Abstract
Microduplications involving the MYT1L gene have mostly been described in series of patients with isolated schizophrenia. However, few reports have been published, and the phenotype has still not been well characterized. We sought to further characterize the phenotypic spectrum of this condition by describing the clinical features of patients with a pure 2p25.3 microduplication that includes all or part of MYT1L. We assessed 16 new patients with pure 2p25.3 microduplications recruited through a French national collaboration (n = 15) and the DECIPHER database (n = 1). We also reviewed 27 patients reported in the literature. For each case, we recorded clinical data, the microduplication size, and the inheritance pattern. The clinical features were variable and included developmental and speech delays (33%), autism spectrum disorder (ASD, 23%), mild-to-moderate intellectual disability (ID, 21%), schizophrenia (23%), or behavioral disorders (16%). Eleven patients did not have an obvious neuropsychiatric disorder. The microduplications ranged from 62.4 kb to 3.8 Mb in size and led to duplication of all or part of MYT1L; seven of these duplications were intragenic. The inheritance pattern was available for 18 patients: the microduplication was inherited in 13 cases, and all parents but one had normal phenotype. Our comprehensive review and expansion of the phenotypic spectrum associated with 2p25.3 microduplications involving MYT1L should help clinicians to better assess, counsel and manage affected individuals. MYT1L microduplications are characterized by a spectrum of neuropsychiatric phenotypes with incomplete penetrance and variable expressivity, which are probably due to as-yet unknown genetic and nongenetic modifiers.
Similar content being viewed by others
Introduction
Microdeletions and single nucleotide variants (SNVs) involving the human MYT1L gene (coding for myelin transcription factor 1-like (MYT1L) and located on chromosome 2 at 2p25.3) have recently been linked to a syndromic presentation (OMIM#616521) consisting of developmental delay (DD), intellectual disability (ID), overweight/obesity, and several dysmorphic features [1, 2]. A better understanding of the mechanism underlying this disorder has prompted researchers to consider that MYT1L haploinsufficiency is responsible for the observed clinical phenotype [2,3,4].
MYT1L is a member of the neural-specific myelin transcription factor 1 (MYT1) family, which is characterized by the presence of two highly conserved clusters of C2HC zinc fingers [5]. In the mouse, the Myt1l transcription factor helps to determine neuronal fate by specifically repressing the expression of non-neuronal genes and negative regulators of neurogenesis (including members of the Notch signaling pathway, such as Hes1) [6, 7]. This role in promoting neuronal differentiation has been proven in vitro by showing that in combination with other transcription factors (Ascl1 and Pou3f2/Brn2), Myt1l can reprogram human and mouse fibroblasts into functional neurons, [8, 9].
Although the phenotype associated with 2p25.3 microdeletions and SNVs varies markedly from one patient to another, some clinical features have been clearly established [2]. In contrast, the published clinical data on individuals with microduplications are often limited. Partial microduplications encompassing MYT1L were first linked to schizophrenia [10] and have also been observed in individuals presenting neuropsychiatric diseases, such as ID and autism spectrum disorder (ASD) [4, 11,12,13,14,15,16,17,18,19,20,21]. A recent study therefore concluded that MYT1L might act as a dosage-sensitive gene involved in neurodevelopmental pathways common to both ID and schizophrenia [22].
Predicting the clinical outcomes of a 2p25.3 microduplication is very challenging - especially in a prenatal diagnostic setting. The high observed phenotypic variability underscores the need to systematically characterize the clinical impact of these rearrangements in large numbers of carriers. To further characterize this phenotypic spectrum, we compared the 16 new patients with 27 previously reported individuals, all of whom presented 2p25.3 microduplications involving all or part of MYT1L.
Material and methods
Members of the Association des Cytogénéticiens de Langue Française (www.eaclf.org) and the AchroPuce networks (https://acpa-achropuce.com) were requested to provide all their clinical and genetic data (including age, phenotype, reason for referral, upstream and downstream breakpoints, and the inheritance pattern) on individuals with a 2p25.3 microduplication involving MYT1L. Through this call for collaboration, 15 new patients diagnosed between 2019 and 2022 (N1 to N15) were recruited. The microduplications were identified with microarrays that differed in their format, resolution, and manufacturer: a Human Genome CGH Microarray 60 K, 150 K, or 180 K (Agilent Technologies, Santa Clara, CA, USA) or an Illumina OmniExpress SNP microarray (Illumina Inc, San Diego, CA, USA).
Microduplications were confirmed by fluorescence in situ hybridization or qPCR. Genomic positions were relative to human genome GRCH37/hg19 Assembly. For all cases, parental blood samples were requested to determine the inheritance pattern. Parental analyses were performed using fluorescence in situ hybridization or chromosomal microarray analysis (CMA). Genetic counseling was offered to the patients and affected families. Prior to inclusion, all the patients provided their written, informed consent to participation in the study.
Additional new cases of 2p25.3 microduplications were retrieved from the Database of Chromosomal Imbalance and Phenotype in Humans using Ensembl Resources (DECIPHER: https://www.deciphergenomics.org/). An e-mail was sent to all the laboratories referenced in DECIPHER, requesting the patients’ details and authorization to use this information. Only one patient (D1, from whom consent was obtained) could be included in the present study. We also searched the PubMed online database (https://pubmed.ncbi.nlm.nih.gov) with the search terms “2p25.3 duplication” and “MYT1L duplication” and identified 27 previously published cases (P1–P27).
All novel variants were submitted to public databases. The variants reported in patients N1-N5 were submitted to ClinVar (http://www.ncbi.nlm.nih.gov/clinvar/), and those reported in patients N6-N15 were submitted to DECIPHER. Statistical analyses were performed using the online tool BiostaTGV (https://biostatgv.sentiweb.fr/). Proportions were compared using the Fisher-Freeman-Halton Exact test. All tests were two-tailed, and the threshold for statistical significance was set to p < 0.05.
Results
A total of 16 new cases were recruited into the study, and 27 cases reported in the literature were reviewed. All harbored pure interstitial microduplications overlapping the 2p25.3 region and involving the MYT1L gene. In the new patients (N1 to N15, and D1), the male:female ratio was 0.8 (7/9) and the age ranged from 6 months to 13 years (mean: 7 years). In the overall cohort, the male:female ratio was 1.6 (20/12) and the mean (range) age at diagnosis (n = 28) was 14 years (6 months to 72 years). The microduplication was diagnosed after the age of 3 years for 22 of the 28 (78%) patients. Of the 16 newly reported patients, 14 were unrelated and two (N9 and N10) were twins. The microduplication was identified during the prenatal period in two cases (N11 and N15).
CMA results
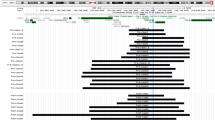
The patients’ tracks are represented in Fig. 1, and the CMA results are described in Table 1. The microduplications were located within the region extending from 859,616 bp to 2,546,048 bp (hg19/GRCh37). The microduplication size ranged from 62.4 Kb to 3.8 Mb, with a mean of 330 Kb. The breakpoints involving the 2p25.3 region were highly variable. Only one large duplication encompassed the entire MYT1L gene and ten adjacent genes. In 17 of the 43 cases (39%), the duplications involved only MYT1L. Seven of these 17 were intragenic, with both of the breakpoints located within the MYT1L gene. The other ten microduplications overlapped partially with the 5’ or 3’ end of MYT1L. In 20 of the 43 cases (46%), the microduplications involved both the MYT1L and PXDN genes. In the largest microduplications (5 out 43, 12%), additional genes were involved (those coding for thyroid peroxidase (TPO) and syntrophin gamma 2 (SNTG2)).
The genes present within this region are annotated and shown at the top. The duplications are represented by horizontal bars labeled according to the associated clinical phenotype. The figure was generated using the UCSC Genome Browser (https://genome.ucsc.edu/).
Inheritance
Data on the inheritance pattern were available for 6 of the 16 new patients. One microduplication was de novo, four were paternally inherited, and one was maternally inherited. All parents with available clinical data (n = 4) were phenotypically normal. Data on the inheritance pattern were available for 12 of the 27 patients in the literature cohort. When considering the 18 patients in the overall cohort with a microduplication and inheritance data, the latter was de novo in five cases (28%), paternally inherited in seven cases, and maternally inherited for the six remaining cases. In two previously reported patients, the microduplication had been transmitted by a healthy mother via germline mosaicism. Clinical data on the parents were available for 11 of the 13 cases with inherited microduplications. In one family, P8 and P9 had inherited the microduplications from their father (P7), who presented with ID, DD, obesity, and behavioral disorders. Lastly, the microduplications were inherited from clinically healthy parents in the remaining nine cases (82%).
Clinical features of the patients
The clinical phenotypes are summarized in Table 2. For the two prenatally diagnosed cases, an invasive procedure was triggered by elevated levels of first trimester maternal serum markers and increased nuchal translucency. For the other (postnatally diagnosed) patients, a routine ultrasound assessment in pregnancy did not yield any specific findings, other than intra-uterine growth restriction for N14.
Six of the 16 newly reported patients (37%) were referred for DD and/or ID. Other prominent clinical findings were ASD in 4 of the 16 cases (25%), growth delay in twins (12%), and congenital malformations in two cases (12%).
When combining our cohort with previously published individuals, the three most common features observed were (i) DD in 14 of the 43 cases (33%), with a significant speech delay and language impairment, (ii) mild-to-moderate ID in 9 of the 43 cases (21%), and (iii) schizophrenia in 10 of the 43 cases (23%), which was the most common feature in patients aged 18 or over. Seven of the 43 cases (16%) were diagnosed with or exhibited features consistent with ASD (including stereotypical movements) in the absence of ID. Three of the 43 cases (7%) had a combination of ASD and ID. Behavioral disorders (including attention deficit and hyperactivity disorder, aggressivity, anxiety, and mood disorders) were reported in 7 of the 43 cases (16%). Other frequently reported features were obesity in 6 of the 43 cases (14%) and microcephaly in 4 (9%). The other features concerned one or two patients only: for details, see Table 2. A neuropsychiatric phenotype was absent in 11 patients (25%). Although most patients had some dysmorphic features (including hypertelorism, up-slanting or down-slanting palpebral fissures, strabismus, bulbous nasal tip, large mouth with downturned oral commissures, full cheeks, and abnormally hemmed ears), none appeared to be specific or distinctive.
Genotype–phenotype correlations
As reported in the literature, microduplications associated to schizophrenia covered a broad interval from intron 1 of PXDN through to intron 21 of MYT1L (chr2:1,737,430–1,836,902, hg19; Fig. 1). The same pattern was observed for microduplications associated with isolated ASD; it covered the same region (chr2:1,742,043–1,834,000, hg19) and disrupted the 3′end of MYT1L (Fig. 1). In order to provide additional insights into genotype–phenotype correlations, we analyzed each microduplication’s composition and position within MYT1L (Table 3). The partial duplication of P4 was not included in this comparison because we did not know which end of MYT1L had been affected. Although microduplications occur throughout the MYT1L gene, a significant correlation between the clinical phenotype and the genomic position was found (Fisher-Freeman-Halton Exact test, p = 0.003). In 26 of the 42 cases (62%), the microduplication was multigenic and thus extended to neighboring genes. Clustering within the 3′ end region of MYT1L was observed in 22 of the 42 cases (52%). In particular, schizophrenia was reported solely in patients with multigenic duplications. Whilst microduplications in probands with isolated ASD involved the MYT1L and PXDN genes, those associated with both ASD and ID tended to be intragenic. In contrast, the 11 microduplications associated with ID and/or DD were randomly distributed throughout the region of interest (chr2:1,617,873–2,546,048, hg19), although six of them (55%) involved only the 5′ end of MYT1L.
Discussion
2p25.3 microduplications are rare, and only 27 cases had been reported previously [16]. Here, we presented clinical and molecular data on a new cohort of 16 individuals harboring a microduplication in the 2p25.3 region, including a patient from the DECIPHER database [23] whose clinical features had not previously been reported. Hence, we were able to compare the new patients’ data with those previously reported in the literature (Tables 1 and 2). To the best of our knowledge, the present new series is the largest yet reported.
The 27 previously published cases were reported to have a clinical phenotype consisting of schizophrenia (37%), ID (18%) and ASD (22%), although 26% of the cases did not have a marked neuropsychiatric disorder [16]. The clinical description of our new cases further expands the phenotypic spectrum associated with 2p25.3 microduplications. The resulting data highlighted a more variable phenotypic outcome. The most frequent clinical features in the 16 newly reported patients were DD and/or ID (37%). Remarkably, developmental and language delays were more common in our new cohort (37%) than in previously reported patients (18%); this highlights the high prevalence of microduplications in the DD/ID population. Some of our clinical findings were consistent with the literature data. For instance, the frequency of ASD in our new cohort was 25%, which supports the putative association between MYT1L microduplications and autism. Our results are in line with previous observations in which psychiatric and neurodevelopmental disorders constituted the main phenotype [4, 10, 16, 21, 24]; however, the clinical feature most frequently reported in the literature (schizophrenia) was not observed in any of the 16 new cases [16]. This discrepancy might be due to differences in the various studies’ inclusion criteria. Most of the previous studies focusing on schizophrenia [10, 14, 19,20,21], which explains why an association between microduplications involving MYT1L and schizophrenia (odds ratio: 15.7; p = 0.001) was identified in large case-control studies of patients of all ages. This association is reportedly stronger for childhood-onset schizophrenia than for adult-onset schizophrenia (odds ratio: 16.6; p = 0.01) [10].
When considering the nine MYT1L-including 2p25.3 microduplications that were inherited from a healthy parent, one can hypothesize that the penetrance is variable. Furthermore, the highly heterogeneous phenotype is suggestive of variable expressivity. This might be due to differences in the breakpoints and duplication sizes, although these genetic features did not appear to be correlated with the clinical severity. For example, patients P17 and P14 (harboring duplications of 110 kb and 3.8 Mb, respectively) had much the same phenotype.
The microduplication size ranged from 62.4 kb to 3.8 Mb, and involved all or part of the critical MYT1L gene [4, 22]. Haplo-insufficiency of MYT1L is not tolerated in humans [25], and the probability of loss of function intolerance score is 1. To date, disease-causing variants in MYT1L have been reported in 62 patients; the phenotype is more homogeneous but the clinical presentations resemble those seen in cases of microduplication syndrome [1]. The most frequent clinical features are behavioral disorders (seen in 98% of cases), DD with language delay (95%), ID (70%), overweight or obesity (58%), and epilepsy (23%) [1]. Furthermore, gene expression profiling of a MYT1L knockout cell line has shown that MYT1L haploinsufficiency can disrupt the expression of critical genes during brain development – suggesting that MYT1L regulates a network of genes involved in the etiology of neurodevelopmental disorders [25]. Chen et al. (2021) generated a Myt1l haploinsufficiency mouse model that mimicked common clinical phenotypes associated with loss-of-function mutations in human MYT1L or 2p25.3 deletions, including obesity, white-matter thinning, microcephaly, hyperactivity, muscle weakness, and social alterations [3].
The phenotypic variability might be explained by a “two-hit” model. The second hit might be another copy number variation, a single-base-pair mutation disrupting a functionally related gene, or even an environmental factor that affects the phenotype. This second hit might be responsible for the development of more severe neurological phenotypes, such as ID/DD, ASD, and schizophrenia [26, 27].
Lastly, one can hypothesize that the phenotype is due to duplication of another gene. However, when considering the 43 patients described in our study and in the literature, duplications in candidate genes that could be responsible for specific clinical findings in this syndrome were found in only 26 cases. Interestingly, 20 of the 43 duplications (particularly those associated with schizophrenia) affected MYT1L and PXDN. This prompts us to wonder whether PXDN is involved in schizophrenia susceptibility. PXDN encodes an extracellular, matrix-associated peroxidase thought to participate in peroxide-driven oxidations, phagocytosis, and immune defense. Many studies have shown that oxidative stress is part of the disease mechanism of schizophrenia [28]. However, there are no literature data on the potential involvement of PXDN in schizophrenia or other neuropsychological features. Some microduplications (n = 6) included additional proximal genes (TPO and SNTG2). Thyroid peroxidase is a key enzyme in thyroid hormone biosynthesis and was reportedly associated with ASD in a study of the contribution of immune-related genes to the pathogenesis of this disease [29]. Syntrophin gamma 2 is a scaffolding protein that interacts with the neuroligins. It has been suggested that SNTG2 is involved in neurodevelopmental disorders, as a partial SNTG2 deletion was identified in a patient with autistic features [30]. However, there were no data on the neuropsychiatric phenotype or the exact age at diagnosis for one of the four patients with microduplications including SNTG2 (P22).
Regarding the mechanism underlying the duplication and given the absence of recurrent breakpoints or flanking segmental duplications, non-allelic homologous recombination can be ruled out [10]. Hence, the involvement of other underlying mechanisms (such as non-homologous end joining, fork stalling and template switching, and microhomology-mediated break-induced replication) could be postulated.
Lastly, our findings reveal that MYT1L duplications are associated with variable, unpredictable phenotypic outcomes. Microduplications are often inherited from a phenotypically normal parent, making it difficult to establish a direct pathogenic effect. In this context, genetic counseling and a clinical prognosis are major challenges for clinicians. The enrichment of this microduplication in patients with neurodevelopmental disabilities and the more frequent occurrence of a second copy number variation in affected carriers suggest that 2p25.3 microduplications involving MYT1L can act as a susceptibility/risk locus for neurodevelopmental impairments.
Data availability
The datasets generated for the current study are included in this article, further inquiries can be directed to the corresponding authors. The variants reported in patients N1–N5 and described in this study have been submitted to the ClinVar repository (SUB12521627, accession numbers SCV002818537-SCV002818541). All the other newly reported variants have been submitted to the DECIPHER database.
References
Coursimault J, Guerrot AM, Morrow MM, Schramm C, Zamora FM, Shanmugham A, et al. MYT1L-associated neurodevelopmental disorder: description of 40 new cases and literature review of clinical and molecular aspects. Hum Genet. 2022;141:65–80.
Windheuser IC, Becker J, Cremer K, Hundertmark H, Yates LM, Mangold E, et al. Nine newly identified individuals refine the phenotype associated with MYT1L mutations. Am J Med Genet A. 2020;182:1021–31.
Chen J, Lambo ME, Ge X, Dearborn JT, Liu Y, McCullough KB, et al. A MYT1L syndrome mouse model recapitulates patient phenotypes and reveals altered brain development due to disrupted neuronal maturation. Neuron. 2021;109:3775–3792.e14.
De Rocker N, Vergult S, Koolen D, Jacobs E, Hoischen A, Zeesman S, et al. Refinement of the critical 2p25.3 deletion region: the role of MYT1L in intellectual disability and obesity. Genet Med Off J Am Coll Med Genet. 2015;17:460–6.
Manukyan A, Kowalczyk I, Melhuish TA, Lemiesz A, Wotton D. Analysis of transcriptional activity by the Myt1 and Myt1l transcription factors. J Cell Biochem. 2018;119:4644–55.
Lee J, Taylor CA, Barnes KM, Shen A, Stewart EV, Chen A, et al. A Myt1 family transcription factor defines neuronal fate by repressing non-neuronal genes. eLife. 2019;8:e46703.
Mall M, Kareta MS, Chanda S, Ahlenius H, Perotti N, Zhou B, et al. Myt1l safeguards neuronal identity by actively repressing many non-neuronal fates. Nature. 2017;544:245–9.
Pang ZP, Yang N, Vierbuchen T, Ostermeier A, Fuentes DR, Yang TQ, et al. Induction of human neuronal cells by defined transcription factors. Nature. 2011;476:220–3.
Vierbuchen T, Ostermeier A, Pang ZP, Kokubu Y, Südhof TC, Wernig M. Direct conversion of fibroblasts to functional neurons by defined factors. Nature. 2010;463:1035–41.
Lee Y, Mattai A, Long R, Rapoport JL, Gogtay N, Addington AM. Microduplications disrupting the MYT1L Gene (2p25.3) are associated with schizophrenia. Psychiatr Genet. 2012;22:206–9.
Braddock A, Del Campo M, Reiff MI, Stein MT. Disruptive behavior, global developmental delay, and obesity in a 5-year-old boy with a chromosome microduplication. J Dev Behav Pediatr JDBP. 2018;39:81–4.
Coe BP, Witherspoon K, Rosenfeld JA, van Bon BWM, Vulto-van Silfhout AT, Bosco P, et al. Refining analyses of copy number variation identifies specific genes associated with developmental delay. Nat Genet. 2014;46:1063–71.
Cooper GM, Coe BP, Girirajan S, Rosenfeld JA, Vu TH, Baker C, et al. A copy number variation morbidity map of developmental delay. Nat Genet. 2011;43:838–46.
International Schizophrenia Consortium. Rare chromosomal deletions and duplications increase risk of schizophrenia. Nature. 2008;455:237–41.
Jakobsson M, Scholz SW, Scheet P, Gibbs JR, VanLiere JM, Fung HC, et al. Genotype, haplotype and copy-number variation in worldwide human populations. Nature. 2008;451:998–1003.
Mansfield P, Constantino JN, Baldridge D. MYT1L: A systematic review of genetic variation encompassing schizophrenia and autism. Am J Med Genet Part B Neuropsychiatr Genet Off Publ Int Soc Psychiatr Genet. 2020;183:227–33.
Meyer KJ, Axelsen MS, Sheffield VC, Patil SR, Wassink TH. Germline mosaic transmission of a novel duplication of PXDN and MYT1L to two male half-siblings with autism. Psychiatr Genet. 2012;22:137–40.
Suktitipat B, Naktang C, Mhuantong W, Tularak T, Artiwet P, Pasomsap E, et al. Copy number variation in Thai population. PloS One. 2014;9:e104355.
Van Den Bossche MJ, Strazisar M, Cammaerts S, Liekens AM, Vandeweyer G, Depreeuw V, et al. Identification of rare copy number variants in high burden schizophrenia families. Am J Med Genet Part B Neuropsychiatr Genet Off Publ. Int Soc Psychiatr Genet. 2013;162B:273–82.
Vrijenhoek T, Buizer-Voskamp JE, van der Stelt I, Strengman E, Genetic Risk and Outcome in Psychosis (GROUP) Consortium, Sabatti C, et al. Recurrent CNVs disrupt three candidate genes in schizophrenia patients. Am J Hum Genet. 2008;83:504–10.
Walsh T, McClellan JM, McCarthy SE, Addington AM, Pierce SB, Cooper GM, et al. Rare structural variants disrupt multiple genes in neurodevelopmental pathways in schizophrenia. Science. 2008;320:539–43.
Stevens SJC, van Ravenswaaij-Arts CMA, Janssen JWH, Klein Wassink-Ruiter JS, van Essen AJ, Dijkhuizen T, et al. MYT1L is a candidate gene for intellectual disability in patients with 2p25.3 (2pter) deletions. Am J Med Genet A. 2011;155A:2739–45.
Firth HV, Richards SM, Bevan AP, Clayton S, Corpas M, Rajan D, et al. DECIPHER: database of chromosomal imbalance and phenotype in humans using ensembl resources. Am J Hum Genet. 2009;84:524–33.
Buizer-Voskamp JE, Muntjewerff JW, Strengman E, Sabatti C, Stefansson H, Vorstman JA, et al. Genome-wide analysis shows increased frequency of CNV deletions in Dutch schizophrenia patients. Biol Psychiatry. 2011;70:655–62.
Blanchet P, Bebin M, Bruet S, Cooper GM, Thompson ML, Duban-Bedu B, et al. MYT1L mutations cause intellectual disability and variable obesity by dysregulating gene expression and development of the neuroendocrine hypothalamus. Stark Z, éditeur. PLOS Genet. 2017;13:e1006957.
Girirajan S, Rosenfeld JA, Coe BP, Parikh S, Friedman N, Goldstein A, et al. Phenotypic Heterogeneity of Genomic Disorders and Rare Copy-Number Variants. N Engl J Med. 2012;367:1321–31.
Girirajan S, Eichler EE. Phenotypic variability and genetic susceptibility to genomic disorders. Hum Mol Genet. 2010;19:R176–87.
Do KQ, Cabungcal JH, Frank A, Steullet P, Cuenod M. Redox dysregulation, neurodevelopment, and schizophrenia. Curr Opin Neurobiol. 2009;19:220–30.
Ramos PS, Sajuthi S, Langefeld CD, Walker SJ. Immune function genes CD99L2, JARID2 and TPO show association with autism spectrum disorder. Mol Autism. 2012;3:4.
Rosenfeld JA, Ballif BC, Torchia BS, Sahoo T, Ravnan JB, Schultz R, et al. Copy number variations associated with autism spectrum disorders contribute to a spectrum of neurodevelopmental disorders. Genet Med. 2010;12:694–702.
Acknowledgements
We thank the patients and their families for their cooperation. This study assessed data generated by the DECIPHER community. A full list of the centers that helped to generate the data is available from https://deciphergenomics.org/about/stats and by e-mail from contact@deciphergenomics.org. Funding for the DECIPHER project was provided by the Wellcome Trust. We declare that those who carried out the original analysis and collection of the data bear no responsibility for the further analysis or interpretation of the data.
Funding
This research did not receive any specific grant from funding agencies in the public, commercial, or not-for-profit sectors.
Author information
Authors and Affiliations
Contributions
MB, FV and BH: contributed to the study conception and design. MB: analyzed the data and wrote the manuscript. JL, GL, FB, MGG, RD: performed the clinical evaluation of the patients. ME, NC, LB, ACT, CSB, JCo: performed the genetic investigations. JCl: participated in the collection of clinical data. FV, BH: did the supervision, review and editing of the manuscript. All authors contributed to data acquisition.
Corresponding authors
Ethics declarations
Ethical approval
In compliance with the Declaration of Helsinki, informed written consent for genetic study was obtained for participated individuals.
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Bouassida, M., Egloff, M., Levy, J. et al. 2p25.3 microduplications involving MYT1L: further phenotypic characterization through an assessment of 16 new cases and a literature review. Eur J Hum Genet 31, 895–904 (2023). https://doi.org/10.1038/s41431-023-01379-9
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41431-023-01379-9
This article is cited by
-
A new impact factor for EJHG in 2022
European Journal of Human Genetics (2023)