structure of a cyanobacterial photosystem I tetramer revealed by cryo-electron microscopy. Nat. Commun. 10, 4929 (2019).
Zheng, L. et al. Structural and functional insights into the tetrameric photosystem I from heterocyst-forming cyanobacteria. Nat. Plants 5, 1087–1097 (2019).
Bolte, K. et al. Complementation of a phycocyanin-bilin lyase from Synechocystis sp. PCC 6803 with a nucleomorph-encoded open reading frame from the cryptophyte Guillardia theta. BMC Plant Biol. 8, 56–56 (2008).
Brouwer, E. M., Ngo, G., Yadav, S., Ladig, R. & Schleiff, E. Tic22 from Anabaena sp. PCC 7120 with holdase function involved in outer membrane protein biogenesis shuttles between plasma membrane and Omp85. Mol. Microbiol. 111, 1302–1316 (2019).
Sauer, M. & Heilemann, M. Single-molecule localization microscopy in eukaryotes. Chem. Rev. 117, 7478–7509 (2017).
Descloux, A., Grußmayer, K. S. & Radenovic, A. Parameter-free image resolution estimation based on decorrelation analysis. Nat. Methods 16, 918–924 (2019).
Paddock, S. W. Confocal laser scanning microscopy. Biotechniques 27, 992–1004 (1999).
Balsera, M., Arellano, J. B., Revuelta, J. L., de las Rivas, J. & Hermoso, J. A. The 1.49 A resolution crystal structure of PsbQ from photosystem II of Spinacia oleracea reveals a PPII structure in the N-terminal region. J. Mol. Biol. 350, 1051–1060 (2005).
Rathner, P. et al. Solution NMR and molecular dynamics reveal a persistent alpha helix within the dynamic region of PsbQ from photosystem II of higher plants. Proteins 83, 1677–1686 (2015).
Jackson, S. A., Fagerlund, R. D., Wilbanks, S. M. & Eaton-Rye, J. J. Crystal structure of PsbQ from Synechocystis sp. PCC 6803 at 1.8 A: implications for binding and function in cyanobacterial photosystem II. Biochem 49, 2765–2767 (2010).
Holm, L. DALI and the persistence of protein shape. Protein Sci. 29, 128–140 (2020).
Tahirov, T. H. et al. Structure of cytochrome c’ from Rhodobacter capsulatus strain St Louis: an unusual molecular association induced by bridging Zn ions. Acta Crystallogr. D. 53, 658–664 (1997).
Arnesano, F. et al. Structural consequences of b- to c-type heme conversion in oxidized Escherichia coli cytochrome b562. Biochem 39, 1499–1514 (2000).
Xie, J. et al. Structural determination of the complement inhibitory domain of Borrelia burgdorferi BBK32 provides insight into classical pathway complement evasion by Lyme disease spirochetes. PLoS Pathog. 15, e1007659 (2019).
Kono, A. et al. Structure and function of LCI1: a plasma membrane CO2 channel in the Chlamydomonas CO2 concentrating mechanism. Plant J. 102, 1107–1126 (2020).
Mikol, V., Kallen, J., Pflügl, G. & Walkinshaw, M. D. X-ray structure of a monomeric cyclophilin A-cyclosporin A crystal complex at 2.1 A resolution. J. Mol. Biol. 234, 1119–1130 (1993).
Peterson, M. R. et al. The three-dimensional structure of a Plasmodium falciparum cyclophilin in complex with the potent anti-malarial cyclosporin A. J. Mol. Biol. 298, 123–133 (2000).
Aldape, R. A. et al. Charged surface residues of FKBP12 participate in formation of the FKBP12–FK506–calcineurin complex. J. Biol. Chem. 267, 16029–16032 (1992).
Vener, A. V., Fulgosi, H., Herrmann, R. G. & Andersson, B. In Photosynthesis: Mechanisms and Effects (ed. Garab, G.) (Springer, 1998).
Hanhart, P. et al. Enzyme activity and structural features of three single-domain phloem cyclophilins from Brassica napus. Sci. Rep. 9, 9368 (2019).
Guskov, A. et al. Cyanobacterial photosystem II at 2.9-A resolution and the role of quinones, lipids, channels and chloride. Nat. Struct. Mol. Biol. 16, 334–342 (2009).
Jordan, P. et al. Three-dimensional structure of cyanobacterial photosystem I at 2.5 A resolution. Nature 411, 909–917 (2001).
Chang, L. et al. Structural organization of an intact phycobilisome and its association with photosystem II. Cell Res. 25, 726–737 (2015).
Zhang, J. et al. Structure of phycobilisome from the red alga Griffithsia pacifica. Nature 551, 57–63 (2017). 2017.
Ma, J. et al. Structural basis of energy transfer in Porphyridium purpureum phycobilisome. Nature 579, 146–151 (2020).
Ido, K. et al. Cross-linking evidence for multiple interactions of the PsbP and PsbQ proteins in a higher plant photosystem II supercomplex. J. Biol. Chem. 289, 20150–20157 (2014).
Cormann, K. U., Bartsch, M., Rögner, M. & Nowaczyk, M. M. Localization of the CyanoP binding site on photosystem II by surface plasmon resonance spectroscopy. Front. Plant Sci. 5, 595 (2014).
Thornton, L. E. et al. Homologs of plant PsbP and PsbQ proteins are necessary for regulation of photosystem ii activity in the cyanobacterium Synechocystis 6803. Plant Cell. 16, 2164–2175 (2004).
Knoppová, J., Yu, J., Konik, P., Nixon, P. J. & Komenda, J. CyanoP is involved in the early steps of photosystem II assembly in the Cyanobacterium Synechocystis sp. PCC 6803. Plant Cell Physiol. 57, 1921–1931 (2016).
Nagarajan, A., Page, L. E., Liberton, M. & Pakrasi, H. B. Consequences of decreased light harvesting capability on photosystem II function in Synechocystis sp. PCC 6803. Life (Basel). 4, 903–914 (2014).
Green, B. R. What happened to the phycobilisome? Biomolecules 9, 748 (2019).
Joshua, S., Bailey, S., Mann, N. H. & Mullineaux, C. W. Involvement of phycobilisome diffusion in energy quenching in cyanobacteria. Plant Physiol. 138, 1577–1585 (2005).
Havaux, M. et al. The chlorophyll-binding protein IsiA is inducible by high light and protects the cyanobacterium Synechocystis PCC 6803 from photooxidative stress. FEBS Lett. 579, 2289–2293 (2005).
Asadulghani et al. Comparative analysis of the hspA mutant and wild-type Synechocystis sp. strain PCC 6803 under salt stress: evaluation of the role of hspA in salt-stress management. Arch. Microbiol. 182, 487–497 (2004).
Nakamoto, H., Suzuki, N. & Roy, S. K. Constitutive expression of a small heat-shock protein confers cellular thermotolerance and thermal protection to the photosynthetic apparatus in cyanobacteria. FEBS Lett. 483, 169–174 (2000).
Nakamoto, H. & Honma, D. Interaction of a small heat shock protein with light-harvesting cyanobacterial phycocyanins under stress conditions. FEBS Lett. 580, 3029–3034 (2006).
Rowland, J. G. et al. Identification of components associated with thermal acclimation of photosystem II in Synechocystis sp. PCC6803. PLoS ONE 5, e10511 (2010).
Rast, A. et al. Biogenic regions of cyanobacterial thylakoids form contact sites with the plasma membrane. Nat. Plants 5, 436–446 (2019).
Altenhoff, A. M. et al. The OMA orthology database in 2018: retrieving evolutionary relationships among all domains of life through richer web and programmatic interfaces. Nucl. Acids Res. 46, D477–D485 (2017).
Lu, S. et al. CDD/SPARCLE: the conserved domain database in 2020. Nuc. Acids Res. 48, D265–D268 (2020).
Tamura, K., Stecher, G., Peterson, D., Filipski, A. & Kumar, S. MEGA6: molecular evolutionary genetics analysis version 6.0. Mol. Biol. Evol. 30, 2725–2729 (2013).
Olmedo-Verd, E., Muro-Pastor, A. M., Flores, E. & Herrero, A. Localized induction of the ntcA regulatory gene in developing heterocysts of Anabaena sp. strain PCC 7120. J. Bacteriol. 188, 6694–6699 (2006).
Elhai, J. & Wolk, C. P. Conjugal transfer of DNA to cyanobacteria. Methods Enzymol. 167, 747–754 (1988).
Pandey, S., Rai, R. & Rai, L. C. Proteomics combines morphological, physiological and biochemical attributes to unravel the survival strategy of Anabaena sp. PCC7120 under arsenic stress. J. Proteom. 75, 921–937 (2012).
Rajaram, H. & Apte, S. K. Nitrogen status and heat-stress-dependent differential expression of the cpn60 chaperonin gene influences thermotolerance in the cyanobacterium Anabaena. Microbiol 154, 317–325 (2008).
Katoh, H., Asthana, R. K. & Ohmori, M. Gene expression in the cyanobacterium Anabaena sp. PCC7120 under desiccation. Microb. Ecol. 47, 164–174 (2004).
Livak, K. J. & Schmittgen, T. D. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods 25, 402–408 (2001).
Rippka, R., Deruelles, J., Waterbury, J. B., Herdman, M. & Stanier, R. Y. Generic assignments, strain histories and properties of pure cultures of Cyanobacteria. Microbiol 111, 1–61 (1979).
Schreiber, U., Schliwa, U. & Bilger, W. Continuous recording of photochemical and non-photochemical chlorophyll fluorescence quenching with a new type of modulation fluorometer. Photosynth. Res. 10, 51–62 (1986).
Misumi, M., Katoh, H., Tomo, T. & Sonoike, K. Relationship between photochemical quenching and non-photochemical quenching in six species of Cyanobacteria reveals species difference in redox state and species commonality in energy dissipation. Plant Cell Physiol. 57, 1510–1517 (2016).
Moslavac, S. et al. Proteomic analysis of the outer membrane of Anabaena sp. strain PCC 7120. J. Proteome Res. 4, 1330–1338 (2005).
Schägger, H. & von Jagow, G.Blue native electrophoresis for isolation of membrane protein complexes in enzymatically active form. Anal. Biochem. 199, 223–231 (1991).
Ladig, R. et al. A high-definition native polyacrylamide gel electrophoresis system for the analysis of membrane complexes. Plant J. 67, 181–194 (2011).
Gray, B. H. & Gantt, E. Spectral properties of phycobilisomes and phycobiliproteins from the blue-green alga-nostoc sp. Photochem. Photobiol. 21, 121–128 (1975).
Rappsilber, J., Ishihama, Y. & Mann, M. Stop and go extraction tips for matrix-assisted laser desorption/ionization, nanoelectrospray, and LC/MS sample pretreatment in proteomics. Anal. Chem. 75, 663–670 (2003).
Chan, K. C. & Issaq, H. J. Fractionation of peptides by strong cation-exchange liquid chromatography. Methods Mol. Biol. 1002, 311–315 (2013).
Trigo, C., Andrade, D. & Vásquez, M. Protein localization in the Cyanobacterium Anabaena sp. PCC7120 Using Immunofluorescence Labeling. Bio-Protoc. 7, e2318 (2017).
Spahn, C., Cella-Zannacchi, F., Endesfelder, U. & Heilemann, M. Correlative super-resolution imaging of RNA polymerase distribution and dynamics, bacterial membrane and chromosomal structure in Escherichia coli. Methods Appl. Fluoresc. 3, 014005 (2015).
Schindelin, J. et al. Fiji: an open-source platform for biological-image analysis. Nat. Meth. 9, 676–682 (2012).
Tokunaga, M., Imamoto, N. & Sakata-Sogawa, K. Highly inclined thin illumination enables clear single-molecule imaging in cells. Nat. Meth. 5, 159–161 (2008).
Spahn, C. K. et al. A toolbox for multiplexed super-resolution imaging of the E. coli nucleoid and membrane using novel PAINT labels. Sci. Rep. 8, 14768 (2018).
Wolter, S. et al. Real-time computation of subdiffraction-resolution fluorescence images. J. Microsc. 237, 12–22 (2010).
Schnitzbauer, J., Strauss, M. T., Schlichthaerle, T., Schueder, F. & Jungmann, R. Super-resolution microscopy with DNA-PAINT. Nat. Protoc. 12, 1198–1228 (2017).
Malkusch, S. & Heilemann, M. Extracting quantitative information from single-molecule super-resolution imaging data with LAMA–LocAlization Microscopy Analyzer. Sci. Rep. 6, 1–4 (2016).
Leong, L. E. C., Walker, P. A. & Porter, A. G. Efficient expression and purification of a protease from the common cold virus, human rhinovirus type 14. J. Cryst. Growth 122, 246–252 (1992).
Kabsch, W. XDS. Acta Cryst. D. 66, 125–132 (2010).
Rossmann, M. G. The molecular replacement method. Acta Cryst. A 46, 73–82 (1990).
McCoy, A. J. et al. Phaser crystallographic software. J. Appl. Crystallogr. 40, 658–674 (2007).
Winn, M. D. et al. Overview of the CCP4 suite and current developments. Acta Cryst. D. 67, 235–242 (2011).
Emsley, P., Lohkamp, B., Scott, W. G. & Cowtan, K. Features and development of Coot. Acta Cryst. D. 66, 486–501 (2010).
Liebschner, D. et al. Macromolecular structure determination using X-rays, neutrons and electrons: recent developments in Phenix. Acta Cryst. D. 75, 861–877 (2019).
Goddard, T. D. et al. UCSF ChimeraX: meeting modern challenges in visualization and analysis. Protein Sci. 27, 14–25 (2018).
Dolinsky, T. J., Nielsen, J. E., McCammon, J. A. & Baker, N. A. PDB2PQR: an automated pipeline for the setup of Poisson-Boltzmann electrostatics calculations. Nucleic Acids Res. 32, W665–W667 (2004).
Campos, B. M. et al. A redox 2-Cys mechanism regulates the catalytic activity of divergent cyclophilins. Plant Physiol. 162, 1311–1323 (2013).
Zhao, Y., Chen, Y., Schutkowski, M., Fischer, G. & Ke, H. Cyclophilin A complexed with a fragment of HIV-1 gag protein: insights into HIV-1 infectious activity. Structure 5, 139–146 (1997).
Favretto, F. et al. The molecular basis of the interaction of Cyclophilin A with α-synuclein. Angew. Chem. Int Ed. Engl. 59, 5643–5646 (2020).
Acknowledgements
We are very thankful for the support and help by Hannah Schätzle during revision of the manuscript. We are grateful to Prof. Werner Kühlbrandt (MPI for Biophysics, Frankfurt, G) for the constant support as well as continuous encouraging and critical discussions. We thank Prof. Klaas Martinus Pos (Goethe University Frankfurt, G) for his constant support. We thank Prof. Claudia Büchel and Dr. Lars Dietzel for their continuous discussions and support. We thank the beam line staff at ID29 and ID23-1 beamlines of the European Synchrotron Radiation Facility (ESRF) for their help during crystal screening and data collection. Molecular graphics and analyses performed with UCSF ChimeraX, developed by the Resource for Biocomputing, Visualization, and Informatics at the University of California, San Francisco, with support from National Institutes of Health R01-GM129325 and the Office of Cyber Infrastructure and Computational Biology, National Institute of Allergy and Infectious Diseases. M.G. and M.H. acknowledge funding by the Deutsche Forschungsgemeinschaft (DFG HE6166/17-1). This work was supported by grants from the Deutsche Forschungsgemeinschaft (DFG SCHL585/7-2) and a fellowship by the DAAD to Shivam Yadav.
Funding
Open Access funding enabled and organized by Projekt DEAL.
Author information
Authors and Affiliations
Contributions
S.Y., L.C.R., and E.S. designed the experiments. S.Y. performed cloning, expression, bacterial and photosynthetic experiments. S.Y. and R.L. performed the mass-spectrometric analysis, M.G. and M.H. designed and performed the super-resolution fluorescence imaging, S.Y. performed crystallization, D.P. and Ö.Y. provided crystallization tools, guided crystallization work and collected the X-ray diffraction data, S.Y., M.C., and Ö.Y. processed the crystallization data, S.Y., E.S., and Ö.Y. analyzed the data. S.Y., Ö.Y., and E.S. co-wrote the paper. All authors discussed the results and commented on the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Nature Communications thanks Thomas Kieselbach and the other, anonymous, reviewers for their contribution to the peer review of this work.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Source data
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Yadav, S., Centola, M., Glaesmann, M. et al. Cyclophilin anaCyp40 regulates photosystem assembly and phycobilisome association in a cyanobacterium. Nat Commun 13, 1690 (2022). https://doi.org/10.1038/s41467-022-29211-w
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41467-022-29211-w
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.