Abstract
Purpose
Noninvasive prenatal screening (NIPS) using cell-free DNA has transformed prenatal care. Belgium was the first country to implement and fully reimburse NIPS as a first-tier screening test offered to all pregnant women. A consortium consisting of all Belgian genetic centers report the outcome of two years genome-wide NIPS implementation.
Methods
The performance for the common trisomies and for secondary findings was evaluated based on 153,575 genome-wide NIP tests. Furthermore, the evolution of the number of invasive tests and the incidence of Down syndrome live births was registered.
Results
Trisomies 21, 18, and 13 were detected in respectively 0.32%, 0.07%, and 0.06% of cases, with overall positive predictive values (PPVs) of 92.4%, 84.6%, and 43.9%. Rare autosomal trisomies and fetal segmental imbalances were detected in respectively 0.23% and 0.07% of cases with PPVs of 4.1% and 47%. The number of invasive obstetric procedures decreased by 52%. The number of trisomy 21 live births dropped to 0.04%.
Conclusion
Expanding the scope of NIPS beyond trisomy 21 fetal screening allows the implementation of personalized genomic medicine for the obstetric population. This genome-wide NIPS approach has been embedded successfully in prenatal genetic care in Belgium and might serve as a framework for other countries offering NIPS.
Similar content being viewed by others
INTRODUCTION
Noninvasive prenatal screening (NIPS) by sequence analysis of cell-free DNA (cfDNA) fragments circulating in the blood of pregnant women enables the detection of fetal aneuploidies.1,2,3 With test sensitivities ranging between 92% and 99% for the viable trisomies 21, 18, and 13 and a specificity of >99%, NIPS outperforms traditional combined first trimester screening (cFTS),4,5,6 reducing the amount of invasive prenatal tests and the concurrent risk of procedure-related pregnancy complications. Hence, different professional societies advocated the implementation of NIPS for high and intermediate risk pregnancies.7,8,9,10 The test was swiftly adopted by the community and has globally transformed prenatal care.11 Nevertheless, implementation, uptake and testing approaches vary largely among and within countries.12
Belgium was the first country to implement and fully reimburse NIPS as a first-tier screening test, offering NIPS virtually for free to all pregnant women. With nationwide implementation of NIPS, reimbursement for serum biochemical assays was phased out and consequently cFTS disappeared from routine pregnancy management.13 Fetal ultrasound is still performed to measure the nuchal translucency (NT) and identify fetal malformations, both indicators for additional genetic counseling and invasive genetic testing.
Prenatal care and prenatal genetic testing monitor the health and aim to improve the outcome for mother and fetus. Despite being the most common genetic cause of birth defects, trisomy 21 constitutes only one of many genetic disorders affecting placental, fetal, and neonatal development and pregnancy outcome. Since all neonatal and invasive prenatal genetic testing in Belgium is performed at one of the eight genetic centers, introduction of NIPS was considered an opportunity to broaden the scope of traditional prenatal testing by implementing personalized genomic medicine for the obstetric population and as a consequence improve overall prenatal care.
The Belgian Advisory Committee on Bioethics advised that secondary findings, i.e., findings beyond the common trisomies, detected during genome-wide NIPS should be reported when clinically significant.14 In case this information may lead to preventive or therapeutic intervention, it is important to share it with the patient and provide genetic counseling. The failure to do so may be construed as serious negligence. This prompted the Belgian Society for Human Genetics to issue guidelines on reporting secondary findings.15 Secondary findings are reported if (1) considered technically valid, (2) there is validated evidence on the associated phenotype, and (3) considered clinically relevant and actionable. Because of the controversy about the validity and clinical utility of NIPS as a screening tool for fetal sex chromosome abnormalities, they are not reported. Since the majority of the cfDNA analyzed by NIPS is of maternal origin, genome-wide NIPS can also reveal maternal copy-number variations (CNVs). According to the Belgian NIPS guidelines and in accordance with the postnatal genomic testing guidelines of the American College of Medical Genetics and Genomics,16 clinically relevant and actionable maternal imbalances are reported.
Here, a consortium consisting of all Belgian genetic centers presents the performance of genome-wide NIPS for both primary and secondary findings as well as the impact on the obstetric practice and on society following the first two years of national implementation.
MATERIALS AND METHODS
NIPS sample processing
Peripheral blood samples were collected in cell-free DNA BCT tubes (Streck, Omaha, NE, USA), cell-free DNA collection tubes (Roche Diagnostics, Germany), or PAXgene blood circulating cfDNA (ccfDNA) tubes (QIAGEN GmbH, Germany) from 12 weeks of gestation onward. Clinical genetic laboratories from the eight Belgian genetic centers performed genome-wide NIPS. Standard centrifugation methods were used for plasma isolation. cfDNA extraction was performed using the QIAamp Circulating Nucleic Acid Kit (Qiagen), the Maxwell HT cfDNA kit (Promega), or the VeriSeq NIPT solution v2 (Illumina) according to the manufacturer’s recommendations. Library preparation for genome-wide shallow genome sequencing was performed using the Kapa HyperPlus preparation kit (Kapa Biosystems); TruSeq ChIP, TruSeq DNA Nano library preparation kit or the VeriSeq NIPT Solution v2 (Illumina); NEXTflex Cell-free DNA-seq kit or NEXTflex Rapid DNA-Seq kit 2.0 (Bioo Scientific). Next-generation sequencing was performed with either the Ion Proton system (ThermoFisher scientific) or the VeriSeq NIPT v2 solution, HiSeq1500, HiSeq2500, HiSeq3000, HiSeq4000, Novaseq6000, NextSeq500 or NextSeq550 sequencer (Illumina). Genome-wide genomic representation profiling and interpretation was performed using the VeriSeq NIPT Assay Control Software v2.0.0 (Illumina) or as previously described.17,18,19
Post-test counseling
Women with a low-risk NIPS result were informed by their obstetric care professional or via the patients’ online medical files. The follow-up procedure for a high-risk result depended on the type of aberration found. In case of a high risk for a common trisomy (21, 18, or 13), women were informed by their obstetrician, followed by invasive prenatal diagnostics if desired. All women at high risk for a secondary finding were referred to and counseled by a clinical geneticist.
Follow-up testing
When cfDNA profiling indicated the presence of a fetal chromosomal abnormality, pregnant women were offered follow-up by standard invasive prenatal diagnosis.
CNV analysis was performed on DNA extracted from chorionic villus sampling (CVS) or amniotic fluid (AF) using the Agilent ISCA 60 K or 44 K array (Agilent), Cytoscan 750 K array (Affymetrix), HumanCytoSNP-12 v2.1 BeadChip kit (Illumina), or by shallow genome sequencing (CNVSeq). Fluorescence in situ hybridization (FISH) was performed following standard procedures. Subsequently, when a discrepancy between the NIPS and the invasive genetic test result was detected, women were requested to donate the placenta upon delivery.
A trisomic embryo can develop into a normal disomic fetus by loss of one of the three chromosomes, a phenomenon called trisomy rescue. In one third of trisomy rescue events this results in uniparental disomy (UPD). UPD testing on amniotic fluid was performed when NIPS indicated a trisomy for chromosomes 6, 7, 11, 14, 15, or 20, as UPD for one of these chromosomes is known to be associated with developmental disorders.20 UPD was tested using the HumanCytoSNP-12 v2.1 BeadChip kit (Illumina), polymorphic short tandem repeat (STR) analysis (Promega), or methylation-specific multiplex ligation-dependent probe amplification (MS-MLPA) probemix ME032-A1 (MRC Holland).
If a maternal constitutional CNV was considered (1) clinically relevant and (2) technically valid based on the NIPS data, this maternal CNV was immediately included in the NIPS report. If the maternal constitutional CNV was classified as clinically relevant but was not considered as technically valid based on the NIPS data, this maternal CNV was first confirmed by an independent analysis on maternal white blood cells, obtained from the stored buffy coats of the NIPS blood sample. Maternal constitutional CNVs were confirmed by FISH (standard protocol), chromosomal microarray analysis, or shallow genome sequencing as previously described.18,21 The timeframe for this confirmation was 1 week on average. The implementation of NIPS and the interpretation of both fetal and maternal incidental findings were approved by the institutional ethics review board and are consented for by both the referring physician and the pregnant women.
RESULTS
NIPS performance in the general obstetric population
Between 1 July 2017 and 30 June 2019, the National Institute for Health and Disability Insurance (RIZIV-INAMI) registered 188,330 NIPS analyses. Since the large majority of NIPS analyses are performed at 12 weeks of gestation and it is known that about 2.5% of 12-week pregnancies result in miscarriage, the estimated number of NIP-screened live births is 183,621.22 Since in 2018, 117,800 and in 2019 115,565 live births were registered, 78.7% of the pregnant women opted for NIPS, a similar percentage when compared to the uptake of cFTS before NIPS.23 The mean maternal age at delivery in 2018 and 2019 was 30.7 years and 30.8 years, respectively.
Of all registered NIP tests, 153,575 pregnant women (81.5%) received NIPS in one of the Belgian genetic centers, consisting of 150,805 (98.2%) singleton pregnancies and 2,770 (1.8%) twin pregnancies. Higher-order pregnancies were not included in this study. A NIPS result could be reported to 99.3% of women. For 0.7% of the pregnancies the result remained inconclusive even after repeated analysis. This failure rate is equal or lower compared with other studies.24
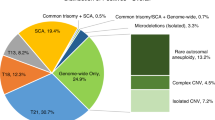
Trisomies 21, 18, and 13 were detected in respectively 494 (0.32%), 115 (0.07%), and 91 (0.06%) cases. Results of invasive testing following a positive NIPS for trisomy 21, 18, and 13 could be collected for 394 (79.8%), 91 (79.1%), and 82 (90.1%) pregnancies and confirmed a fetal trisomy in 364, 77, and 36 cases, respectively. Of note, there were no false positives among 11 dizygotic twin pregnancies for which invasive follow-up data were available. This results in positive predictive values (PPVs) of 92.4% for trisomy 21, 84.6% for trisomy 18, and 43.9% for trisomy 13 (Table 1). These results are largely consistent with published data for both high-risk and general obstetric populations.4,25
In case of a discrepancy between the noninvasive and the invasive test, women were requested to participate in a follow-up study allowing placental analysis after delivery to determine whether confined placental mosaicism (CPM) could explain the false positive outcome. Placental tissue could be obtained for respectively 5, 3, and 16 apparent false positive trisomies 21, 18, and 13; placental mosaicism was observed in 3, 1, and 8 cases. This result indicates that at least half of the false positive NIPS results can be explained by CPM and are not caused by technical issues.
False negative results for trisomies 21, 18, and 13 were reported in 4, 2, and 0 cases, respectively. Because all prenatal/neonatal cytogenetic testing in Belgium is performed in one of the genetic centers involved in this study, it is unlikely a false negative is missed. One of the four trisomy 21 false negatives presented in only 7% of placental cells, while the trisomy was found in 100% of fetal cells as well as in 75% of umbilical cord cells. One of the two false negatives for trisomy 18 was shown to be mosaic, with trisomy 18 present in about 60% of the placental cells. Placental follow-up data was not available for the remaining four false negatives. There were no false negatives among 2,029 dichorionic diamniotic (DCDA) twin samples showing that NIPS has a high sensitivity for twin pregnancies as well.
Genomic medicine for the obstetric population
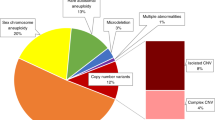
A total of 350 (0.23%) pregnancies were reported with a rare autosomal fetal aneuploidy, of which 339 pregnancies presented with a rare autosomal trisomy (RAT) (Fig. 1). As in other cohorts,25,26,27 the most frequent RAT was trisomy 7 (39.2%), followed by trisomy 20 (11.5%), 8 (8.8%), 16 (8.3%), 22 (6.2%), and 15 (5.3%). In 78.5% (266/339) of RATs, an invasive genetic test result was obtained. Eleven RATs turned out positive in the fetus: trisomy 2 (n = 1), 8 (n = 3), 9 (n = 1), 16 (n = 4), and 22 (n = 2), resulting in a positive predictive value of 4.1%. The remaining 255/266 (95.9%) RATs showed no fetal imbalance following invasive or postnatal genetic testing. Based on the ratio between the theoretical versus the observed chromosomal z-scores in function of fetal fraction, the test allows to predict CPM.28 This analysis suggested that the large majority of false positive RATs are the result of CPM and this was strengthened by placental or chorionic villus biopsy: for 51 of the apparently false positive RATs with follow-up, placental mosaicism was confirmed in over half of the cases (28/51; 54.9%).
UPD testing was performed in 64 pregnancies: trisomy 7 (n = 43), 11 (n = 2), 14 (n = 9), 15 (n = 5), and 20 (n = 5). Three UPDs were detected: 1 for trisomy 7 and 2 for trisomy 15. Hence, UPD was observed in 4.7%.
Eleven pregnancies presented with a monosomy: monosomy 13 (n = 1), 15 (n = 1), 16 (n = 2), 18 (n = 4) and 21 (n = 3). For six monosomies, an invasive prenatal test was performed; none was confirmed.
In 0.07% (109/153,575) of the pregnancies, NIPS was suggestive of the presence of a fetal segmental imbalance. We observed 11 potential genomic disorders, 14 whole chromosomal arm gains and 1 loss, 9 interstitial gains and 30 losses, 17 terminal segmental gains and 19 losses, 5 likely unbalanced translocations (both a terminal deletion and terminal duplication), and 3 complex rearrangements (Table S1, Figure S1). Fetal or postnatal follow-up data was available for 92 cases (84.4%). Of those, 43 were confirmed to be true positives, resulting in a PPV of 47%. Two imbalances that were not confirmed in the fetus were shown to be present in the placenta. Eight additional imbalances turned out not to be fetal but (mosaic) maternal in origin, as shown by fluorescent in situ hybridization (FISH) or chromosomal microarray on DNA extracted from the maternal white blood cells.
Clinically relevant maternal CNVs were reported in 0.32% of the cases (Fig. 2). NIPS profiles reminiscent of the presence of maternal cancers29 were detected and confirmed in 12 cases (0.008%). Other reported maternal secondary findings included heterozygosity for pathogenic CNVs related to highly penetrant dominant or X-linked disorders such as cancer predisposition syndromes or Duchenne muscular dystrophy,30 respectively.
Impact on the number of invasive tests
The high specificity of NIPS has significantly reduced the number of amniocenteses and CVS procedures worldwide.31,32,33 Also in Belgium, a decline in invasive obstetric procedures has been registered by the RIZIV-INAMI between 2013, the last year before the nonreimbursed NIPS was introduced in Belgium, and 2018, the first full year following generalized introduction of reimbursed NIPS. During this period, the absolute number of invasive obstetric procedures performed dropped from 6,279 to 3,047 (Fig. 3). When normalized based on the number of live births, a number linearly related with the number of pregnancies, this corresponds to a decrease of 52%. Although the overall number of invasive tests significantly decreased, the actual figure outnumbers the number of invasive tests predicted on the basis of the trisomy 21 incidence by 440.23 This can be explained by the invasive follow-up of inconclusive results and secondary findings as well as improved ultrasound performance.
With the introduction of NIPS as a first-tier screening test, invasive tests because of advanced maternal age or abnormal cFTS largely ceased to exist. The increased specificity of NIPS compared with cFTS screening is clearly reflected by the drop in the relative ratio of invasive tests performed in case of advanced maternal age or abnormal cFTS in the pre-NIPS era (before November 2013) from 52.8% to 26.2% of invasive follow-up performed for a positive NIPS result in the first-tier NIPS era (after July 2017). The ratio of invasive tests performed for indications not eligible for NIPS (e.g., ultrasound anomalies, infections, familial history) increased from 47.2% pre-NIPS to 73.8%.
Incidence of Down syndrome live births
One of the unanswered questions arising from the widespread implementation of NIPS for trisomy 21 detection is its effect on terminations of pregnancy and hence the number of children born with Down syndrome.34,35 We collected all neonatal trisomy 21 genetic test results and observed a decline from 77 trisomy 21 live births (0.06% of live births) in 2014 to 52 trisomy 21 live births (0.04% of live births) in 2018. Since there are no plausible biological/demographic changes in the general obstetric population that could point to a significant impact on the ratio of the frequency of trisomy 21–related spontaneous miscarriages to the trisomy 21 incidence during pregnancy over this four-year period, this decline might be explained by a combination of the decreased number of false negatives with NIPS compared with cFTS and the choice for pregnancy termination for the large majority of women with a trisomy 21 NIPS outcome.
DISCUSSION
Several professional societies raised awareness that broadening the scope of NIPS requires careful considerations of proper counseling of the pregnant women.36 The Belgian context provides a unique setting: genetic counseling is fully reimbursed for all and the geneticists as test providers collaborate closely with the obstetric community. When a fetal or maternal genetic lesion is detected, the obstetrician is informed and the pregnant woman is invited for genetic counseling. This end-to-end multidisciplinary approach creates a sense of control and confidence among doctors and patients on the national implementation of genome-wide NIPS as a first-tier screening test for all pregnant women as standard of care.
While the association of trisomy 16 with a higher risk of adverse pregnancy outcome is well established,37 this association remains controversial for the other RATs.25,26,37 When a RAT is detected in CVS, it is attributable to CPM in 97% of the cases.38 The low PPV for RATs confirms that most RATs are confined to the placenta. CPM in which both mesenchymal and cytotrophoblast cells contain the RAT has been proven to be associated with complications in pregnancy.39 Genome-wide NIPS studies suggest a higher incidence of adverse pregnancy outcomes when a RAT is detected.26 Overall, the clinical consequences of most RATs are poorly mapped and broad scale reporting may require larger-scale studies on associated obstetric complications to determine which RATs need follow-up and could provide obstetric relevant information.25,26 Equally, the follow-up of segmental imbalances requires further refinement. For known balanced translocation carriers, the detection of the associated unbalanced translocation in the fetus is virtually diagnostic. For larger segmental imbalances resulting in more severe developmental disorders as compared with Down syndrome, an invasive test seems warranted despite the lower PPV.
There are clinical and societal challenges remaining that need to be addressed. The emotional distress caused by reporting secondary findings requires attention. All pregnant women undergoing NIPS are informed during pretest counseling that, in rare cases, genome-wide NIPS can also reveal other clinically relevant findings that will be reported. There are concerns that women may be insufficiently prepared for the eventual diagnosis of a fetus with a serious disorder or a personal health issue of which they were unaware.36 Monitoring the understanding of couples about the test, the impact on societal pressure for taking a NIPS test, the free choice for a pregnancy interruption after a positive NIPS, and the acceptance of children with Down syndrome40,41 would be valuable. To improve future reporting, better national pregnancy and birth registries would be welcomed.
The implementation of NIPS as a first-tier screening test remains restricted to a handful of countries,12 due to the predicted higher cost for health care and reduced PPV. This two-year experience with national publicly funded first-tier NIPS has not only demonstrated a high PPV for trisomy 21 detection in the general population, but also met expectations when it comes to the predicted cost efficiency for the health-care system.23 Broadening of the scope by leveraging genomic medicine beyond trisomy 21 screening provides additional societal and health economic benefits. With further reductions in sequencing costs and increasing resolution, the societal expectations to detect fetal and maternal genetic variants in cfDNA are likely to increase. We expect that the development of consensus guidelines among professional societies and thoughtful follow-up of both primary and secondary findings by genetic professionals provides a proper framework for future prenatal genetic care.
Data availability
This clinical utility study presents aggregated clinical data. All data are presented within the paper.
References
Vermeesch, J. R., Voet, T. & Devriendt, K. Prenatal and pre-implantation genetic diagnosis. Nat. Rev. Genet. 17, 643–656 (2016).
Fan, H. C., Blumenfeld, Y. J., Chitkara, U., Hudgins, L. & Quake, S. R. Noninvasive diagnosis of fetal aneuploidy by shotgun sequencing DNA from maternal blood. Proc. Natl. Acad. Sci. U. S. A. 105, 16266–16271 (2008).
Chiu, R. W. K. et al. Noninvasive prenatal diagnosis of fetal chromosomal aneuploidy by massively parallel genomic sequencing of DNA in maternal plasma. Proc. Natl. Acad. Sci. U. S. A. 105, 20458–20463 (2008).
Gil, M. M., Accurti, V., Santacruz, B., Plana, M. N. & Nicolaides, K. H. Analysis of cell-free DNA in maternal blood in screening for aneuploidies: updated meta-analysis. Ultrasound Obstet. Gynecol. 50, 302–314 (2017).
Taylor-Phillips, S. et al. Accuracy of non-invasive prenatal testing using cell-free DNA for detection of Down, Edwards and Patau syndromes: a systematic review and meta-analysis. BMJ Open 6, e010002 (2016).
Badeau, M. et al. Genomics-based non-invasive prenatal testing for detection of fetal chromosomal aneuploidy in pregnant women. Cochrane Database Syst. Rev. 11, CD011767 (2017).
Gregg, A. R. et al. Noninvasive prenatal screening for fetal aneuploidy, 2016 update: a position statement of the American College of Medical Genetics and Genomics. Genet. Med. 18, 1056–1065 (2016).
Committee opinion no. 640: cell-free DNA screening for fetal aneuploidy. Obstet. Gynecol. 126, e31–e37 (2015).
Society for Maternal-Fetal Medicine (SMFM) Publications Committee. SMFM statement: clarification of recommendations regarding cell-free DNA aneuploidy screening. Am. J. Obstet. Gynecol. 213, 753–754 (2015).
Benn, P. et al. Position statement from the Chromosome Abnormality Screening Committee on behalf of the Board of the International Society for Prenatal Diagnosis. Prenat. Diagn. 35, 725–734 (2015).
Bianchi, D. W. & Chiu, R. W. K. Sequencing of circulating cell-free DNA during pregnancy. N. Engl. J. Med. 379, 464–473 (2018).
Gadsbøll, K. et al. Current use of noninvasive prenatal testing in Europe, Australia and the USA: a graphical presentation. Acta Obstet. Gynecol. Scand. 99, 722–730 (2020).
Van Elslande, J. et al. The sudden death of the combined first trimester aneuploidy screening, a single centre experience in Belgium. Clin. Chem. Lab. Med. 57, e294–e297 (2019).
FPS Public Health. Opinion no. 66 of 9 May 2016—non-invasive prenatal testing (NIPT). https://www.health.belgium.be/en/opinion-no-66-non-invasive-prenatal-testing-nipt (2016).
Belgium Society of Human Genetics prenatal working group. Belgian Guidlines for managing incidental findings detected by NIPT. https://www.college-genetics.be/assets/recommendations/fr/guidelines/BELGIAN%20GUIDELINES%20FOR%20MANAGING%20INCIDENTAL%20FINDINGS%20DETECTED%20BY%20NIPT%20(2019).pdf.
Kalia, S. S. et al. Recommendations for reporting of secondary findings in clinical exome and genome sequencing, 2016 update (ACMG SF v2.0): a policy statement of the American College of Medical Genetics and Genomics. Genet. Med. 19, 249–255 (2017).
Bayindir, B. et al. Noninvasive prenatal testing using a novel analysis pipeline to screen for all autosomal fetal aneuploidies improves pregnancy management. Eur. J. Hum. Genet. 23, 1286–1293 (2015).
Dheedene, A. et al. Implementation of non-invasive prenatal testing by semiconductor sequencing in a genetic laboratory. Prenat. Diagn. 36, 699–707 (2016).
Raman, L., Dheedene, A., De Smet, M., Van Dorpe, J. & Menten, B. WisecondorX: improved copy number detection for routine shallow whole-genome sequencing. Nucleic Acids Res. 47, 1605–1614 (2019).
Engel, E. A fascination with chromosome rescue in uniparental disomy: Mendelian recessive outlaws and imprinting copyrights infringements. Eur. J. Hum. Genet. 14, 1158–1169 (2006).
Brison, N. et al. Accuracy and clinical value of maternal incidental findings during noninvasive prenatal testing for fetal aneuploidies. Genet. Med. 19, 306–313 (2017).
Ammon Avalos, L., Galindo, C. & Li, D.-K. A systematic review to calculate background miscarriage rates using life table analysis. Birth Defects Res. A Clin. Mol. Teratol. 94, 417–423 (2012).
Gyselaers, W., Hulstaert, F. & Neyt, M. Contingent non-invasive prenatal testing: an opportunity to improve non-genetic aspects of fetal aneuploidy screening. Prenat. Diagn. 35, 1347–1352 (2015).
Yaron, Y. The implications of non-invasive prenatal testing failures: a review of an under-discussed phenomenon. Prenat. Diagn. 36, 391–396 (2016).
van der Meij, K. R. M. et al. TRIDENT-2: national implementation of genome-wide noninvasive prenatal testing as a first-tier screening test in the Netherlands. Am. J. Hum. Genet. 105, 1091–1101 (2019).
Pertile, M. D. et al. Rare autosomal trisomies, revealed by maternal plasma DNA sequencing, suggest increased risk of feto-placental disease. Sci. Transl. Med. 9, eaan1240 (2017).
Scott, F. et al. Rare autosomal trisomies: Important and not so rare. Prenat. Diagn. 38, 765–771 (2018).
Brison, N. et al. Predicting fetoplacental chromosomal mosaicism during non-invasive prenatal testing. Prenat. Diagn. 38, 258–266 (2018).
Amant, F. et al. Presymptomatic identification of cancers in pregnant women during noninvasive prenatal testing. JAMA Oncol. 1, 814–819 (2015).
Brison, N. et al. Maternal copy-number variations in the DMD gene as secondary findings in noninvasive prenatal screening. Genet. Med. 21, 2774–2780 (2019).
Robson, S. J. & Hui, L. National decline in invasive prenatal diagnostic procedures in association with uptake of combined first trimester and cell-free DNA aneuploidy screening. Aust. N. Z. J. Obstet. Gynaecol. 55, 507–510 (2015).
Larion, S. et al. Association of combined first-trimester screen and noninvasive prenatal testing on diagnostic procedures. Obstet. Gynecol. 123, 1303–1310 (2014).
Platt, L. D. et al. Impact of noninvasive prenatal testing in regionally dispersed medical centers in the United States. Am J Obstet. Gynecol. 211, 368.e1–7 (2014).
van Schendel, R. V. et al. Attitudes of pregnant women and male partners towards non-invasive prenatal testing and widening the scope of prenatal screening. Eur. J. Hum. Genet. 22, 1345–1350 (2014).
Hill, M. et al. Has noninvasive prenatal testing impacted termination of pregnancy and live birth rates of infants with Down syndrome? Prenat. Diagn. 37, 1281–1290 (2017).
Dondorp, W. et al. Noninvasive prenatal testing for aneuploidy and beyond: challenges of responsible innovation in prenatal screening. Eur. J. Hum. Genet. 23, 1438–1450 (2015).
Grati, F. R. et al. Outcomes in pregnancies with a confined placental mosaicism and implications for prenatal screening using cell-free DNA. Genet. Med. 22, 309–316 (2020).
Malvestiti, F. et al. Interpreting mosaicism in chorionic villi: results of a monocentric series of 1001 mosaics in chorionic villi with follow-up amniocentesis. Prenat. Diagn. 35, 1117–1127 (2015).
Toutain, J., Goutte-Gattat, D., Horovitz, J. & Saura, R. Confined placental mosaicism revisited: impact on pregnancy characteristics and outcome. PLoS One 13, e0195905 (2018).
van Schendel, R. V. et al. What do parents of children with Down syndrome think about non-invasive prenatal testing (NIPT)? J. Genet. Couns. 26, 522–531 (2017).
Crombag, N. M., Page-Christiaens, G. C., Skotko, B. G. & de Graaf, G. Receiving the news of Down syndrome in the era of prenatal testing. Am. J. Med. Genet. A 182, 374–385 (2020).
Acknowledgements
Part of this work was funded by KULeuven funding (C1/018).
Author information
Authors and Affiliations
Contributions
Conceptualization: K.V.D.B., K.D., J.R.V. Investigation & methodology: K.V.D.B, N.B. Formal analysis: L.L., N.B. Visualization: V.G. Data curation: M.B., B.B., F.B. L.B., A.D.L., MD., J.D., A.Dheedene., A.Duquenne., N.F., A.F., J.G., K.J., S.J., D.L., A.M., B.M. C.M. L.P., B.P. E.S., E.V, V.B., G.S., Y.S. Supervision: K.V.D.B., K.D., J.R.V. Validation: N.B., K.V.D.B. Writing—original draft, review, and editing: K.V.B., L.L., N.B., K.J., K.D., J.R.V.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
About this article
Cite this article
Van Den Bogaert, K., Lannoo, L., Brison, N. et al. Outcome of publicly funded nationwide first-tier noninvasive prenatal screening. Genet Med 23, 1137–1142 (2021). https://doi.org/10.1038/s41436-021-01101-4
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41436-021-01101-4
This article is cited by
-
Supporting patient decision-making in non-invasive prenatal testing: a comparative study of professional values and practices in England and France
BMC Medical Ethics (2024)
-
Prenatal diagnosis of a trisomy 7 mosaic case: CMA, CNV-seq, karyotyping, interphase FISH, and MS-MLPA, which technique to choose?
BMC Pregnancy and Childbirth (2024)
-
Clinical outcomes of screen-positive genome-wide cfDNA cases for trisomy 20: results from the global expanded NIPT Consortium
Molecular Cytogenetics (2024)
-
Should non-invasive prenatal testing (NIPT) be used for fetal sex determination? Perspectives and experiences of healthcare professionals
European Journal of Human Genetics (2024)
-
Population screening for 15q11-q13 duplications: corroboration of the difference in impact between maternally and paternally inherited alleles
European Journal of Human Genetics (2024)