Abstract
Purpose
Patient care involving genetics is challenging for nongenetics health-care providers. Clinical decision support (CDS) tools are a potential solution because they provide patient-specific risk assessments and/or management recommendations. This systematic review synthesized evidence on whether using CDS tools resulted in appropriate changes in genetics-related patient management made by nongenetics health-care providers.
Methods
A comprehensive search in MEDLINE, Embase, and CINAHL yielded 2,239 unique articles. Two independent reviewers screened abstracts and full texts for quantitative, qualitative, and mixed-methods articles on management changes by nongenetics clinicians using a CDS tool as part of patient care. Effect sizes were calculated for quantitative studies and all articles were analyzed together using narrative synthesis. Twenty articles were included.
Results
In 12/16 quantitative studies, CDS tools slightly increased appropriate changes in management, but study design appeared to affect the statistical significance of the effect. The qualitative data in the four remaining studies reaffirmed that CDS tools facilitated management decisions but raised questions about their effect on patient outcomes.
Conclusion
Our review assessed clinical utility of CDS tools, finding that they slightly increase appropriate management changes by nongenetics providers. Future studies on CDS tools should explicitly evaluate decision making and patient outcomes.
Similar content being viewed by others
INTRODUCTION
Genetic tests are increasingly being used across medical disciplines, yet evidence indicates that nongenetics specialists have limited proficiency and confidence in deciding when to order genetic tests, refer to genetics specialists, or change treatment or surveillance based on genetic risk factors.1,2 These challenges will only grow with the volume of information generated by genome sequencing and the evolving evidence base linking genetic variants to human disease.3 New approaches are needed to support nongenetics specialists in managing genetic test results especially in light of the limited genetics specialist workforce.4
Clinical decision support (CDS) tools have been suggested as a solution to support the management of genetic tests in the practices of nongenetics clinicians and potentially prevent harmful or unnecessary interventions.5 CDS tools guide assessments or recommendations at the point of care that are appropriate based on clinical management guidelines, best practices, and/or research evidence.6 Thus, using CDS tools may have clinical utility because the tools can influence changes in management, facilitating the use of genetic testing to inform clinical decision making.7 CDS tools can be administered by paper or computer, can comprise standalone tools or be integrated into the clinical workflow/electronic health records, and can notify providers to log in or alert them about assessments/recommendations.5
In 2011, Welch and Kawamoto conducted a systematic review about genetic CDS tools but the effect of these tools on clinical decision making was not rigorously assessed or available at the time.8 Since then, the use and evaluation of genetic CDS tools have proliferated, yet this new evidence has not been comprehensively reviewed to assess the utility of genetic CDS on clinical management decisions.9 We conducted a mixed-methods systematic review to synthesize evidence on the clinical utility of CDS tools in clinical management decisions by nongenetics clinicians, compared with standard clinical care without CDS.
MATERIALS AND METHODS
A convergent mixed-methods systematic review was conducted to integrate quantitative and qualitative evidence about changes in management when using genetic CDS tools.10 A segregated approach was taken in which quantitative and qualitative evidence were synthesized separately and then brought together through narrative synthesis.11 The protocol was registered on PROSPERO (ID: CRD42020129821). Manuscript preparation was guided by the Preferred Reporting Items for Systematic Reviews and Meta-Analyses (PRISMA) standards.12
Literature search strategy
A professional librarian (E.U.) searched MEDLINE, MEDLINE-in-Process, MEDLINE Epubs Ahead of Print, and Embase Classic+Embase databases (OvidSP, and CINAHL (EBSCOHost)) from the year these databases were created to 7 March 2019. Both subject headings and textword terms were used to search for CDS tools and genetic testing, including family history assessment (Supplementary appendix 1). Reference lists of included articles and relevant systematic reviews were hand-searched for additional references.
Study selection
References were reviewed against the following inclusion criteria: primary English peer-reviewed research article studying humans, comparison of a genotype- or family history–related CDS tool to standard clinical care, CDS use by nongenetics clinicians in a patient care context, and outcomes reporting changes in management (Supplementary appendix 2). These inclusion criteria were designed to capture data on genetic CDS tools use by nongenetics clinicians within clinical practice, therefore papers solely using hypothetical patient data were excluded. Peer-review ensured that included articles met a publication standard. Screening non-English articles was not feasible.
Each stage of literature screening was conducted by two independent reviewers (A.S. and L.E.O./S.S./C.M.) using Rayyan and a piloted screening form.13 Titles and abstracts were screened to assess whether the population and intervention met inclusion criteria. Any conflicting decisions were discussed and resolved or moved to full text review. Full text review of included or undecided abstracts was conducted such that articles included in the final systematic review met all inclusion criteria, with exclusion reasons recorded on Rayyan. Undecided articles were discussed until consensus was reached or resolved by a third reviewer (J.C.C.).
Data extraction
Quantitative and qualitative data extraction was completed independently for all included articles based on piloted forms. Data extraction items for all studies included general study characteristics, the population, CDS intervention, and comparison with standard clinical care. Genetic CDS tools were defined as tools providing assessments or recommendations based on the patient’s genotype, gene expression profile, or family history. The comparison was defined as assessments or recommendations made without the use of a CDS tool.
Quantitative and qualitative primary and secondary outcome data were extracted separately. For quantitative studies and mixed-methods (quantitative component only), data extraction included effect size calculation (e.g., number of clinicians who changed medications using a genetic CDS tool) while for qualitative studies and mixed-methods (qualitative component only), relevant themes and illustrative quotes, observations, or other data were extracted.10 The primary outcome was defined as a change in management by the clinician, i.e., change in medication, treatment (e.g., surgery), screening, referral for genetic testing or to a genetics clinic, monitoring, or surveillance. A change was determined to be appropriate by the reviewers if the study reported that guidelines, clinical or epidemiological studies were the basis of the tool’s recommendations.14 Secondary outcomes were changes in the risk assessment of patients’ disease by clinicians, changes in patient communication (discussion with patients about disease risk prior to making management decisions), clinician use of the CDS tool, and clinician satisfaction with the CDS tool. These outcomes were included because changes in assessed level of risk and increases in patient communication may indicate a preliminary step toward changes in clinical management, and clinician use and satisfaction with the CDS tool could influence whether the tool is used to make management changes.
Data analysis and synthesis
Consistent with a convergent, segregated design, quantitative and qualitative synthesis were first conducted separately before being synthesized together.11 For quantitative synthesis, effect sizes of CDS tools were calculated using aggregate participant data and described using narrative synthesis. Risk ratios (RR) were calculated for primary outcomes from randomized controlled trials (RCTs). Relative risk reduction (RRR) was calculated for other study designs and for secondary outcomes.15 To infer statistical significance and precision for RRs and RRRs, 95% confidence intervals (CIs) were calculated.16,17 Since clinician use and satisfaction only applied to the intervention arm, proportions were calculated as the summary measure. Quantitative meta-analysis and subgroup analysis were considered to aggregate effects of multiple tools but were not feasible or meaningful due to overall heterogeneity of interventions, study designs, and outcomes and small/unreported sample sizes.18 Qualitative synthesis was conducted separately following a meta-aggregation approach.19 The qualitative findings (themes and illustrative supporting data) of included studies were aggregated into categories and further grouped into synthesized findings based on their similarity in meaning. Finally, the qualitative and quantitative syntheses were compared and synthesized using a narrative approach.11
Critical appraisal
Risk of bias of individual studies was assessed by two independent reviewers (A.S. and L.E.O./S.S./C.M.) for all included articles at the study level using the Joanna-Briggs Institute (JBI) quality assessment checklists as appropriate for the study design.20 The JBI checklists provided a consistent way to assess studies of different designs. No studies were excluded from the review based on quality. Reporting bias (publication bias and selective outcome reporting bias) was assessed through query of a trial database (clinicaltrials.gov) comparing trial registrations relevant to the review, their publication status, and reported outcomes.21 Grading of recommendations, assessment, development, and evaluations (GRADE) was not used to assess confidence in cumulative evidence since it is not advised for mixed-methods reviews.10
RESULTS
Included studies
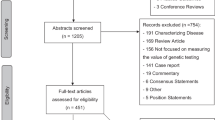
The literature search yielded 2,239 unique articles. Two thousand one hundred fifty-three articles were excluded in title and abstract review. The remaining 86 articles underwent full text review, leaving 12 quantitative, two mixed-methods, and two qualitative articles that met all inclusion criteria. Hand searching the reference lists of the included articles resulted in the inclusion of four more quantitative articles. Thus, 20 articles were ultimately included in this review.17,18,19,20,21,22,23,24,25,26,27,28,29,30,31,32,33,34,35,36 The study selection process and exclusion reasons are displayed in Fig. 1.
Study and patient characteristics
A table summarizing the 20 included articles is presented in Table 1. Since three tools (Genomic Prescribing System,23,35 YouScript,34,36 and Genomic Classifier25,27) were covered by two articles each, the included articles spanned 17 CDS tools. Most studies (15/20) were conducted in the United States, with 2 from the United Kingdom, 2 from Spain, and 1 from Australia. Eight studies were conducted in an academic health setting, four were conducted in community settings, and two were multisite studies covering both academic and community settings. As for type of genetic information used, eight CDS tools used genotype as their input, seven used family history, one used tumor genotype, and one used viral genotype. Most quantitative studies (9/16) followed a quasi-experimental design, 5 studies were RCTs, and 2 were cohort studies. Two were qualitative studies. The final two were mixed-methods studies for which only the qualitative components were found to be relevant to this review.
Patient clinical indications varied. Eight studies targeted primary care patients. Other articles studied patient indications such as cancer (n = 8 across prostate, breast, ovarian, colorectal), polypharmacy (n = 2), mental health (n = 3), cardiomyopathy-related disorders (n = 1), and prenatal care (n = 1). Patient demographics also varied: nine studies targeted adults of any age or sex, while others targeted specifically women (n = 4 breast or ovarian cancer and prenatal care), men (n = 2 prostate cancer), children (n = 1), or the elderly (n = 1).
Provider characteristics
The articles covered a wide range of clinicians, with many articles including more than one type of clinician. Most tools targeted physicians such as family physicians (n = 10), psychiatrists (n = 3), and cardiologists (n = 3). Only 7/16 quantitative articles reported the number of clinicians using the CDS tool. This was one source of heterogeneity since other studies reported management changes with respect to the number of patients or number of genetic test results.
Intervention characteristics
The clinical focus of the CDS tools was mainly cancer (n = 8) or pharmacogenetics (n = 7), with the remaining tools focusing on thrombophilia, cystic fibrosis, or cardiomyopathy. With respect to pharmacogenetics, tools typically targeted mental health or polypharmacy, with one tool targeting human immunodeficiency virus and another targeting a variety of conditions including antiplatelet therapy. With respect to cancer, the tools covered breast, prostate, ovarian, and colorectal cancer, as well as hereditary cancer syndromes.
Most (14/17) tools were computerized. Three of 17 tools in this review (YourHealthSnapshot,28 GeneInsight Clinic37 and one unnamed CDS30) generated active alerts that automatically provided clinicians with decision support. Six of 17 tools were integrated into the clinical workflow: five were integrated with the electronic medical record (EMR) (name of two tools not reported,26,30 PREDICT,38 MyFamily,39 and Genomic Prescribing System,23 [which was initially a standalone tool]); and the other (MeTree40) was integrated through study staff who printed and manually inserted reports into patient charts.
Management changes
In 12/16 quantitative studies, CDS tools slightly increased changes in management compared with standard of care (Table 1), but study design appeared to affect whether these changes were statistically significant. Effect sizes were statistically significant in 7/9 quasi-experimental studies, but only in 1/3 RCTs (Table 2). Effect sizes were not meta-analyzed due to the heterogeneity of study designs and reported outcomes.18 Of the remaining four studies, one quasi-experimental study reported mixed results because management changes were seen in only one of the two targeted clinical conditions (Pregnancy and Health Profile24). The three final studies did not clearly describe management changes in comparison with a standard of care (RetroGram,33 cohort evaluating YouScript,36 YourHealthSnapshot28).
Pharmacogenetic CDS tools were associated with a variety of management changes including switching the patient’s medication, adjusting the dose of their existing medication, and reducing polypharmacy (the use of multiple medications). For four of these tools, the changes were reported as appropriate because they were based on evidence such as guidelines or FDA information.29,33,34,35,36 However, two tools did not clearly report a clinical evidence base for the changes so it was unclear if the changes suggested by these tools were clinically appropriate.22,41 The qualitative findings confirmed management changes as a result of pharmacogenetic CDS tools, but identified concerns about whether the unestablished clinical utility of these tools justified costlier alternative medications (Table 3).23,38 In addition, clinicians raised concerns about the clinical utility of the CDS tool given that many drugs they commonly prescribed had limited pharmacogenetic information in the tool.
Family history CDS tools such as MeTree40 and Pregnancy and Health Profile24 were associated primarily with increase in referrals to medical genetics/genetic counseling, as well as an increased rate of screening for cancer or carrier status. These changes were considered to be appropriate because they were suggested by the tool based on guidelines and published studies. Participants described one family history tool (MyFamily39) as facilitating decisions about screening or referrals by providing a dedicated opportunity to elicit thorough and accurate family history information (Table 3).
Of the two remaining tools, one classified risk of tumor metastasis and statistically significantly increased appropriate cancer treatment recommendations.25,27 The other tool focused on updated genetic test results (e.g., variant reclassifications) and was qualitatively reported to influence cascade genetic testing decisions.37
Therefore, use of CDS tools seemed to be associated with increased changes in management (most of which were justified as appropriate using evidence such as guidelines), with statistical significance potentially impacted by study design. The nature of the management changes included changes in medication choice, dose or number, referrals to medical genetics/genetic counseling, increased rate of screening, testing, and treatment.
Risk assessment and patient communication
CDS tools statistically significantly improved identification of those at increased disease risk, such as those with more serious drug therapy problems (YouScript34) or those at higher risk for cancer or thrombophilia (MeTree40). Most CDS tools statistically significantly improved documentation of information such as family history or race/ethnicity, which was used to detect those at higher risk and provide risk estimates (e.g., risk of hemoglobinopathies due to African, Asian, Mediterranean, Middle Eastern, and/or Hispanic ancestry) (Supplementary appendix 3). Almost all participants using MyFamily39 described the tool as improving the accuracy of family history and thus potentially improving risk assessment (Supplementary appendix 4). Despite interest in using the MyFamily tool to identify those at low or average risk and delay their need for screening, evidence from another family history CDS tool (MeTree40) found no statistically significant improvement in identifying those who required average risk management. Furthermore, one MyFamily participant raised the concern that documentation of risk information might be less accurate when using a CDS tool since the patient providing the information might unwittingly self-censor potentially important risk information.
CDS tools did not measurably improve patient communication about disease risk that could result in management changes. CDS tools did not consistently improve the number of patients who had discussions and reminders about screening or genetic testing (Supplementary appendix 3). Some clinicians described a lack of preparedness to have detailed discussions about the patient’s genetic risk despite using CDS tools (PREDICT38) (Supplementary appendix 4). Clinicians asked for patient/family correspondence and educational materials to better prepare them for patient communication about genetic disease risk.37,38 On the other hand, clinicians using other CDS tools described them as facilitating communication with patients and other clinicians by engaging patients in a consistent, targeted discussion about family history and reducing the time to communicate results with other clinicians.37,39
Therefore, CDS tools seemed to have a statistically significant improvement of risk assessment in terms of identification of those at high risk of disease and increased accurate documentation of those risks, though not identification of those at average risk. CDS tools did not statistically significantly improve patient communication about the patient’s genetic risk. While some clinicians felt that the CDS tools facilitated communication, other clinicians reported feeling unprepared for discussions. Participants brought up that patient education materials as part of a CDS intervention would improve patient communication.
Clinician use and satisfaction
Use of the CDS tools varied widely: studies reported 14.6% to 83.3% use among clinicians who had access to the tools.25,32 Clinicians described several reasons for why they did not use CDS tools: most commonly lack of time, as well as determining that the genetic information was not appropriate to use during a visit and expecting no new information from the tool (Supplementary appendix 4).23 For CDS tools that used active alerts, clinicians accessed approximately 55% of the alerts.30,35 Clinicians requested that alerts be less frequent and preferred not to be notified by email of low level alerts.37 Clinician satisfaction with CDS tools was found to be very high (~95%) (Supplementary appendix 3),29,36 particularly when the CDS tool was helpful in improving workflow (Supplementary appendix 4).37
Therefore, the proportion of clinicians using the CDS tools varied widely, but clinicians accessed approximately half of tool alerts. Lower use may be primarily due to lack of time. Clinician satisfaction using CDS tools was found to be very high and could be because CDS tools improved workflow.
Critical appraisal
Risk of bias for quantitative studies is presented by study design in Supplementary appendix 5. Overall, the quantitative articles were of moderate to weak quality.
Of the ten quasi-experimental studies, eight were single-arm studies and ran the risk of bias from not including a control group. Six quasi-experimental studies did not include multiple outcome measurements and three did not analyze the differences in loss to follow-up between groups. The reliability of outcome measurement was unclear for three studies. One study also noted some significant demographic differences between the comparison groups. Three were industry-sponsored and did not report any strategies employed to potentially mitigate this bias (Genomic Classifier,25,27 Neuropharmagen22).
Regarding the five RCTs, one displayed a relatively high risk of bias because it did not adequately address 6 of the 13 JBI checklist items. Four of five RCTs did not blind the participants or health-care providers to treatment assignment. Four RCTs also did not clearly report allocation concealment methods. Three RCTs (YouScript,34 RetroGram,33 CNSDose41) were industry-sponsored—of these, only the YouScript trial described mitigating bias by conducting the post hoc analysis without funding.
The remaining cohort study compared intervention participants with historically matched controls, but no strategies were used to address incomplete follow-up and it was unclear how management changes were measured for the matched controls since these outcomes were not reported. It was also industry-sponsored but described that data analysis was conducted through an unrestricted university research grant (YouScript36).
Risk of bias is presented for qualitative studies and the qualitative component of mixed method studies in Supplementary appendix 6. Overall, these articles were of moderate quality. None of the studies reported the philosophical perspective, so it was not possible to determine congruity between the philosophical perspective and the research methodology. No study theoretically or culturally located the researcher or addressed the influence of the researcher on the research and vice versa. One study also did not state the qualitative research methodology used.39
Regarding publication bias, the query of the trials database revealed 14 relevant registrations. Among these, only one was a trial without an identifiable publication over 5 years after completion. Therefore, publication bias on this topic was assessed to be low.
Regarding selective outcome reporting bias, outcomes in publications generally matched the outcomes proposed in methods sections or in protocols where available. None of the studies in the review were missing outcomes, although three were missing pertinent details on some secondary outcomes such as measures at all timepoints or statistical significance. However, only two studies included in the review had published protocols, so assessment of their selective outcome reporting bias was limited. Of the six published studies from the database query, two were missing a report of a secondary outcome measure. Among the 30 studies from the database query and from this review, 4 studies did not completely report outcomes but were ongoing or only completed recently. Thus, selective outcome reporting bias was assessed to be low.
DISCUSSION
Nongenetics providers are gatekeepers to genetics services and may use genetic information in screening or treatment decisions, but experience barriers such as lack of awareness, training, or time.42 CDS tools can potentially improve and support uptake of genetics services but it is unknown whether they actually influence management decisions. This study systematically assessed the clinical utility of genetic CDS tools for nongenetics providers. This study found that genetic CDS tools appear to slightly increase management changes made by nongenetics providers, including changes in medication choice, dose, or number; increased referrals to medical genetics/genetic counseling; and increased rate of screening and genetic testing, and changes in treatment (in this review, namely cancer treatment). The majority of these management changes were appropriate per guidelines and clinical/epidemiological studies. Therefore, our findings support the use of CDS tools to assist nongenetics providers in making appropriate clinical decisions for genetics-related care. Future research should include rigorous collection of specific management changes and patient outcomes when using CDS tools. This data is necessary to evaluate whether CDS tools are effective at influencing management decisions at the point of care and at ultimately improving health outcomes.
We also found that genetic CDS tools statistically improved risk assessment: identification of those at high genetic risk through more accurate documentation of family history or race/ethnicity. However, although some clinicians described CDS tools as facilitating patient communication about genetic risk, statistical improvements were not observed and patient education materials were requested to aid discussions. Nevertheless, clinicians were highly satisfied with the tools. Thus, CDS tools seem to have utility in practice because they promote management changes, most of which are appropriate, and improve risk assessment by nongenetics providers. There is a role for CDS tool designers to develop patient education materials to further enhance the ability of CDS tools to aid nongenetics providers when communicating with patients about potential management decisions.
The nature of genetic CDS tools in our review is consistent with findings from previous reviews in that they tend to focus on cancer and pharmacogenetics.8 This may be because the evidence base for these conditions is closer to readiness for use in practice. Nevertheless, our review provides novel insights about clinical utility of these tools. Our review also found that few genetic CDS tools incorporated active alerts and workflow integration, which are previously identified predictors of CDS tool success.43 Even within tools with active alerts, our review showed that alerts were only used about half the time. This might be due to alert fatigue, which is commonly reported in CDS tools; designers of genetic CDS tools should thus limit disruptive alerts to serious management changes.44 Unlike most CDS tools today, most genetic CDS tools in this review were not integrated with the EMR to minimize workflow disruption. This may be because of particular challenges with integrating genetic information into EMRs. Often genetics involves complex logic, no standard vocabulary, and reports that are not in a computable format.45 To maximize the possible benefit genetic CDS tools could have on management, future research and tool design should explore judicious use of active alerts and using an external web service such as a cloud rather than directly integrating with the EMR.
Before a CDS tool can influence clinicians’ management, it must be accessed. We found that providers may not use genetic CDS tools because they lacked time or considered the information irrelevant for that particular visit. This adds to previously reported reasons for lack of use, which include lack of computer literacy, lack of awareness of tools that are not part of their typical workflow, and disruption of face-to-face interaction with patients.44 Future studies should continue to report reasons for lack of use to evaluate and address any potential barriers upstream of management. Targeting these reasons for lack of use, such as through education about their existence, benefit, evidence base, and integration within a clinical workflow, can improve the tools’ uptake and subsequently influence clinical management. For CDS tools available outside of an institution or research study, continuing education and dissemination through professional organizations should be considered to help increase awareness of these tools. Studies should also report the number of providers using the tool. Conclusions about management changes may be limited if only a small number of providers have access to the CDS tool since their response may not be representative if the tool is implemented more widely.
Finally, it is important to note that genetic CDS tools cannot and are not intended to replace assessment and management by genetics experts. Rather, these tools have utility in supporting nongeneticists to appropriately refer to genetics experts for testing and management. Our review demonstrates that genetic CDS tools aid nongeneticists by identifying those at higher risk and increasing appropriate referrals, making efficient use of the limited number of geneticists and genetic counselors.
This review had several limitations. Effect sizes could not be meta-analyzed due to the heterogeneity of study designs and reported outcomes. In addition, only studies on actual patients were included. Since many genetic conditions are relatively rare in nongeneticists’ practices, research using CDS tools for specific genetic disorders is challenging. Including studies on hypothetical patients would likely have broadened the review but these were excluded because management changes based on a limited number of hypothetical cases may not reflect overall clinical utility when encountering a variety of real patients. Another limitation was that there were few comparative studies evaluating the effect of CDS tools in comparison with standard of care. Within these comparative studies, management changes were often not the primary outcome and were not always reported in the comparison group. This limited the data that could be used to calculate effect sizes. In addition, the conclusions about the secondary outcomes are less robust since they were not reported in all included studies. Finally, the included studies were found to be of weak to moderate quality.
In conclusion, this is the most comprehensive synthesis of qualitative and quantitative evidence of genetic CDS tools used by nongenetics clinicians, which found that these tools slightly increase appropriate management changes compared with standard of care, but this increase is not consistently statistically significant. While these tools do improve risk assessment of those at increased risk and have high clinician satisfaction, there is mixed evidence about whether they are used often or improve patient communication about genetic risk that could precede management decisions. Thus, genetic CDS tools are promising as an aid to nongenetics providers at the point of care but further evidence is required. Further research should continue to collect data on clinical management and patient outcomes to enable robust evaluation and meta-analysis about the clinical utility of these tools.
Data availability
Data relevant to this systematic review is available upon request.
References
Mikat-Stevens, N. A., Larson, I. A. & Tarini, B. A. Primary-care providers’ perceived barriers to integration of genetics services: a systematic review of the literature. Genet. Med. 17, 169–176, https://doi.org/10.1038/gim.2014.101 (2015).
Carroll, J.C., Allanson, J. & Morrison, S. et al. Informing integration of genomic medicine into primary care: an assessment of current practice, attitudes, and desired resources. Front. Genet. 10, 1189, https://doi.org/10.3389/fgene.2019.01189 (2019).
Krier, J. B., Kalia, S. S. & Green, R. C. Genomic sequencing in clinical practice: applications, challenges, and opportunities. Dialogues Clin. Neurosci. 18, 299–312 (2016).
Berberich, A. J., Ho, R. & Hegele, R. A. Whole genome sequencing in the clinic: empowerment or too much information? CMAJ. 190, E124–E125, https://doi.org/10.1503/cmaj.180076 (2018).
Overby, C. L., Kohane, I. & Kannry, J. L. et al. Opportunities for genomic clinical decision support interventions. Genet. Med. 15, 817–823, https://doi.org/10.1038/gim.2013.128 (2013).
Sim, I. et al. Clinical decision support systems for the practice of evidence-based medicine. J. Am. Med. Inform. Assoc. 8, 527–534, http://www.ncbi.nlm.nih.gov/pubmed/11687560 (2001).
Grosse, S. D. & Khoury, M. J. What is the clinical utility of genetic testing? Genet. Med. 8, 448–450, https://doi.org/10.1097/01.gim.0000227935.26763.c6 (2006).
Welch, B. M. & Kawamoto, K. Clinical decision support for genetically guided personalized medicine: a systematic review. J. Am. Med. Inform. Assoc. 20, 388–400, https://doi.org/10.1136/amiajnl-2012-000892 (2013).
Bombard, Y., Bach, P. B. & Offit, K. Translating genomics in cancer care. J. Natl. Compr. Canc. Netw. 11, 1343–1353, http://www.ncbi.nlm.nih.gov/pubmed/24225968 (2013).
Lizarondo. L. et al. Mixed methods systematic reviews. In: Aromataris E, Munn Z, eds. Joanna Briggs Institute Reviewer’s Manual. The Joanna Briggs Institute; Adelaide, Australia; 2019. Accessed May 19, 2020. https://reviewersmanual.joannabriggs.org/.
Hong, Q. N., Pluye, P., Bujold, M. & Wassef, M. Convergent and sequential synthesis designs: implications for conducting and reporting systematic reviews of qualitative and quantitative evidence. Syst. Rev. 6, 61, https://doi.org/10.1186/s13643-017-0454-2 (2017).
Moher, D., Liberati, A., Tetzlaff, J. & Altman, D. G. Preferred reporting items for systematic reviews and meta-analyses: the PRISMA statement. BMJ. 339, 332–336, https://doi.org/10.1136/bmj.b2535 (2009).
Ouzzani, M., Hammady, H., Fedorowicz, Z. & Elmagarmid, A. Rayyan—a web and mobile app for systematic reviews. Syst. Rev. 5, 210, https://doi.org/10.1186/s13643-016-0384-4 (2016).
Vassy, J. L., Bates, D. W. & Murray, M. F. Appropriateness: a key to enabling the use of genomics in clinical practice? Am. J. Med. 129, 551–553, https://doi.org/10.1016/j.amjmed.2016.02.010 (2016).
Mirzazadeh, A., Malekinejad, M. & Kahn, J. G. Relative risk reduction is useful metric to standardize effect size for public heath interventions for translational research. J. Clin. Epidemiol. 68, 317–323, https://doi.org/10.1016/j.jclinepi.2014.11.013 (2015).
Azzopardi, D. Group comparison calculator. http://www.neoweb.org.uk/Additions/compare.htm (2019).
Andrade, C. Understanding relative risk, odds ratio and related terms: as simple as it can get. J. Clin. Psychiatry. 76, e857–e861 (2015).
Borenstein M, Hedges LV, Higgins JPT, Rothstein HR When does it make sense to perform a meta-analysis? In: Introduction to Meta-Analysis. John Wiley and Sons; Chichester, West Sussex, United Kingdom; 2009:357-64. https://doi.org/10.1002/9780470743386.
Lockwood, C. et al. Chapter 2: Systematic reviews of qualitative evidence. In: Aromataris E, Munn Z, eds. Joanna Brigg’s Institute Reviewer’s Manual. The Joanna Briggs Institute; Adelaide, Australia; 2017. Accessed May 21, 2020. https://wiki.joannabriggs.org/display/MANUAL/Chapter+2%3A+Systematic+reviews+of+qualitative+evidence.
Tufanaru C, Munn Z, Aromataris E, Campbell JHL Chapter 3: Systematic reviews of effectiveness. In: Aromataris E, Munn Z, eds. Joanna Briggs Institute Reviewer’s Manual. The Joanna Briggs Institute; Adelaide, Australia; 2017. Accessed April 25, 2019. https://reviewersmanual.joannabriggs.org/.
Balshem, H. et al. Finding Grey Literature Evidence and Assessing for Outcome and Analysis Reporting Biases When Comparing Medical Interventions: AHRQ and the Effective Health Care Program Methods Guide for Comparative Effectiveness Reviews.; Rockville, Maryland, United States; 2013. Accessed October 14, 2020. www.effectivehealthcare.ahrq.gov/reports/final.cfm.
Blasco-Fontecilla, H. Clinical utility of pharmacogenetic testing in children and adolescents with severe mental disorders. J. Neural Transm. (Vienna). 126, 101–107, https://doi.org/10.1007/s00702-018-1882-4 (2019).
Borden, B. A. et al. Assessment of provider-perceived barriers to clinical use of pharmacogenomics during participation in an institutional implementation study. Pharmacogenet. Genomics 29, 31–38, https://doi.org/10.1097/FPC.0000000000000362 (2019).
Edelman, E. A. et al. Implementation of an electronic genomic and family health history tool in primary prenatal care. Am. J. Med. Genet. C. 166, 34–44 (2014).
Michalopoulos, S. N. et al. Influence of a genomic classifier on post-operative treatment decisions in high-risk prostate cancer patients: results from the PRO-ACT study. Curr. Med. Res. Opin. 30, 1547–1556, https://doi.org/10.1185/03007995.2014.919908 (2014).
Petzel, S. V., Vogel, R. I., McNiel, J., Leininger, A., Argenta, P. A. & Geller, M. A. Improving referral for genetic risk assessment in ovarian cancer using an electronic medical record system. Int. J. Gynecol. Cancer 24, 1003–1009, https://doi.org/10.1097/IGC.0000000000000148 (2014).
Badani, K. et al. Impact of a genomic classifier of metastatic risk on postoperative treatment recommendations for prostate cancer patients: A report from the DECIDE study group. Oncotarget. 4, 600–609, https://doi.org/10.18632/oncotarget.918 (2013).
Baer, H. J. et al. Use of a web-based risk appraisal tool for assessing family history and lifestyle factors in primary care. J. Gen. Intern. Med. 28, 817–824, https://doi.org/10.1007/s11606-013-2338-z (2013).
Hall-Flavin, D. K. et al. Utility of integrated pharmacogenomic testing to support the treatment of major depressive disorder in a psychiatric outpatient setting. Pharmacogenet. Genomics 23, 535–548, https://doi.org/10.1097/FPC.0b013e3283649b9a (2013).
Scheuner, M. T. et al. A cancer genetics toolkit improves access to genetic services through documentation and use of the family history by primary-care clinicians. Genet. Med. 16, 60–69, https://doi.org/10.1038/gim.2013.75 (2014).
Emery, J. et al. The GRAIDS Trial: a cluster randomised controlled trial of computer decision support for the management of familial cancer risk in primary care. Br. J. Cancer 97, 486–493, https://doi.org/10.1038/sj.bjc.6603897 (2007).
Wilson, B. J. et al. Cluster randomized trial of a multifaceted primary care decision-support intervention for inherited breast cancer risk. Fam. Pract. 23, 537–544, https://doi.org/10.1093/fampra/cml026 (2006).
Tural, C. et al. Clinical utility of HIV-1 genotyping and expert advice: the Havana trial. AIDS. 16, 209–218, https://doi.org/10.1097/00002030-200201250-00010 (2002).
Kim, K., Magness, J. W., Nelson, R., Baron, V. & Brixner, D. I. Clinical utility of pharmacogenetic testing and a clinical decision support tool to enhance the identification of drug therapy problems through medication therapy management in polypharmacy patients. J. Manag. Care Spec. Pharm. 24, 1251 (2018).
O’Donnell, P. H. K. et al. Pharmacogenomics-based point-of-care clinical decision support significantly alters drug prescribing. Clin. Pharmacol. Ther. 102, 859–869, https://doi.org/10.1002/cpt.709 (2017).
Brixner, D. A. et al. The effect of pharmacogenetic profiling with a clinical decision support tool on healthcare resource utilization and estimated costs in the elderly exposed to polypharmacy. J. Med. Econ. 19, 213–228, https://doi.org/10.3111/13696998.2015.1110160 (2016).
Klinkenberg-Ramirez, S. et al. Evaluation: a qualitative pilot study of novel information technology infrastructure to communicate genetic variant updates. Appl. Clin. Inform. 7, 461–476, https://doi.org/10.4338/ACI-2015-11-RA-0162 (2016).
Unertl, K. M., Field, J. R., Price, L. & Peterson, J. F. Clinician perspectives on using pharmacogenomics in clinical practice. Per. Med. 12, 339–347, https://doi.org/10.2217/PME.15.10 (2015).
Doerr, M., Edelman, E., Gabitzsch, E., Eng, C. & Teng, K. Formative evaluation of clinician experience with integrating family history-based clinical decision support into clinical practice. J. Pers. Med. 4, 115–136, https://doi.org/10.3390/jpm4020115 (2014).
Orlando, L. A. et al. Clinical utility of a Web-enabled risk-assessment and clinical decision support program. Genet. Med. 18, 1020–1028, https://doi.org/10.1038/gim.2015.210 (2016).
Singh, A. B. Improved antidepressant remission in major depression via a pharmacokinetic pathway polygene pharmacogenetic report. Clin. Psychopharmacol. Neurosci. 13, 150–156, https://doi.org/10.9758/cpn.2015.13.2.150 (2015).
Gottesman, O. et al. The CLIPMERGE PGx program: clinical implementation of personalized medicine through electronic health records and genomics-pharmacogenomics. Clin. Pharmacol. Ther. 94, 214–217, https://doi.org/10.1038/clpt.2013.72 (2013).
Kawamoto, K., Houlihan, C. A., Balas, E. A. & Lobach, D. F. Improving clinical practice using clinical decision support systems: a systematic review of trials to identify features critical to success. BMJ. 330, 765, https://doi.org/10.1136/bmj.38398.500764.8F (2005).
Sutton, R. T., Pincock, D., Baumgart, D. C., Sadowski, D. C., Fedorak, R. N. & Kroeker, K. I. An overview of clinical decision support systems: benefits, risks, and strategies for success. npj Digit. Med. 3, 1–10, https://doi.org/10.1038/s41746-020-0221-y (2020).
Williams, M. S. et al. Genomic information for clinicians in the electronic Health record: lessons learned from the Clinical Genome Resource Project and the Electronic Medical Records and Genomics Network. Front. Genet. 10, 1059, https://doi.org/10.3389/fgene.2019.01059 (2019).
Acknowledgements
This project is partially funded by an Early Career Award from the Ontario Ministry of Research and Innovation (ER17-13-045). A.S. was supported by an Ontario Graduate Scholarship from the University of Toronto. Y.B. was supported by a New Investigator Award from the Canadian Institutes of Health Research (CIHR). We thank Andrea Tricco, Lusine Abrahamyan, and Petros Pechlivanoglou for their advice and feedback over the course of this study.
Author information
Authors and Affiliations
Contributions
Conceptualization: A.S., Y.B., J.C.C. Formal analysis: A.S., Y.B., J.C.C. Funding acquisition: Y.B., A.S. Investigation: A.S., E.U., L.E.O., C.M., S.S. Methodology: A.S., Y.B., J.C.C. Project administration: A.S., Y.B., J.C.C. Supervision: Y.B., J.C.C. Visualization: A.S. Writing—original draft: A.S., Y.B., J.C.C. Writing—review & editing: A.S., Y.B., J.C.C., E.U., L.E.O., C.M., S.S.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
About this article
Cite this article
Sebastian, A., Carroll, J.C., Oldfield, L.E. et al. Effect of genetics clinical decision support tools on health-care providers’ decision making: a mixed-methods systematic review. Genet Med 23, 593–602 (2021). https://doi.org/10.1038/s41436-020-01045-1
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41436-020-01045-1
This article is cited by
-
Implementation of pharmacogenomic clinical decision support for health systems: a cost-utility analysis
The Pharmacogenomics Journal (2022)