Abstract
Purpose
The purpose of this document is to provide updated guidance for the genetic evaluation of cardiomyopathy and for an approach to manage secondary findings from cardiomyopathy genes. The genetic bases of the primary cardiomyopathies (dilated, hypertrophic, arrhythmogenic right ventricular, and restrictive) have been established, and each is medically actionable; in most cases established treatments or interventions are available to improve survival, reduce morbidity, and enhance quality of life.
Methods
A writing group of cardiologists and genetics professionals updated guidance, first published in 2009 for the Heart Failure Society of America (HFSA), in a collaboration with the American College of Medical Genetics and Genomics (ACMG). Each recommendation was assigned to teams of individuals by expertise, literature was reviewed, and recommendations were decided by consensus of the writing group. Recommendations for family history, phenotype screening of at-risk family members, referral to expert centers as needed, genetic counseling, and cardiovascular therapies, informed in part by phenotype, are presented in the HFSA document.
Results
A genetic evaluation of cardiomyopathy is indicated with a cardiomyopathy diagnosis, which includes genetic testing. Guidance is also provided for clinical approaches to secondary findings from cardiomyopathy genes. This is relevant as cardiomyopathy is the phenotype associated with 27% of the genes on the ACMG list for return of secondary findings. Recommendations herein are considered expert opinion per current ACMG policy as no systematic approach to literature review was conducted.
Conclusion
Genetic testing is indicated for cardiomyopathy to assist in patient care and management of at-risk family members.
Similar content being viewed by others
Introduction
Continued rapid progress has been made in understanding the genetic basis of cardiomyopathy. This work, which describes the content, approach, and expertise needed for a cardiomyopathy genetic evaluation, was first developed in a guideline statement in 2008 and published in 2009 for the Heart Failure Society of America (HFSA).1 This has now been updated by a writing group organized by the American College of Medical Genetics and Genomics (ACMG) and the HFSA to serve as a practice resource (ACMG) and as a revised guideline statement (HFSA).2
This collaboration of cardiovascular and genetics professionals mirrors a recent proliferation of specialized cardiovascular genetics clinics.3 Most commonly cardiologists, adult or pediatric, with special interest or training in cardiovascular genetics, team up with genetics professionals, usually board-eligible or board-certified genetic counselors and/or clinical geneticists, ideally with cardiovascular expertise, to provide state-of-the-art genetics services to the many patients and families with cardiomyopathy. This growth has been triggered by improvements in technology for clinical genetic testing, resulting in the availability of large clinical genetic testing panels, where numerous genes of interest can be sequenced quickly and efficiently and accurately using continually developing massively parallel DNA sequencing technologies. This growth also recognizes the critical importance of integrated expert phenotypic information with final clinical recommendations in light of burgeoning sequence information.4
This collaboration also speaks to the recent prominence of cardiovascular genetics and genomics brought about by the emergence of clinical exome sequencing and the ACMG recommendation, first in 20135 and updated in 2016,6 to return relevant and actionable secondary findings. Of the 59 medically actionable genes cited in 2016, 30 (51%) had cardiovascular phenotypes, and 16 (27%) were genes that included cardiomyopathy phenotypes. By request from the ACMG, we also provide guidance for secondary findings derived from cardiomyopathy genes.
The rationale for the inclusion of cardiomyopathy genes in the ACMG secondary findings list, and the basis for the clinical screening, counseling, and molecular recommendations contained herein, are because the cardiomyopathies are medically actionable: well-established treatments or interventions are available to improve survival, reduce morbidity, and enhance quality of life.7,8 Cardiomyopathies may present late in their course with advanced disease, which includes heart failure, heart block and/or life-threatening arrhythmias including sudden cardiac death, and thromboembolic events, including stroke from atrial arrhythmias or ventricular thrombus. Thus, the rationale to identify genetic risk is compelling, so that those found to be at-risk can undergo interval screening to detect the earliest manifestations of the cardiomyopathy phenotype. The first evidence of a phenotype then permits earlier interventions,8 including lifestyle modifications, drugs to slow or halt disease progression or to prevent thromboembolism, and procedures, drugs, or devices to reduce the risk of sudden cardiac death.7 Identification of at-risk individuals, whether affected but asymptomatic or those clinically unaffected may also have implications for genetic counseling and reproductive decision-making.
Cardiovascular physicians are expert at assessing the nuances of cardiomyopathy phenotypes or subphenotypes, an essential contribution to cardiovascular genetics care. As in 2009,1 our current approach continues to be stratified by cardiomyopathy phenotype, as clinical and genetic data collection, analysis, and decision-making for the cardiomyopathies remain anchored by phenotypic categories.
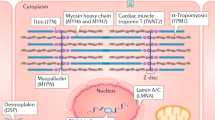
Genetic cardiomyopathy has substantial complexity, as shown by overlap in phenotype as well as an overlap of genes.9 The primary cardiomyopathies covered in this document include hypertrophic cardiomyopathy (HCM), dilated cardiomyopathy (DCM), arrhythmogenic right ventricular cardiomyopathy (ARVC), and restrictive cardiomyopathy (RCM). Despite this complex interplay of genes, variants, and phenotypes, current knowledge when combined with expert phenotyping and the sensitivity and specificity of current genetic testing is sufficient to effectively conduct genetic cardiomyopathy evaluations. We caution, however, that variant interpretation must be thoughtful, rigorous, and leverage the most up-to-date approaches, as not all variants identified by genetic testing will be clinically significant or disease-causing. Key resources include use of the most recent ACMG/AMP guidance,6,10 now being augmented by ClinGen, a National Human Genome Research Institute–sponsored initiative to curate genes and variants and place them into ClinVar, a publicly accessible database,11,12 and other large publicly accessible reference databases.
Materials and methods
The writing group was established conjointly with the ACMG and HFSA between 2013 and 2015. The approaches to creating, curating, and approving practice guidelines or practice resources for the HFSA and ACMG, respectively, have been outlined in each publication, and the material covered in this and the companion document2 are congruent with one another. Differences in scope, including supplemental materials, are denoted and cross-referenced.
The writing group was comprised of a panel of experts, board-certified cardiologists, and genetics professionals with experience and expertise in genetic cardiomyopathies, with a goal to revise a prior HFSA publication in a conjoint effort with a new document for the ACMG. Each author was screened for relevant conflicts of interest and all conflicts shown were considered nonsubstantial to influence the document. Dr. Vatta was included in the writing group prior to his employment with a for-profit genetic testing company; following his employment potential conflicts of interest regarding genetic testing indications were managed by his recusal from pertinent discussions.
The genetic recommendations presented herein are best considered as expert opinion. Major topic areas were divided between author groups based on background and expertise. Each conducted nonsystematic literature reviews (no established date ranges, no search strings), and with no formal method of grading of evidence, as is now recommended by the ACMG.13 We clarify that the project and writing group were assembled in 2013 and 2014, and began work in 2015 prior to this ACMG recommendation.
As discussed in the companion document,2 no large randomized, placebo-controlled studies meeting usual criteria for highest levels of evidence (e.g., see7 for usual cardiovascular guideline standards) are available in the genetic cardiomyopathy literature. Selected reviews that cite primary references to substantiate the expert opinion are provided.14,15,16,17,18,19,20,21 Numbered guidance provided herein have been labeled “Recommendations” as per the ACMG and the ACMG approach to production of practice resource documents.
This document has been limited to recommendations regarding genetic testing and secondary findings of cardiomyopathy genes. Recommendations relevant to the conduct a full genetic evaluation are provided in the companion document,2 and include (1) obtaining a family history of at least three generations; (2) phenotypic screening for cardiomyopathy in at-risk family members, which includes serial phenotype screening recommendations, and types of testing, by cardiomyopathy phenotype; (3) referral of patients to expert centers as needed; (4) genetic counseling of patients and families; and (5) therapy based upon phenotype, including drugs, devices, and special clinical recommendations by gene. Additional background on cardiomyopathy is provided in the companion publication.2
Recommendation 1. Genetic testing is recommended for patients with cardiomyopathy
-
(a)
Genetic testing is recommended for the most clearly affected family member.
-
(b)
Cascade genetic testing of at-risk family members is recommended for pathogenic and likely pathogenic variants.
-
(c)
In addition to routine newborn screening tests, specialized evaluation of infants with cardiomyopathy is recommended, and genetic testing should be considered.
Genetic testing is recommended to determine if a pathogenic variant can be identified to facilitate patient management and family screening. The identification of at-risk family members is critical because the first presentation may be sudden death. Cascade genetic screening identifies asymptomatic affected family members and presymptomatic carriers of pathogenic variants.22 Institution of therapy in asymptomatic affected individuals improves outcomes and decreases hospitalization and death due to heart failure.23,24 Preliminary studies indicate that treatment of presymptomatic carriers of pathogenic variants may improve outcome as well although larger studies are needed.25 Genetic testing and cascade screening for HCM have been shown to be cost-effective in Australia and the United States.26,27 The identification of a molecular cause may also lead to critical gene-specific cardiac or extracardiac management recommendations. For example, cardiac hypertrophy seen in LAMP2, PRKAG2, PTPN11, and RAF1 pathogenic variant carriers can represent a genocopy of hypertrophy seen with sarcomeric pathogenic variants; yet LAMP2, PRKAG2, PTPN11, and RAF1 patients have different clinical courses and management needs.28,29 In sarcomeric carriers, genotype status is associated with long-term outcomes, including all-cause mortality.30,31 In DCM, there is evidence for prognostication value of genetic testing32,33,34,35 and management implications for specific genetic findings, such as consideration of implantable cardioverter defibrillator (ICD) for primary prevention in carriers of LMNA pathogenic variants.36 In ARVC, ICD placement for primary prevention in asymptomatic male carriers of a malignant pathogenic variant showed significant impact on long-term clinical outcome.37
Testing should ideally be initiated on the person in a family with the most definitive diagnosis and most severe manifestations. This approach will maximize the likelihood of obtaining diagnostic results and detecting whether multiple pathogenic variants may be present and contributing to variable disease expression or severity.
Nomenclature follows the ACMG/AMP approach10 for calling variants as pathogenic (P), likely pathogenic (LP), variants of uncertain significance (VUS), likely benign, and benign. The indications for genetic testing include guiding patient management and facilitating family screening and reproductive risk assessment.
Test selection: genes and gene panels
Since the 2009 HFSA guideline,1 the number of genes known that harbor rare variants that cause cardiomyopathy has increased, the number of clinical laboratories performing high-volume cardiovascular genetic testing has expanded, and the number, type, and technologies available for gene-based sequencing have been in constant evolution. While the 2009 HFSA guideline suggested that “genetic testing should be considered,” additional data on the importance of genetic testing for prognostication and management as well as cascade screening and risk stratification of relatives support the current genetic testing recommendation. Furthermore, the cost for most large genetic panels is substantially lower than it was in 2009, with expectations for continued decline.38 Nevertheless, genetic testing is probabilistic in nature and interpretation of genetic variation will continue to be refined as additional sequencing information becomes available from both affected and unaffected individuals.
These recommendations do not address molecular testing in prenatal, newborn screening, or in vitro fertilization settings.
We also note ongoing challenges of variant interpretation in non-Caucasian, non-northern European populations, as most genetic testing, and hence repositories of known pathogenic variants, has previously been conducted principally in the Caucasian/northern European population. The recent development of very large population databases (e.g., ExAC, http://exac.broadinstitute.org, or gnomAD, http://gnomad.broadinstitute.org) now provides limited numbers of reference alleles from non-European cohorts, which has greatly assisted variant interpretation. However, genetic test interpretation of variant alleles from ethnic groups not represented or represented in low numbers in reference data sets has become extremely challenging, and must be approached with considerable caution.
A variety of resources are publicly available that provide additional relevant information (e.g., GeneReviews, http://www.ncbi.nlm.nih.gov/books/NBK1116), on individual genes (e.g., Online Mendelian Inheritance in Man, http://www.omim.org), specific genetic variants and their population frequencies (e.g., dbSNP, http://www.ncbi.nlm.nih.gov/snp; ExAC browser, http://exac.broadinstitute.org; Genome Aggregation Database [gnomAD], http://gnomad.broadinstitute.org/; Exome Variant Server, http://evs.gs.washington.edu/EVS; or 1000 Genomes, http://www.1000genomes.org), and information for the interpretation of these variants (e.g., ClinVar, http://www.ncbi.nlm.nih.gov/clinvar and ClinGen, http://www.clinicalgenome.org).
We also note that large insertion/deletion variants (e.g., >25 nucleotides) and other structural changes in DNA, referred to as copy-number variants, in a preliminary study represent <1% of cardiomyopathy cases,39 although structural variants have received minimal investigation in the cardiomyopathies and may have greater relevance than is currently understood.
Whom to test
To yield the most conclusive, informative results, diagnostic genetic testing is optimally initiated on a confirmed affected individual. Furthermore, as there are sometimes multiple genetic variants contributing to disease in a single family, the testing should ideally be initiated on the person who is most likely to harbor the disease-causing variant or variants. This is frequently the individual in the family with the most severe disease and/or the earliest disease onset. This is a well-established principle in clinical genetics, as selecting the individual with the most evident disease increases the likelihood of finding a genetic cause. If the ideal person for initiation of genetic testing in a family is unavailable or unwilling to proceed, then comprehensive genetic testing should be considered for another unequivocally affected family member.
When to test
The timing for ordering genetic testing in a patient with cardiomyopathy has not been studied. Because results may guide management, we recommend genetic testing at the time a new cardiomyopathy diagnosis is made, but it can be conducted at any time following diagnosis. Education and counseling regarding genetic testing options are a key component of the process. For those who have had genetic testing in the past, retesting may be appropriate if the previous testing produced negative or inconclusive results and the test’s detection rate has improved. This latter point is particularly relevant for DCM as the gene panels have rapidly expanded (e.g., TTN40,41,42 and others) and are anticipated to continue.
Genetic testing for the cardiomyopathies may best be viewed as continuously evolving, as new genes, and hence larger panels with greater sensitivity, continue to emerge. Although no data are available, we suggest that repeat genetic testing is reasonable if test sensitivity has increased by 5–10%. An alternative approach is to tailor retesting if particular characteristics of the patient’s phenotype are consistent with a newly identified gene. Further, the genetics provider involved in a patient’s care should periodically revisit results as variants may be reclassified over time.43,44,45 Such reclassification includes upgrading variants from VUS to likely pathogenic or pathogenic, as additional probands and affected family members with the phenotype of interest are found to carry the variant. Conversely, some variants, previously considered pathogenic, are downgraded to a VUS, or likely benign or benign, as larger data sets from expanded ethnicities become available.
How to test
With the development of next-generation sequencing (NGS), panels incorporating dozens of genes relevant to the phenotype have become the norm, as they are technically feasible and less costly.38 As a result, clinical genetic testing panels for these disorders are changing rapidly. Molecular genetic testing for multiple genes using a multigene panel is now the standard of practice for cardiovascular genetic medicine. Furthermore, multigene panel genetic testing is recommended over a serial single-gene testing approach due to the genetically heterogeneous nature of cardiomyopathy. Genetic testing and cascade screening have been shown to be cost-effective.26,27
Large gene panels for cardiomyopathy may include genes that cause genetic syndromes associated with cardiomyopathy (e.g., Fabry disease, Danon disease, Alström syndrome), neuromuscular conditions associated with cardiomyopathy (e.g., limb girdle muscular dystrophies) or metabolic conditions. These large gene panels have the advantage of increasing the likelihood of identifying a molecular etiology, especially in patients with mixed phenotypes or those who lack pathognomonic features.46,47 Considerable overlap of genes among different types of cardiomyopathy (and other phenotypes) is also well established.9 Panels also increase the likelihood of identifying individuals who carry disease-causing variants in multiple genes, and this knowledge is extremely important for appropriate targeted testing of family members.
With larger gene panels, the likelihood of identifying a VUS increases in proportion to the number of genes tested, increasing the complexity of the interpretation and genetic counseling. Importantly, the strength of evidence for gene–disease pairs on current panels differs, with some well-established genes having a wealth of information regarding disease-causing variants while more recently identified genes have much less information available. The latter case increases the likelihood of a variant being classified as a VUS. The composition of gene panels varies by testing lab. It is critical that the ordering physician has an understanding of the uses, benefits, and limitations of specific test types to select the most appropriate test for their patient (Supplemental Table). Addition of TTN and BAG3 to DCM panels increased genetic testing yield by more than 10%,40,41,42 but for HCM, recent studies have shown that expanded panels do not currently increase sensitivity.27 Thus the decision to order a panel that includes a larger number of genes should be based on the specifics of the patient’s medical history, physical exam findings, and family history.
HCM
The level of evidence for testing in HCM is based on studies showing a high diagnostic yield of genetic testing in children and adults and prognostic value of genotype status.27,30,31,48,49 HCM is considered a disease of the sarcomere, and variations in genes encoding sarcomeric proteins, in which there is low tolerance for genetic variation are common causes.50 The diagnostic yield of HCM testing is approximately 30–60% (Table 1). The yield of testing is higher in individuals who have a known family history of HCM.27,49 Pathogenic variants in MYH7 and MYBPC3 account for approximately 80% of all cases for which a molecular diagnosis is achieved.51,52 Beyond sarcomeric genes, core genes to screen in patients with HCM include GLA, PRKAG2, and LAMP2.
Infants and children with HCM may require more specialized evaluation and diagnostic testing because of the rate of syndromic conditions and inborn errors of metabolism associated with HCM at these ages.53,54,55 Consultation with a geneticist is indicated.
DCM
Evidence indicates that clinical genetic testing can identify the cause of DCM in families with autosomal dominant inheritance in approximately 25-40% of cases, whereas in isolated cases of DCM, the yield of testing is commonly estimated at 10–25%.56,57,58,59 Core genes to be tested in individuals with DCM include genes encoding sarcomeric and cytoskeletal proteins (Table 1), although DCM testing panels typically carry several dozen genes, some with uncertain significance. In most cases, all HCM and ARVC genes are included in DCM panels because of gene/phenotype overlap.
Protein-truncating variants in TTN (TTNtv) represent the most common genetic testing finding in DCM, ranging from 10 to 20% of cases.40,41,42 While many commercial testing laboratories will adjudicate all TTNtv’s, whether singleton or familial, as pathogenic or likely pathogenic, variant interpretation is challenging due to the large size of the gene and the frequency of truncating TTN variants in reference populations.40,41,60,61 Most studies have not been family-based, where segregation could be evaluated, but some nonsegregation of TTNtv’s has been identified.60,61,62 Further, recent cardiac magnetic resonance data of normal individuals from a population-based study showed a small but significant decrement in LV function with TTNtv’s in constitutive cardiac exons,61 suggesting that in some cases a TTNtv may function as a risk allele.
The LMNA gene is the second most commonly identified cause of DCM with a diagnostic yield of 5.5%, and gene-specific management recommendations are available.63,64 More recently identified genetic causes of DCM such as BAG3, a chaperone regulator, and RBM20, a protein required for RNA splicing, identify novel molecular mechanisms for disease,65,66 and are each identified in approximately 2% of DCM cases. DCM is a common complication of neuromuscular disease such as Duchenne or Becker muscular dystrophy. Genetic testing is important in mothers of individuals with Duchenne or Becker to determine carrier status because carrier females may develop DCM in the third to fifth decade of life.67 As in HCM, infants and children with DCM may require additional diagnostic evaluation.
ARVC
The genetic basis of ARVC was initially identified as a disease of the desmosome.68 Genetic testing of PKP2, DSP, DSG2, DSC2, JUP, TMEM43, and PLN resulted in a molecular diagnosis in 63% of patients who fulfilled Task Force criteria for ARVC.21 Digenic inheritance and compound heterozygosity are frequent69 and, combined with decreased penetrance that is a feature of ARVC, may significantly complicate genetic counseling. ARVC overlaps with arrhythmogenic left ventricular cardiomyopathy, sometimes more broadly referred to as arrhythmogenic cardiomyopathy.70 This reflects genetic and phenotypic overlap among these forms of cardiomyopathy. Accordingly, genetic testing for ARVC using a larger cardiomyopathy panel may identify nondesmosomal genes with pathogenic variants. Similarly, desmosome gene pathogenic variations have also been identified in patients diagnosed with DCM.71 Exercise has a well-established role in the pathogenesis of desmosomal cardiomyopathies, and recognition of a desmosome gene mutation can help to determine optimal exercise recommendations.72
RCM
Genetic causes of RCM continue to be identified, but because RCM is a relatively rare form of cardiomyopathy, numbers remain limited. A recent study identified a pathogenic variant in 60% of subjects, primarily occurring in genes known to cause HCM.73 Family members were frequently identified with HCM or HCM with restrictive physiology. Cardiac amyloidosis resulting from pathogenic variants in TTR needs to be differentiated from other forms of RCM due to the age demographic in which this occurs, the slowly progressive nature of this disease, and therefore different management strategies.74,75 The TTR allele p.Val142Ile (commonly referred to as Val122Ile based on nomenclature for the circulating protein after N-terminal peptide cleavage) has been found in 10% of African Americans older than age 65 with severe congestive heart failure.76 Substantial recent progress with amyloidosis, both in imaging strategies, including cardiac magnetic resonance and pyrophosphate scanning, and therapeutic interventions in ongoing clinical trials, provide new incentives for genetic diagnosis.77 Hemochromatosis is uncommon but easily excluded with iron studies, such as percent saturation of transferrin, and if present can be treated with iron removal.78
Left ventricular noncompaction
The left ventricular noncompaction (LVNC) phenotype may be observed in conjunction with all other cardiomyopathy phenotypes,2 so considerations related to genetic testing should always be directed by findings of a cardiomyopathy (or other cardiovascular) phenotype.2,79,80 Genetic testing is not recommended when the LVNC phenotype is identified serendipitously in asymptomatic individuals with otherwise normal cardiovascular structure and function.2,81
Special circumstances
A genetic etiology should be considered and a genetic evaluation conducted in cases of peripartum cardiomyopathy, as rare variants in genes known to cause DCM have been identified in these patients,82,83,84 and TTN truncating variants are present at rates similar to those found in the DCM population.84 In cases of sudden death with an autopsy diagnosis of cardiomyopathy, genetic testing may facilitate risk stratification of family members.85,86
Interpretation of genetic testing
Genetic testing results are probabilistic rather that determinative, and thus rely on strength of evidence, both for and against, of specific variants causing or contributing to disease. New guidelines have attempted to standardize and increase the stringency of interpretation, with greater clarity regarding the criteria for strength of evidence and the weighting of multiple sources of information that need to be incorporated to arrive at the interpretation.10 Despite this, the interpretations provided for a given variant may differ between clinical genetic testing laboratories.87,88 In addition, updates and revisions of the laboratory interpretation may occur as more information is obtained from larger cohorts, sometimes leading to reissuing of a clinical report with changed interpretation by diagnostic laboratories.
Because of their probabilistic nature, results of genetic testing must always be interpreted in the context of the patient’s medical and family history.44 For example, family history information and the segregation of a putative disease-causing variant within the family may be important information to guide clinical interpretation, especially in cases where novel genetic variants are identified. Also, family studies have noted more than one pathogenic variant in up to 10% of families with ARVC.89 Two or more variants have been seen in 3–5% of HCM cases,90,91,92 particularly if onset is early or severe.48 Although not reported systematically, digenic inheritance has been suggested to occur at even higher frequency with DCM.56
The diagnostic yield of genetic testing for each subtype of cardiomyopathy is much less than 100% (Table 1) and a negative genetic test result (in this setting including VUS and likely benign or benign variants) does not rule out a genetic cause. Such an uninformative result in a proband simply indicates that the genetic testing performed was unable to identify the specific cause of disease in the given family. In these circumstances, an uninformative genetic testing result cannot be used for predictive, cascade genetic testing in unaffected relatives. Rather, family screening using phenotypic evaluations is recommended.2 Larger panels, better coverage of the relevant genes, analysis for deletions, duplications, and rearrangements in the genes of interest, or exome sequencing in families with multiple living affected individuals may identify a genetic etiology.
Finally, the recent availability of and much greater focus on extensive genetic testing panels should not diminish or distract from the critical importance of expert phenotyping of patients and families, and the relevance of highly insightful phenotype and gene–variant correlations. Current genetics practice suggests that results provided by molecular genetics laboratories drive clinical decision-making, specifically actionability, in a genetic evaluation. In the Family Management section below, this guidance states that a VUS cannot be used for predictive testing, which the writing group firmly supports. However, we acknowledge that compelling clinical data, for example, the pregenetic test specification of a disease gene highly likely to harbor a disease-associated variant of interest, seldom impacts the clinician’s decision of whether a variant classified as a VUS by a laboratory report is actionable. More specifically, cardiovascular genetics experts have become quite sanguine, for example, at specifying the pretest likelihood of identifying a LMNA variant based upon phenotype and/or family data. However, finding a novel missense or nonsense variant in any gene, even with such a pretest specification, cannot be classified with current ACMG rules as likely pathogenic (or pathogenic), and thus actionable, unless data regarding the same variant is available from multiple probands and/or affected family members. While we propose no solution to this present conundrum, we do acknowledge its existence. Efforts to accumulate extensive catalogs of expertly adjudicated phenotype and variant information, such as the ClinGen effort,11 may eventually partially mitigate this situation.
Considerations of family management
Predictive genetic testing
Risk stratification in family members is an important and valuable reason for genetic testing. If a pathogenic or likely pathogenic variant is identified in the index patient initially tested, opportunities emerge for the predictive testing of at-risk family members. As noted above, variants of uncertain significance (VUS) are not useful to conduct predictive genetic testing.
Negative cascade genetic testing in an at-risk family member
If genetic testing is negative in an at-risk phenotype-negative family member for a pathogenic or likely pathogenic variant present in the proband, that family member’s risk of developing the cardiomyopathy is substantially reduced. In this situation, the need for serial phenotype screening after a baseline clinical evaluation in such a genotype-negative family member in most cases is unnecessary, and the family member can be discharged from serial clinical phenotype screening. However, the strength of the recommendation to release a family member from ongoing interval phenotype screening is based upon the strength of the evidence that the variant indeed is the cause of disease in the family under care. In most cases this evidence must be assembled from prior patients and families, usually in publicly accessible databases or the medical literature, and/or from evidence gathered and assessed from the family under care. The family member should be counseled that their risk has been substantially reduced, but is not reduced to zero, with the caveat that if they develop relevant symptoms, phenotype screening should be reconsidered because of the possibility that one or more yet undetected variants may be at play.
Positive cascade genetic testing in an at-risk family member
On the other hand, if a pathogenic or likely pathogenic variant is identified in an asymptomatic, at-risk phenotype-negative family member, the confidence is much greater to infer risk for that individual. They should be counseled on the presenting signs and symptoms of the specific cardiomyopathy, any associated reduced penetrance and variable expressivity, and the rationale and frequency of the recommended clinical surveillance (reviewed at recommendation 2).
Leveraging family-based segregation information to impact variant analysis
Some variants detected with cardiomyopathy genetic testing will be novel, that is, variants that have not been previously reported in publicly accessible databases, and will meet other usual criteria for pathogenicity. However, even if the variant is of the type that is known to be disease-causing and has occurred in a well-established gene associated with the cardiomyopathy phenotype in the family, such novel variants will often be adjudicated as VUSs because of lack of prior case or family data. In this circumstance, searching for segregation of the variant in question with the cardiomyopathy phenotype in additional family members can provide additional valuable information. Depending upon the size of the pedigree, the number of individuals tested, and the genetic testing results, such information may help reclassify a variant attribution from VUS to a pathogenic or benign attribution. The ClinGen initiative11 proposes to rectify this issue by aggregating all available disease-associated variants into ClinVar, a publicly accessible database utilizing a standardized curation approach tailored after the ACMG/AMP recommendations,10 and all professionals with any access to genetic data relevant to the cardiomyopathies are urged to contribute to this important database. However, because of the numbers of genes involved in the cardiomyopathies, many variants in the near term will likely be curated as VUSs. For example, in one HCM study, the cardiomyopathy with the largest disease-specific databases and where ~80% of pathogenic variants can be identified in two genes, MYPBC3 or MYH7, in one recent study 30% and 35% of variants were novel, respectively, for these two genes. In other well-established HCM genes 76% of variants were unique.93
The corollary of the above is that if the VUS does not segregate with affected family members, the likelihood that the VUS is relevant for the family phenotype is reduced. However, this analysis must encompass the growing reality of bilineal or multivariant disease, which has been postulated to be more common in DCM9,56 and ARVC.94
In most clinical situations, sequencing a VUS is not undertaken in family members who have completed clinical screening and have been shown to be free of the phenotype (negative clinical phenotype screening), as genetic information will not inform variant pathogenicity. One important exception to this is parental sequencing to confirm the possibility of de novo occurrence of a variant. A second exception to this includes sequencing older unaffected family members, who are highly informative when assessing the penetrance of a variant. Application of this principle depends greatly upon the age of onset of the phenotype in the family (infant, pediatric, early adult, late adult), the clarity and severity of the phenotype, as well as the gene involved and disease mechanisms.
Finally, as noted above, variant calls may change. The most problematic is when a previously called variant, deemed pathogenic or likely pathogenic, is downgraded to a VUS. In this circumstance, recommendations for the clinical surveillance screening of at-risk family members change. Most importantly, a genotype-negative family member must now be counseled that they remain at risk for the family phenotype, and hence need to reengage in clinical screening. The proband and any family members who tested positive for the variant, now downgraded to a VUS, must also be counseled that future genetic reevaluation may be appropriate. All clinicians participating in genetic evaluations must be aware of the implications of changes in variant calls, and the family members should be counseled regarding these possibilities during the initial genetic evaluation and the need for possible future contact. Given the seeming recent increase in downgrading to a VUS, this highly impactful change in variant status carries great potential for unintended clinical errors if not identified and communicated effectively to the relevant family unit.
Recommendation 2. Focused cardiovascular phenotyping is recommended when pathogenic or likely pathogenic variants in cardiomyopathy genes, designated for reporting of secondary findings by the ACMG, are identified in an individual.
-
(a)
If a cardiovascular phenotype is identified as would be predicted by currently available knowledge of the gene/variant pair, all usual approaches described in this document for a genetic evaluation, including family-based approaches, are recommended.
-
(b)
If no cardiovascular disease phenotype is identified in the individual, recommendations for surveillance screening at intervals should be considered.
-
(c)
If no cardiovascular phenotype is identified in the individual, cascade evaluation of at-risk relatives may be considered, tempered by the strength of evidence supporting the pathogenicity of the variant, the usual age of onset of the gene/variant pair, and pedigree information (e.g., the ages of at-risk family members, other previously known cardiovascular clinical data in the pedigree, and related information).
Across specialties genetic testing is moving toward use of large gene panels, exome sequencing, and potentially genome sequencing. These tests may be performed for a wide variety of indications and diseases that do not include a cardiac phenotype. Individuals who undergo genetic testing for a disease that does not involve the heart may have a genetic variant discovered that may predispose that individual to a cardiomyopathy. This discovery may occur in two ways. First, the gene, known to confer risk from high-penetrance variants that are medically actionable, may be intentionally analyzed as recommended by the ACMG. Variants identified from intentional analysis are termed secondary findings. Second, a variant is identified incidentally or accidentally through the analysis of genes related to the original phenotype for which the test was performed. These are termed incidental findings.
The ACMG has developed guidelines to manage secondary findings, which were first published in 20135 and updated in 2016.6 The ACMG guidance directs the reporting only of known pathogenic (KP) or expected pathogenic (EP) variants,6 the former defined as “Sequence variation is previously reported and is a recognized cause of the disorder” and the latter as “Sequence variation is previously unreported and is of the type which is expected to cause the disorder.” These definitions were taken from the ACMG 2008 guidance for variant interpretation,95 which was updated by the ACMG/AMP in 201510 with modified nomenclature of “pathogenic” (P) and “likely pathogenic” (LP). The latter attributions (P, LP) are now nearly universally used in clinical genetic testing laboratories in the United States. This nomenclature is also used in ClinGen,11,12 the ClinGen Cardiovascular Clinical Domain Working Group,96 and this document. Despite possible subtle differences of KP/EP and P/LP, because the P and LP attributions are used for the other recommendation in this document, for simplicity and parsimony these attributions will also be used in this section.
Thus, variants in the ACMG-listed cardiomyopathy genes (Table 1) that have been identified as secondary findings and adjudicated as P or LP are considered medically actionable. In those cases, cardiac phenotyping should be conducted in the individuals who carry those variants, assuming that the individual has not opted out of notification.
Greater difficulty in determining whether a variant is medically actionable may occur for incidental findings reported by the diagnostic laboratory that fall outside the ACMG guidelines. Incidental findings may be classified as pathogenic, likely pathogenic, variants of uncertain significance, likely benign, or benign, with specific criteria for the strength of assertion.10
The single most important analysis for determining if a specific incidental finding is actionable rests on the strength of evidence for disease causality of the gene/variant pair. Identifying a variant in a gene previously observed in multiple cases or families, including at times functional data confirming a damaging effect, can have substantial evidentiary strength, and such variants may be able to be classified as pathogenic or likely pathogenic. Such evidence forms the basis of the ACMG recommendations and informs sections (a), (b), and (c) of this recommendation. For HCM, where ~80% of genetic cause, when found, is within two genes (MYBPC3, MYH7), a greater likelihood exists that prior case data may be available. However, in contrast to HCM, the gene ontology for DCM is much more extensive, as most genes contribute only a small fraction to the totality of known genetic cause, and many reported variants remain private. The number of genes considered relevant for ARVC is smaller than either DCM or HCM, but because it is much less common than HCM or DCM, many ARVC variants will also remain private. Overall it is likely that most cardiomyopathy variants identified as incidental findings, even those for HCM, will remain VUSs because of lack of prior data, or lack of the requisite genetic data to assess segregation in large and well-phenotyped families with multiple affected individuals.
Item C of this recommendation suggests thoughtful and cautiously implemented, cascade clinical (phenotype) screening of putatively at-risk family members may be considered even if the clinical phenotype screening was negative in the individual (proband) who completed genetic analysis. This statement recognizes the possibility that the proband may be younger than the usual age of onset of the cardiovascular phenotype. It also recognizes the utility and necessity of gathering clinical phenotype data in an extended family to help interpret the genetic information in cascade testing if phenotypes are encountered in the family members predicted by the gene/variant pair.
We also recognize that at times a novel variant will be identified in an established, well-curated97 gene known to have other variants of high risk, and the variant will be recognized as the type that is expected to be pathogenic, but because it is novel it may be appropriately adjudicated as a VUS. In select situations within the context of expert evaluation and known limitations summing the integrated risk derived from molecular genetics and clinical knowledge of the gene/variant pair (recommendation 1), a personal and family history, pedigree analysis, and phenotyping of the individual harboring such a VUS may be considered. The rationale for this comment results directly from the significant risk of morbidity and mortality noted above that may devolve from such cardiomyopathy genes and variants. If phenotype evidence is found to support a disease association in the individual, the remainder of these recommendations would become operative, including consideration of pedigree expansion to help establish or refute the pathogenicity of the variant, and to better discern the overall risk incurred to the individual and the family.
A distinct limitation is that we are unaware of published outcomes data to support, validate, or refute the above guidance, which can only be considered as expert opinion. This emphasizes the need for well-designed rigorous studies examining outcomes of phenotyping and family studies following secondary or incidental findings of variants relevant for the cardiomyopathies.
Change history
01 May 2019
In the original version of this Article, the Statement on Conflict of Interest originally submitted by the authors was not published. Furthermore, in the Acknowledgements section we neglected to state that the article is an abbreviated version of “Genetic Evaluation of Cardiomyopathy—a Heart Failure Society of America Practice Guideline,” published in Journal of Cardiac Failure. The PDF and HTML versions of the Article have now been corrected.
References
Hershberger RE, Lindenfeld J, Mestroni L, et al. Genetic evaluation of cardiomyopathy—a Heart Failure Society of America practice guideline. J Card Fail. 2009;15:83–97.
Hershberger RE, Givertz MM, Ho CY, et al. Genetic evaluation of cardiomyopathy—a Heart Failure Society of America practice guideline. J Card Fail. 2018;24:281–302.
Mital S, Musunuru K, Garg V, et al. Enhancing literacy in cardiovascular genetics: a scientific statement from the American Heart Association. Circ Cardiovasc Genet. 2016;9:448–67.
Furqan A, Arscott P, Girolami F, et al. Care in specialized centers and data sharing increase agreement in hypertrophic cardiomyopathy genetic test interpretation. Circ Cardiovasc Genet. 2017;10:e001700. https://doi.org/10.1161/CIRCGENETICS.116.001700, Accessed 8 June 2018.
Green RC, Berg JS, Grody WW, et al. ACMG recommendations for reporting of incidental findings in clinical exome and genome sequencing. Genet Med. 2013;15:565–74.
Kalia SS, Adelman K, Bale SJ, et al. Recommendations for reporting of secondary findings in clinical exome and genome sequencing, 2016 update (ACMG SFv2.0): a policy statement of the American College of Medical Genetics and Genomics. Genet Med. 2016;19:249–55.
Yancy CW, Jessup M, Bozkurt B, et al. 2013 ACCF/AHA guideline for the management of heart failure: a report of the American College of Cardiology Foundation/American Heart Association Task Force on Practice Guidelines. Circulation. 2013;128:1810–52.
Bennette CS, Gallego CJ, Burke W, et al. The cost-effectiveness of returning incidental findings from next-generation genomic sequencing. Genet Med. 2015;17:587–95.
Hershberger RE, Hedges DJ, Morales A. Dilated cardiomyopathy: the complexity of a diverse genetic architecture. Nat Rev Cardiol. 2013;10:531–47.
Richards S, Aziz N, Bale S, et al. Standards and guidelines for the interpretation of sequence variants: a joint consensus recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Genet Med. 2015;17:405–23.
Rehm HL, Berg JS, Brooks LD, et al. ClinGen—the Clinical Genome Resource. N Engl J Med. 2015;372:2235–42.
ClinGen (Clinical Genome Resource). the NHGRI (National Human Genome Research Institute), Bethesda, MD. http://clinicalgenome.org/, Accessed date 17 May 2018.
Bradley L, Scheuner MT, Smith W, et al. American College of Medical Genetics and Genomics—protocol manual for evidence-based guideline development. American College of Medical Genetics and Genomics, Bethesda, MD, 2014.
Gersh BJ, Maron BJ, Bonow RO, et al. 2011 ACCF/AHA guideline for the diagnosis and treatment of hypertrophic cardiomyopathy: a report of the American College of Cardiology Foundation/American Heart Association Task Force on Practice Guidelines. Circulation. 2011;124:e783–831.
Cirino AL, Ho C. Familial hypertrophic cardiomyopathy overview. In: Pagon RA, Bird TD, Dolan CR, Stephens K, editors. GeneReviews. University of Washington: Seattle, WA; 1993.
Burkett EL, Hershberger RE. Clinical and genetic issues in familial dilated cardiomyopathy. J Am Coll Cardiol. 2005;45:969–81.
Hershberger RE, Morales A, Siegfried JD. Clinical and genetic issues in dilated cardiomyopathy: a review for genetics professionals. Genet Med. 2010;12:655–67.
Corrado D, Wichter T, Link MS, et al. Treatment of arrhythmogenic right ventricular cardiomyopathy/dysplasia: an International Task Force consensus statement. Circulation. 2015;132:441–53.
Iyer VR, Chin AJ. Arrhythmogenic right ventricular cardiomyopathy/dysplasia (ARVC/D). Am J Med Genet C Semin Med Genet. 2013;163C:185–97.
Dunn KE, Ashley EA. Arrhythmogenic Right Ventricular Cardiomyopathy: Toward a Modern Clinical and Genomic Understanding. Circ Cardiovasc Genet. 2015;8:421–4.
Groeneweg JA, Bhonsale A, James CA, et al. Clinical presentation, long-term follow-up, and outcomes of 1001 arrhythmogenic right ventricular dysplasia/cardiomyopathy patients and family members. Circ Cardiovasc Genet. 2015;8:437–46.
Miller EM, Wang Y, Ware SM. Uptake of cardiac screening and genetic testing among hypertrophic and dilated cardiomyopathy families. J Genet Couns. 2013;22:258–67.
The SOLVD Investigators. Effect of enalapril on mortality and the development of heart failure in asymptomatic patients with reduced left ventricular ejection fractions. N Engl J Med. 1992;327:685–91.
Colucci WS, Kolias TJ, Adams KF, et al. Metoprolol reverses left ventricular remodeling in patients with asymptomatic systolic dysfunction: the REversal of VEntricular Remodeling with Toprol-XL (REVERT) trial. Circulation. 2007;116:49–56.
Ho CY, Lakdawala NK, Cirino AL, et al. Diltiazem treatment for pre-clinical hypertrophic cardiomyopathy sarcomere mutation carriers: a pilot randomized trial to modify disease expression. JACC Heart Fail. 2015;3:180–8.
Ingles J, McGaughran J, Scuffham PA, et al. A cost-effectiveness model of genetic testing for the evaluation of families with hypertrophic cardiomyopathy. Heart. 2012;98:625–30.
Alfares AA, Kelly MA, McDermott G, et al. Results of clinical genetic testing of 2,912 probands with hypertrophic cardiomyopathy: expanded panels offer limited additional sensitivity. Genet Med. 2015;17:880–8.
Aggarwal V, Dobrolet N, Fishberger S, et al. PRKAG2 mutation: an easily missed cardiac specific non-lysosomal glycogenosis. Ann Pediatr Cardiol. 2015;8:153–6.
Thevenon J, Laurent G, Ader F, et al. High prevalence of arrhythmic and myocardial complications in patients with cardiac glycogenosis due to PRKAG2 mutations. Europace. 2017;19:651–9.
van Velzen HG, Vriesendorp PA, Oldenburg RA, et al. Value of genetic testing for the prediction of long-term outcome in patients with hypertrophic cardiomyopathy. Am J Cardiol. 2016;118:881–7.
Li Q, Gruner C, Chan RH, et al. Genotype-positive status in patients with hypertrophic cardiomyopathy is associated with higher rates of heart failure events. Circ Cardiovasc Genet. 2014;7:416–22.
van Rijsingen IA, van der Zwaag PA, Groeneweg JA et al. Outcome in phospholamban R14del carriers: results of a large multicentre cohort study. Circ Cardiovasc Genet. 2014;7:455–65.
Ortiz-Genga MF, Cuenca S, Dal Ferro M, et al. Truncating FLNC mutations are associated with high-risk dilated and arrhythmogenic cardiomyopathies. J Am Coll Cardiol. 2016;68:2440–51.
van Rijsingen IA, Arbustini E, Elliott PM, et al. Risk factors for malignant ventricular arrhythmias in lamin a/c mutation carriers a European cohort study. J Am Coll Cardiol. 2012;59:493–500.
Kayvanpour E, Sedaghat-Hamedani F, Amr A, et al. Genotype-phenotype associations in dilated cardiomyopathy: meta-analysis on more than 8000 individuals. Clin Res Cardiol. 2017;106:127–39.
Meune C, Van Berlo JH, Anselme F, et al. Primary prevention of sudden death in patients with lamin A/C gene mutations. N Engl J Med. 2006;354:209–10.
Hodgkinson KA, Howes AJ, Boland P, et al. Long-term clinical outcome of arrhythmogenic right ventricular cardiomyopathy in individuals with a p.S358L mutation in TMEM43 following implantable cardioverter defibrillator therapy. Circ Arrhythm Electrophysiol. 2016;9.
Wilson KD, Shen P, Fung E, et al. A rapid, high-quality, cost-effective, comprehensive and expandable targeted next-generation sequencing assay for inherited heart diseases. Circ Res. 2015;117:603–11.
Ceyhan-Birsoy O, Pugh TJ, Bowser MJ, et al. Next generation sequencing-based copy number analysis reveals low prevalence of deletions and duplications in 46 genes associated with genetic cardiomyopathies. Mol Genet Genom Med. 2016;4:143–51.
Golbus JR, Puckelwartz MJ, Fahrenbach JP, et al. Population-based variation in cardiomyopathy genes. Circ Cardiovasc Genet. 2012;5:391–9.
Herman DS, Lam L, Taylor MR, et al. Truncations of titin causing dilated cardiomyopathy. N Engl J Med. 2012;366:619–28.
Burke MA, Cook SA, Seidman JG, et al. Clinical and mechanistic insights into the genetics of cardiomyopathy. J Am Coll Cardiol. 2016;68:2871–86.
Walsh R, Thomson KL, Ware JS, et al. Reassessment of Mendelian gene pathogenicity using 7,855 cardiomyopathy cases and 60,706 reference samples. Genet Med. 2016;19:192–203.
Ingles J, Semsarian C. Conveying a probabilistic genetic test result to families with an inherited heart disease. Heart Rhythm. 2014;11:1073–8.
Ingles J, Zodgekar PR, Yeates L, et al. Guidelines for genetic testing of inherited cardiac disorders. Heart Lung Circ. 2011;20:681–7.
Arad M, Maron BJ, Gorham JM, et al. Glycogen storage diseases presenting as hypertrophic cardiomyopathy. N Engl J Med. 2005;352:362–72.
Long PA, Evans JM, Olson TM. Exome sequencing establishes diagnosis of Alstrom syndrome in an infant presenting with non-syndromic dilated cardiomyopathy. Am J Med Genet A. 2015;167A:886–90.
Morita H, Rehm HL, Menesses A, et al. Shared genetic causes of cardiac hypertrophy in children and adults. N Engl J Med. 2008;358:1899–908.
Ingles J, Sarina T, Yeates L, et al. Clinical predictors of genetic testing outcomes in hypertrophic cardiomyopathy. Genet Med. 2013;15:972–7.
Pan S, Caleshu CA, Dunn KE, et al. Cardiac structural and sarcomere genes associated with cardiomyopathy exhibit marked intolerance of genetic variation. Circ Cardiovasc Genet. 2012;5:602–10.
Teekakirikul P, Kelly MA, Rehm HL, et al. Inherited cardiomyopathies: molecular genetics and clinical genetic testing in the postgenomic era. J Mol Diagn. 2013;15:158–70.
Walsh R, Buchan R, Wilk A, et al. Defining the genetic architecture of hypertrophic cardiomyopathy: re-evaluating the role of non-sarcomeric genes. Eur Heart J. 2017;38:3461–8.
Cox GF, Sleeper LA, Lowe AM, et al. Factors associated with establishing a causal diagnosis for children with cardiomyopathy. Pediatrics. 2006;118:1519–31.
Kindel SJ, Miller EM, Gupta R, et al. Pediatric cardiomyopathy: importance of genetic and metabolic evaluation. J Card Fail. 2012;18:396–403.
Ware SM. Evaluation of genetic causes of cardiomyopathy in childhood. Cardiol Young. 2015;25(suppl 2):43–50.
Haas J, Frese KS, Peil B, et al. Atlas of the clinical genetics of human dilated cardiomyopathy. Eur Heart J. 2015;36:1123–35a.
Lakdawala NK, Funke BH, Baxter S, et al. Genetic testing for dilated cardiomyopathy in clinical practice. J Card Fail. 2012;18:296–303.
Pugh TJ, Kelly MA, Gowrisankar S, et al. The landscape of genetic variation in dilated cardiomyopathy as surveyed by clinical DNA sequencing. Genet Med. 2014;16:601–8.
Morales A, Hershberger RE. The rationale and timing of molecular genetic testing for dilated cardiomyopathy. Can J Cardiol. 2015;31:1309–12.
Roberts AM, Ware JS, Herman DS, et al. Integrated allelic, transcriptional, and phenomic dissection of the cardiac effects of titin truncations in health and disease. Sci Transl Med. 2015;7:270ra6.
Schafer S, de Marvao A, Adami E, et al. Titin-truncating variants affect heart function in disease cohorts and the general population. Nat Genet. 2017;49:46–53.
Norton N, Li D, Rampersaud E, et al. Exome sequencing and genome-wide linkage analysis in 17 families illustrates the complex contribution of TTN truncating variants to dilated cardiomyopathy. Circ Cardiovasc Genet. 2013;6:144–53.
Arbustini E, Pilotto A, Repetto A, et al. Autosomal dominant dilated cardiomyopathy with atrioventricular block: a lamin A/C defect-related disease. J Am Coll Cardiol. 2002;39:981–90.
Parks SB, Kushner JD, Nauman D, et al. Lamin A/C mutation analysis in a cohort of 324 unrelated patients with idiopathic or familial dilated cardiomyopathy. Am Heart J. 2008;156:161–9.
Norton N, Li D, Reider MJ, et al. Genome-wide studies of copy number variation and exome sequencing identify rare variants in BAG3 as a cause of dilated cardiomyopathy. Am J Hum Genet. 2011;88:273–82.
Brauch KM, Karst ML, Herron KJ, et al. Mutations in ribonucleic acid binding protein gene cause familial dilated cardiomyopathy. J Am Coll Cardiol. 2009;54:930–41.
McCaffrey T, Guglieri M, Murphy AP, et al. Cardiac involvement in female carriers of Duchenne or Becker muscular dystrophy. Muscle Nerve. 2016;55:810–8.
Awad MM, Calkins H, Judge DP. Mechanisms of disease: molecular genetics of arrhythmogenic right ventricular dysplasia/cardiomyopathy. Nat Clin Pract Cardiovasc Med. 2008;5:258–67.
Quarta G, Muir A, Pantazis A, et al. Familial evaluation in arrhythmogenic right ventricular cardiomyopathy: impact of genetics and revised task force criteria. Circulation. 2011;123:2701–9.
Saffitz JE. Arrhythmogenic cardiomyopathy: advances in diagnosis and disease pathogenesis. Circulation. 2011;124:e390–2.
Elliott P, O’Mahony C, Syrris P, et al. Prevalence of desmosomal protein gene mutations in patients with dilated cardiomyopathy. Circ Cardiovasc Genet. 2010;3:314–22.
James CA, Bhonsale A, Tichnell C, et al. Exercise increases age-related penetrance and arrhythmic risk in arrhythmogenic right ventricular dysplasia/cardiomyopathy-associated desmosomal mutation carriers. J Am Coll Cardiol. 2013;62:1290–7.
Gallego-Delgado M, Delgado JF, Brossa-Loidi V, et al. Idiopathic restrictive cardiomyopathy is primarily a genetic disease. J Am Coll Cardiol. 2016;67:3021–3.
Rapezzi C, Lorenzini M, Longhi S, et al. Cardiac amyloidosis: the great pretender. Heart Fail Rev. 2015;20:117–24.
Ton VK, Mukherjee M, Judge DP. Transthyretin cardiac amyloidosis: pathogenesis, treatments, and emerging role in heart failure with preserved ejection fraction. Clin Med Insights Cardiol. 2014;8:39–44.
Buxbaum JN, Ruberg FL. Transthyretin V122I (pV142I)* cardiac amyloidosis: an age-dependent autosomal dominant cardiomyopathy too common to be overlooked as a cause of significant heart disease in elderly African Americans. Genet Med. 2017;19:733–42.
Maurer MS, Elliott P, Comenzo R, et al. Addressing common questions encountered in the diagnosis and management of cardiac amyloidosis. Circulation. 2017;135:1357–77.
Fleming RE, Ponka P. Iron overload in human disease. N Engl J Med. 2012;366:348–59.
Arbustini E, Weidemann F, Hall JL. Left ventricular noncompaction: a distinct cardiomyopathy or a trait shared by different cardiac diseases? J Am Coll Cardiol. 2014;64:1840–50.
Probst S, Oechslin E, Schuler P, et al. Sarcomere gene mutations in isolated left ventricular noncompaction cardiomyopathy do not predict clinical phenotype. Circ Cardiovasc Genet. 2011;4:367–74.
Miller EM, Hinton RB, Czosek R, et al. Genetic testing in pediatric left ventricular noncompaction. Circ Cardiovasc Genet. 2017;10:e001735.
Morales A, Painter T, Li R, et al. Rare variant mutations in pregnancy-associated or peripartum cardiomyopathy. Circulation. 2010;121:2176–82.
van Spaendonck-Zwarts KY, van Tintelen JP, van Veldhuisen DJ, et al. Peripartum cardiomyopathy as a part of familial dilated cardiomyopathy. Circulation. 2010;121:2169–75.
Ware JS, Li J, Mazaika E, et al. Shared genetic predisposition in peripartum and dilated cardiomyopathies. N Engl J Med. 2016;374:233–41.
Cann F, Corbett M, O’Sullivan D, et al. Phenotype-driven molecular autopsy for sudden cardiac death. Clin Genet. 2017;91:22–29.
Semsarian C, Ingles J, Wilde AA. Sudden cardiac death in the young: the molecular autopsy and a practical approach to surviving relatives. Eur Heart J. 2015;36:1290–6.
Tsuchiya KD, Shaffer LG, Aradhya S, et al. Variability in interpreting and reporting copy number changes detected by array-based technology in clinical laboratories. Genet Med. 2009;11:866–73.
Van Driest SL, Wells QS, Stallings S, et al. Association of arrhythmia-related genetic variants with phenotypes documented in electronic medical records. JAMA. 2016;315:47–57.
Xu T, Yang Z, Vatta M, et al. Compound and digenic heterozygosity contributes to arrhythmogenic right ventricular cardiomyopathy. J Am Coll Cardiol. 2010;55:587–97.
Van Driest SL, Vasile VC, Ommen SR, et al. Myosin binding protein C mutations and compound heterozygosity in hypertrophic cardiomyopathy. J Am Coll Cardiol. 2004;44:1903–10.
Richard P, Charron P, Carrier L, et al. Hypertrophic cardiomyopathy: distribution of disease genes, spectrum of mutations, and implications for a molecular diagnosis strategy. Circulation. 2003;107:2227–32.
Ingles J, Doolan A, Chiu C, et al. Compound and double mutations in patients with hypertrophic cardiomyopathy: implications for genetic testing and counselling. J Med Genet. 2005;42:e59.
Ross SB, Bagnall RD, Ingles J, et al. Burden of Recurrent and Ancestral Mutations in Families With Hypertrophic Cardiomyopathy. Circ Cardiovasc Genet. 2017;10.
Bauce B, Nava A, Beffagna G, et al. Multiple mutations in desmosomal proteins encoding genes in arrhythmogenic right ventricular cardiomyopathy/dysplasia. Heart Rhythm. 2010;7:22–9.
Richards CS, Bale S, Bellissimo DB, et al. ACMG recommendations for standards for interpretation and reporting of sequence variations: revisions 2007. Genet Med. 2008;10:294–300.
ClinGen Cardiovascular Domain Working Group. https://www.clinicalgenome.org/working-groups/clinical-domain/sub-groups/cardiovascular/, Accessed 8 June 2018.
Strande NT, Riggs ER, Buchanan AH, et al. Evaluating the clinical validity of gene-disease associations: an evidence-based framework developed by the Clinical Genome Resource. Am J Hum Genet. 2017;100:895–906.
Acknowledgements
We acknowledge the assistance of Natalia Leach as the liaison person for the PP&G; also David Miller, Natalia Leach, and Ed Esplin for detailed review of the draft manuscript. This article is an abbreviated version of “Genetic Evaluation of Cardiomyopathy—a Heart Failure Society of America Practice Guideline,” published in Journal of Cardiac Failure, and on which the American College of Medical Genetics and Genomics and the Heart Failure Society of America collaborated.
Author information
Authors and Affiliations
Consortia
Corresponding author
Ethics declarations
Disclosure
R.E.H. has served as a consultant for Array Biopharma. C.Y.H. has served as a consultant and has received research support from MyoKardia. D.P.J. has served as a consultant for Array Biopharma, Eidos Therapeutics, Glaxo Smith Kline, Invitae, MyoKardia, and Pfizer. M.R.G.T. has served as a consultant for Array Biopharma, Guidepoint Global, and Wellpoint, Inc., and has served as a speaker for GeneDx. M.V. is employed by Invitae Corporation. The other authors declare no conflicts of interest.
Additional information
Disclaimer
This practice resource is designed primarily as an educational resource for medical geneticists and other clinicians to help them provide quality medical services. Adherence to this practice resource is completely voluntary and does not necessarily assure a successful medical outcome. This practice resource should not be considered inclusive of all proper procedures and tests or exclusive of other procedures and tests that are reasonably directed to obtaining the same results. In determining the propriety of any specific procedure or test, the clinician should apply his or her own professional judgment to the specific clinical circumstances presented by the individual patient or specimen. Clinicians are encouraged to document the reasons for the use of a particular procedure or test, whether or not it is in conformance with this practice resource.
Clinicians also are advised to take notice of the date this practice resource was adopted, and to consider other medical and scientific information that becomes available after that date. It also would be prudent to consider whether intellectual property interests may restrict the performance of certain tests and other procedures.
Electronic supplementary material
Rights and permissions
About this article
Cite this article
Hershberger, R.E., Givertz, M., Ho, C.Y. et al. Genetic evaluation of cardiomyopathy: a clinical practice resource of the American College of Medical Genetics and Genomics (ACMG). Genet Med 20, 899–909 (2018). https://doi.org/10.1038/s41436-018-0039-z
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41436-018-0039-z
Keywords
This article is cited by
-
The outcome of genetic and non-genetic pediatric cardiomyopathies
The Egyptian Heart Journal (2024)
-
Actionability and familial uptake following opportunistic genomic screening in a pediatric cancer cohort
European Journal of Human Genetics (2024)
-
Mechano-energetic efficiency in patients with hypertrophic cardiomyopathy with and without sarcomeric mutations
Journal of Cardiovascular Translational Research (2024)
-
Genetic heterogeneity of cardiomyopathy and its correlation with patient care
BMC Medical Genomics (2023)
-
Cardiac splicing as a diagnostic and therapeutic target
Nature Reviews Cardiology (2023)