Abstract
We previously reported lncRNA HAR1A as a tumor suppressor in non-small cell lung cancer (NSCLC). However, the delicate working mechanisms of this lncRNA remain obscure. Herein, we demonstrated that the ectopic expression of HAR1A inhibited the proliferation, epithelial-mesenchymal transition (EMT), migration, and invasion of NSCLC cells and enhanced paclitaxel (PTX) sensitivity in vitro and in vivo. We identified the oncogenic protein annexin 2 (ANXA2) as a potential interacting patterner of HAR1A. HAR1A overexpression enhanced ANXA2 ubiquitination and accelerated its degradation via the ubiquitin–proteasome pathway. We further uncovered that HAR1A promoted the interaction between E3 ubiquitin ligase TRIM65 and ANXA2. Moreover, the ANXA2 plasmid transfection could reverse HAR1A overexpression-induced decreases in proliferation, migration, and invasion of NSCLC cells and the activity of the NF-κB signaling pathway. Finally, we found that HAR1A loss in NSCLC might be attributed to the upregulated METTL3. The m6A modification levels of HAR1A were increased in cancer cells, while YTHDF2 was responsible for recognizing m6A modification in the HAR1A, leading to the disintegration of this lncRNA. In conclusion, we found that METTL3-mediated m6A modification decreased HAR1A in NSCLC. HAR1A deficiency, in turn, stimulated tumor growth and metastasis by activating the ANXA2/p65 axis.
Similar content being viewed by others
Introduction
Long non-coding RNAs (lncRNAs) are referred to as transcribed RNA molecules consisting of more than 200 nucleotides and lacking protein-coding potential. Although used to be thought of as transcriptional noise or artefactual transcripts, lncRNAs are critical for gene expression by extensively participating in and controlling transcription, splicing, post-translational modification, stability, and translation [1]. Accumulating evidence has highlighted systematical alterations in RNA processing during carcinogenesis, leading to aberrant expression levels of various RNAs such as mRNA, miRNA, tRNA, circRNA, and lncRNAs [2]. Dysregulated lncRNAs can act as oncogenes or tumor suppressors to contribute to tumorigenesis, metastasis, drug resistance, and progression [2]. Elevated LncRNA LINC00942 conferred gastric cancer (GC) chemoresistance by activating the MSI2/c-Myc axis to induce stemness and suppress apoptosis of GC cells [3]. Oncogenic lncRNA SNHG20 was increased in NSCLC tissues and associated with prognosis, which epigenetically inhibited p21 expression to promote tumor cell growth [4]. Tumor-suppressive lncRNA DRAIC was reduced in castration-resistant advanced prostate cancer (PC), which inactivated nuclear factor-κB (NF-κB) to inhibit tumor progression [5]. In recent years, lncRNAs have shown potential as diagnostic markers and therapy monitoring in cancer [6, 7]. Moreover, ncRNAs are potential therapeutic targets for cancer due to their complex spatial structures and tumor-promoting and -suppressing functions [8]. Therefore, it is imperative to identify and characterize the regulation and functional significance of novel lncRNAs to understand the fundamental biological processes of tumorigenesis and advance cancer treatments.
lncRNA Highly Accelerated Region 1A (HAR1A) is a large intergenic non-coding RNA. Our pan-cancer analysis revealed that the expression of HAR1A was downregulated in a broad spectrum of cancers, including lung adenocarcinoma (LUAD), lung squamous carcinoma (LUSC), bladder urothelial carcinoma (BLCA), breast invasive carcinoma (BRCA), uterine corpus endometrial carcinoma (CESC), liver hepatocellular carcinoma (LIHC), prostate adenocarcinoma (PRAD), and stomach adenocarcinoma (STAD) [9]. In vitro and in vivo studies demonstrated that HAR1A suppressed NSCLC growth [9]. Moreover, lncRNA HAR1A was found to retard the progression of oral cancer by inhibiting the ALPK1/BRD7/myosin IIA axis [10]. However, The molecular mechanisms underlying the HAR1A’s tumor-suppressing functions are largely unclear. This study aimed to interrogate further the biological functions of HAR1A and the molecular mechanisms behind it.
Annexin A2 (ANXA2) is a member of the annexin family, comprising calcium-dependent phospholipid-binding proteins. ANXA2 is overexpressed in several types of cancer, such as hepatocellular carcinoma (HCC) [11], breast cancer [12], and NSCLC [13], and contributes to cancer development and progression. For instance, ANXA2 was activated via sequential phosphorylation by S6K1 and K63-linked polyubiquitination by FBXW10 in HCC tissues from male patients [14]. Activated ANXA2 transferred to the cell membrane to activate the KRAS/MEK/ERK pathway, promoting HCC proliferation and lung metastasis [14]. These findings suggest that ANXA2 may hold promise to be a potential biomarker and therapeutic target for cancer. Moreover, the regulation of ANXA2 on many signaling pathways has been documented, including the β-catenin [15], ERK [14, 16], and NF-κB [17, 18].
N6-methyladenosine (m6A) RNA modification is an epigenetic regulatory mechanism for coding and ncRNAs, which plays essential roles in almost all vital biological and pathological processes, including malignant transformations. m6A modifications are catalyzed by a m6A methyltransferase complex (MTC), comprising the methyltransferase-like 3 (METTL3), METTL14, WTAP, and other regulatory components. Elevated METTL3 has been seen in NSCLC and is involved in cancer progress [19,20,21]. Therefore, we also explore whether HAR1A is subjected to METTL3-dependent m6A modification.
Here, we demonstrated that HAR1A was downregulated by METTL3-mediated m6A modification. HAR1A, in part, controlled proliferation, migration, invasion, and EMT of NSCLC cells by boosting the interaction between ANXA2 and its E3 ligase TRIM65 and thereby disintegrating oncogenic ANXA2 to inactivate the NF-κB signaling pathway. Thereby, we identified a novel fine-tuning mechanism interpreting how the loss of HAR1A promoted cancer development and how HAR1A was decreased in NSCLC.
Results
lncRNA HAR1A increases NSCLC cells’ response to paclitaxel
We first determined the endogenous expression levels of HAR1A in the indicated cell lines. Compared to normal epithelial cells, several cancer cell lines exhibited decreased expression of HAR1A (Supplementary Fig. 1a). A549 and H1299 cells were infected with lentiviral vectors expressing HAR1A to establish stable cell lines (Supplementary Fig. 1b). The proliferation of A549 and H1299 cells was significantly inhibited by HAR1A overexpression (Supplementary Fig. 1c).
Next, the wound healing assay indicated that elevated expression of HAR1A also decelerated the migration of NSCLC cells. In contrast to the control group, the created gaps remained unfilled post 48 h (Fig. 1A). Consistently, immunocytostaining and RT-qPCR results demonstrated that cells overexpressing HAR1A showed an increase in E-cadherin and decreases in N-cadherin and vimentin, suggesting a reversal of epithelial-mesenchymal transition (EMT) related to reduced migration ability of cells (Fig. 1B, C). We also used siRNA to knock down HAR1A in the A549 and H1299 cells (Supplementary Fig. 1d). Silencing HAR1A exhibited opposite effects on cancer cells. The downregulation of HAR1A significantly prompted the growth and migration of NSCLC cells (Supplementary Fig. 1e and Fig. 1D). Moreover, enforced expression of the lncRNA sensitized NSCLC cells to paclitaxel (PTX) as indicated by IC50 (Fig. 2A). PTX treatment hindered proliferation and induced apoptosis of NSCLC cells. The enforced expression of HAR1A enhanced the cytotoxicity of PTX on NSCLC cells as revealed by colony formation, Edu, and TUNEL assays, and similar results were observed (Fig. 2B–D).
A Representative images and quantifications indicating scratching assay of HAR1A-overexpressing A549 and H1299 stable cells and respective empty vector-expressing cells. B Immunofluorescent staining was performed to detect the expression of epithelial-mesenchymal transition (EMT) biomarkers, including E-cadherin, N-cadherin, and vimentin. Fluorescence intensity (100%) was measured to represent the expression levels of EMT biomarkers. C The mRNA levels of these EMT biomarkers were also evaluated using RT-qPCR. D The wound healing assay indicated that gaps almost disappeared in siHAR1A cells compared with unfilled gaps in siNC cells at 36h after scratching, suggesting that silencing HAR1A promoted NSCLC cell migration. *p < 0.05; **p < 0.01; ***p < 0.001; ****p < 0.0001.
A A549 and H1299 cells were infected lentivirus (LV) empty vectors and LV overexpressing HAR1A (LV-HAR1A) and incubated with increasing concentrations of PTX. Cell viability was checked after 24 h using CCK8 assay. IC50 was calculated in both A549 and H1299 cells. The following experiments were performed using LV-vector and LV-HAR1A cells in the presence or absence of PTX. B HAR1A further aggravated PTX’s colony formation-inhibiting effects on NSCLC cells, suggesting that HAR1A might have acted as a chemosensitizer. C Edu incorporation assays were used to monitor dividing cells. With the percentage of EdU-labeling cells used as an estimate for proliferation rate, PTX exhibited significantly stronger inhibitory effects on LV-HAR1A NSCLC cells than on vector control cells. D TUNEL assay revealed that PTX treatment triggered significantly more cells to undergo apoptosis in LV-HAR1A NSCLC cells than in vector control cells. *p < 0.05; **p < 0.01; ***p < 0.001; ****p < 0.0001.
lncRNA HAR1A suppresses NSCLC growth and metastasis in vivo
Given our in vitro findings, we subcutaneously implanted stable HAR1A-overexpressing H1299 cells into female BALB/c nude mice to assess the effects of HAR1A and PTX tumor growth (Fig. 3A). We found that paclitaxel treatment or HAR1A overexpression significantly decreased tumor volume and weight compared to the control group (Fig. 3B–D). Paclitaxel administration further shrank tumor xenografts arising from HAR1A-overexpressing H1299 cells (Fig. 3B–D). EdU assay confirmed fewer Edu-positive cells in tumors with paclitaxel treatment or HAR1A overexpression alone and the least proliferating cells in the combination group, compared with the control group (Fig. 3E). TUNEL assay also verified more apoptotic cells in the combination group than others (Fig. 3F). These results suggest that paclitaxel and overexpression of HAR1A synergistically inhibited tumor growth. We next investigated whether HAR1A regulated tumor metastasis using a tail vein injection model (Fig. 3G). Five weeks post-injection, the lungs were excised, and gross analysis demonstrated that the lung surface was occupied by many metastatic tumor nodules in the control group, compared to relatively smooth surface lung morphology in the mice injected with cells stably expressing HAR1A (Fig. 3H). HE and IHC stains indicated tumor nodules in the lung and Ki67+ cells in the nodules (Fig. 3H). HAR1A-overexpressing H1299 cells led to significantly fewer nodules in the lung of host mice than vector-expressing cells (Fig. 3I). Moreover, EdU and TUNEL assays revealed significantly few EdU+ proliferative cells and more TUNEL+ apoptotic cells in metastasis nodules from stable HAR1A-expressing cells-injected mice than control mice (Fig. 3J, K).
Impacts of PTX and HAR1A overexpression on tumor growth of PTX-treated xenograft mice. A The workflow of in vivo tumor growth study. The effect of PTX on the growth of tumors originated from stable cells infected with lentiviruses containing HAR1A cDNA (HAR1A) or empty lentivirus vectors (vector). Paclitaxel was administered at a dose of 15 mg/kg once every four days. B Representative images of mice bearing subcutaneous tumor xenografts for indicated groups and resected tumors from corresponding mice. C Tumor growth curves (volume) in mice subcutaneously injected with LV-HAR1A or LV-vector stably infected H1299 cells with or without PTX inventions. D Tumor weight in mice injected with H1299 stable LV-HAR1A or LV-vector cells at day 28 after PTX treatment. E, F EdU and TUNEL assays were performed to detect proliferation and apoptosis in metastatic nodes, respectively, followed by quantifications. G The workflow of in vivo tumor metastasis study. H, I Ex vivo mouse lungs elucidated that reduced lung metastasis in mice injected with HAR1A-overexpressing NSCLC cells, compared with mice receiving vector cells. HE and Ki67 stains showed metastatic nodes and proliferating cells. J, K EdU and TUNEL assays on metastatic nodes and quantifications. *p < 0.05; **p < 0.01; ***p < 0.001; ****p < 0.0001.
lncRNA HAR1A interacts with the oncogenic protein ANXA2
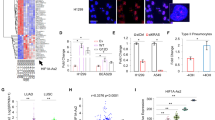
We next questioned how HAR1A inhibits NSCLC growth and metastasis. RT-qPCR results showed that HAR1A was mainly localized in the cell nucleus, but a small amount of this lncRNA was also seen in the cytoplasm (Fig. 4A). RNA pull-down assays, followed by mass spectrometry, were performed to identify candidate proteins interacting with HAR1A. Numerous HAR1A-binding proteins were enriched and immunoprecipitated (Fig. 4B). Among them, myosin IC (MYO1C) and annexin 2 (ANXA2) were found to be the most abundant protein among all identified proteins (Table S2), and they have been reported to be associated with carcinogenesis [22,23,24]. The peptide mass fingerprinting of ANXA2 is displayed in Fig. 4C. When we used HAR1A probe to perform RNA pull-down experiments, ANXA2 could be readily detected in the precipitates by western blotting in both cell lines, while only a weak band was observed for MYO1C in H1299 cells (Fig. 4D). RIP experiments using anti-ANXA2 antibodies were performed to validate the interaction between HAR1A and ANXA2 in the whole lysis of A549 and H1299 cells. As shown in Fig. 4E, RT-qPCR could amplify HAR1A in ANXA2 immunoprecipitates. FISH and immunocytostaining validated the colocalization of ANXA2 in two NSCLC cell lines (Fig. 4F). When we examined whether HAR1A regulated ANXA2 expression to transmit signaling, we found that ectopic expression of HAR1A did not significantly alter mRNA levels but reduced protein levels of ANXA2 (Fig. 4G, H). Interestingly, GSEA suggested that HAR1A-related genes are greatly enriched in the ubiquitin-mediated proteolysis and proteasome pathways (Fig. 4I), implying the potential roles of HAR1A in the ubiquitination and degradation of proteins.
A RT-qPCR was performed to quantify the mRNA levels of HAR1A, GAPDH, and U6 in nuclear and cytoplasmic fractions of A549 and H1299 cells. B RNA pull-downs were performed using biotinylated antisense and sense sequences of HAR1A. Co-precipitated proteins were resolved in SDS-PAGE and visualized with silver staining. C Identification of ANAX2 in precipitants of RNA pull-down assay by LC/MS. D Following the HAR1A RNA pull-down assay, western blot results showed the presence of ANXA2 among the pulled-down proteins. E RIP analysis with anti-ANXA2 antibody was conducted in A549 and H1299 cells to show co-precipitated HAR1A using RT-qPCR. F The representative photograph showing fluorescence in situ hybridization (FISH) with a probe against HAR1A and immunofluorescent staining with anti-ANXA2 antibody in A549 and 1299 cells. Red, green, and blue represent biotin-labeled probe against HAR1A, immunofluorescent staining of ANXA2, and DAPI staining of the nucleus, respectively. Yellow in the merged image indicates the colocalization of HAR1A and ANXA2 in the cells. G RT-qPCR analysis of the ANXA2 mRNA levels in LV-vector and LV-HAR1A treated A549 and H1299 cells. H Immunoblotting to examine ANXA2 levels in A549 and H1299 stable cells overexpressing HAR1A or control cells expressing LV-vector. I Gene set enrichment analysis (GSEA) to explore the downstream pathway of HAR1A. *p < 0.05; **p < 0.01; ***p < 0.001; ****p < 0.0001.
HAR1A facilitates TRIM65-mediated ANXA2 ubiquitination and degradation
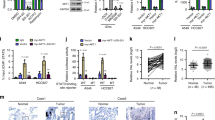
The results of the cycloheximide (CHX) chase assay demonstrated that ANXA2 in HAR1A overexpression cells exhibited a shorter half-life when compared with control cells, whereas the half-life of ANXA2 was much longer in HAR1A-knockdown cells than in the control cells (Fig. 5A). Moreover, MG132 could block HAR1A-mediated ANXA2 degradation (Fig. 5B). Finally, we found that transfection of HAR1A enhanced the ubiquitination levels of ANXA2 (Fig. 5C). These results suggest that HAR1A may promote ANXA2 degradation by upregulating its ubiquitination levels. Using the online tool UbiBrowser2.0 (http://ubibrowser.bio-it.cn/ubibrowser_v3/), we retrieved 20 E3 ligases potentially mediating ubiquitination modification of ANXA2, such as MARCHF2, WWP2, NEDD4L, CBLL1, and TRIM65 (Fig. 5D). Among them, tripartite motif containing 65 (TRIM65) was reported to ubiquitylate ANXA2 in urothelial carcinoma of the bladder (UCB) [25]. Furthermore, of the predicted 20 E3 ubiquitin ligases, only TRIM65 was identified in the mass spectrometry results. Consequently, we examined whether HAR1A functions as a scaffold to facilitate interactions between ANXA2 and TRIM65, thereby regulating ANXA2 stability. We then investigated whether TRIM65 mediates ubiquitination and degradation of ANXA2 and whether HAR1A is involved in this process. First, using an RNA pull-down assay, we showed that HAR1A binds to TRIM65 in A549 and H1299 cells (Fig. 5E). We also verified the interaction through RIP in the two cell lines (Fig. 5F). Second, ANXA2 could be immunoprecipitated using an anti-TRIM65 antibody, while we further confirmed that TRIM65 was physically co-immunoprecipitated with ANXA2 using an anti-ANXA2 antibody (Fig. 5G). Ultimately, we also investigated the effects of HAR1A on TRIM65-ANXA2 interaction. Co-IP experiments revealed that anti-TRIM65 antibody enriched more ANXA2 in cells overexpressing HAR1A than in control cells (Fig. 5H), suggesting HAR1A overexpression boosted TRIM65’s interaction with ANXA2. These results indicated that HAR1A enhanced the interaction between ANXA2 and TRIM65 to facilitate TRIM65-mediated ANXA2 ubiquitination, consequently accelerating its degradation.
A Cells transfected with siNC or si-HAR1A, as well as cells with stable HAR1A overexpression, were treated with cycloheximide (CHX) for the specified periods. Subsequently, cell lysates were analyzed by western blot to examine the half-life of ANXA2 protein. Protein band intensities were quantified using Image J software. B Western blot was performed to check the effects of proteasome inhibitor MG132 on the ANXA2 expression levels. C HA-Ub and Flag-ANXA2 plasmids were co-introduced into stable A549 and H1299 LV-vector and LV-HAR1A cells. Cells were treated with MG132. Immunoprecipitation was performed with cell lysates using an anti-Flag antibody and western blot followed with an anti-Flag or anti-ubiquitin antibody. D Predicated E3 ligases for ANAX2 using the UbiBrowser tool. E HAR1A RNA pull-down assay and western blot showed the interaction between HAR1A and TRIM65. F RIP analysis coupled with RT-qPCR indicated that TRIM65 could precipitate HAR1A in A549 and H1299 cells. G Co-immunoprecipitation (Co-IP) of ANXA2 and TRIM65 in A549 and H1299 cells. Western blot with anti-ANXA2 antibody was used to probe the ANXA2 in the anti-TRIM65 antibody-mediated immunoprecipitates. Reverse co-IP was also performed to testify the immunoprecipitation of TRIM65 with anti-ANXA2 antibody. H Co-IP revealed that HAR1A facilitated the interaction between ANXA2 and TRIM65. *p < 0.05; **p < 0.01; ***p < 0.001; ****p < 0.0001.
HAR1A abrogates NSCLC cells’ aggressive behaviors by downregulating ANXA2
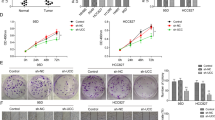
Since HAR1A decreases ANXA2 by shortening its half-life, we investigated whether ANXA2 is indispensable for HAR1A-induced inhibition on the proliferation and migration of A549 and H1299 cells. We first restored the expression of ANXA2 in stable HAR1A-overexpressing NSCLC cells by transfecting ANXA2 plasmids. We found that HAR1A-induced decreases in migration and invasion of NSCLC cells were partially recovered by the overexpression of ANXA2 (Fig. 6A). Moreover, CCK8 assay demonstrated that the re-expression of ANXA2 reversed HAR1A’s inhibitory effects on the proliferation of A549 and H1299 cells (Fig. 6B). These results substantiated that HAR1A suppresses the proliferation, migration, and invasion of cancer cells by reducing ANXA2 expression. Gene set enrichment analysis (GSEA) indicated that it may regulate NF-kB (Fig. 6C). Additionally, several publications have implied that ANXA2 functions by activating the NF-kB signaling pathway [18, 26, 27]. Our western blot results elucidated that HAR1A overexpression led to a reduction in the levels of p65 and its phosphorylated form; however, simultaneous re-expression of ANXA2 could reactivate the NF-kB signaling pathway suppressed by HAR1A (Fig. 6D). Since the oncogenic role of the NF-kB signaling pathway is well documented, it is biologically plausible to infer that HAR1A may exert tumor-suppressing functions by repressing the ANXA2/NF-kB axis. We also collected paired normal and tumor tissues from 8 patients with lymph node stage N0-2 NSCLC (Fig. 6E). RT-qPCR analysis showed a significant decrease of HAR1A in tumors versus normal tissues (Fig. 6F), and N1-2 tumors versus N0 tumors (Fig. 6G). Western blot results showed ANXA2 was significantly increased in N1-2 tumors compared with N0 tumors (Fig. 6H), which was also validated using immunohistochemistry staining (Fig. 6I). Moreover, K–M plotter indicated that high levels of HAR1A and ANXA2 in NSCLC were associated with OS in the opposite direction (Fig. 6J, K).
Stable A549 and H1299 LV-HAR1A cells were further transfected with ANXA2 plasmids. A Representative image of crystal violet-stained A549 and H1299 cells with modified expression of HAR1A and ANXA2 in the transwell assays. Quantification of migrated and invaded cells indicated that transfection of ANXA2 plasmids partially counteracted HAR1A-induced repression of tumor cell migration and invasion. B CCK8 was used to measure proliferation of A549 and H1299 cells transduced with LV-vector, LV- HAR1A+ANXA2 plasmid, and LV- HAR1A+empty plasmid. C Gene set enrichment analysis (GSEA) showed the downstream pathway of ANXA2. D Western blot to determine the protein levels of p65, p-p65, and ANXA2 in A549 and H1299 cells with indicated gene manipulations. Tumor samples were collected from eight NSCLC patients, along with matched normal tissues. E Chest computed tomography imaging of the eight NSCLC patients. The red circles highlight primary lung tumors and the blue arrows indicate regional lymph nodes with pathologically proven metastasis. According to the 8th edition of AJCC TNM staging, N category was defined as N0 (no metastatic lymph node), N1 (metastasis in ipsilateral peribronchial and/or ipsilateral hilar lymph nodes), N2 (metastasis in ipsilateral mediastinal and/or subcarinal lymph nodes). F RT-qPCR was used to measure HAR1A in normal tissues versus tumors. G Comparsion of HAR1A in N0 versus N1-2 tumors. H Western blot to compare the ANXA2 levels between N0 and N1-2 tumors. I ANXA2 immunohistochemistry staining in NSCLC tumor samples. J, K Kaplan–Meier survival curves for lung cancer patients dichotomized by the levels of HAR1A or ANXA2, using the K–M plotter. *p < 0.05; **p < 0.01; ***p < 0.001; ****p < 0.0001.
METTL3-mediated m6A modifications expedites HAR1A disintegration
Finally, we questioned how the expression levels of HAR1A declined in NSCLC compared with normal lung tissues. Dysregulation of RNA m6A modification has been well known to be implicated in carcinogenesis. Methyltransferase Like 3 (METTL3), installing m6A in RNAs, is highly expressed in NSCLC and promotes cancer progress [19,20,21]. Therefore, we scrutinized whether METTL3 regulates HAR1A expression. Both gain and loss function analysis showed that METTL3 negatively regulated HAR1A (Fig. 7A, B). Moreover, METTL3 overexpression speeded up HAR1A degradation with time (Fig. 7C), denoting that METTL3 might destabilize HAR1A through m6A modification. Indeed, meRIP-qRCR experiments showed that m6A-modified HAR1A was enhanced in A549 and H1299 cells in comparison to HBE cells (Fig. 7D). By screening the known m6A readers, including YTHDF1-3, we found that the knockdown of YTHDF2 leads to upregulation of HAR1A (Fig. 7E). Consistently, YTHDF2 depletion prevented HAR1A from degrading (Fig. 7F). Finally, RIP experiments using anti-YTHDF2 antibodies confirmed the binding of YTHDF2 to HAR1A (Fig. 7G).
A, B RT-qPCR was performed to investigate the effects of overexpression and knockdown of METTL3 on the HAR1A levels in A549 and H1299 cells. C After inhibiting RNA synthesis with actinomycin D, HAR1A degraded faster in NSCLC cells with METTL3 overexpression than in control vector cells at different times. D m6A RIP coupled with RT-qPCR showed that HAR1A was subjected to m6A modification, and significantly more m6A-modified HAR1A RNAs were enriched in A549 and H1299 than in HBE cells. E Cells were transduced with siRNAs targeting YTHDF1, YTHDF2, YTHDF3, and scramble controls. siYTHDF2 treatment reduced HAR1A levels as shown by RT-qPCR. F In cells where RNA synthesis was blocked with actinomycin D, YTHDF2 siRNA slowed down the degradation of HAR1A. G RIP assay, followed by RT-qPCR, revealed the precipitation of YTHDF2 with HAR1A. H Schematic diagram of molecular mechanisms. *p < 0.05; **p < 0.01; ***p < 0.001; ****p < 0.0001.
Discussion
Lung cancer is the top cause of cancer-related mortality. NSCLC is the most predominant type of lung cancer. Despite revolutionary advances in treatment modalities, including surgery, radiation, chemotherapy, targeted therapies, and immunotherapies, the prognosis of NSCLC remains unsatisfying. Due to the lack of reliable early-detection biomarkers, most patients could not be diagnosed until the development of locally advanced or widely metastatic tumors. Therefore, further dissecting the molecular landscape of tumors, particularly ncRNAs, has the promise of discovering novel cancer-related biomarkers for early diagnosis and prognosis prediction and novel molecular targets for developing efficacious therapeutic strategies.
As far as we know, only up to 2% of human genomic DNA is protein-coding, whereas ncRNAs may constitute 70% of the human genome. In contrast to limited protein, tons of functional lncRNAs warrant to be identified, which may have great potential to be biomarkers or druggable targets for cancer [6,7,8]. Tremendous lncRNAs are deregulated in cancers, and monitoring circulating lncRNA is minimally invasive [6]. For instance, lncRNA H19 showed promise for being an early diagnosis marker of gastric cancer. The levels of plasma H19 significantly declined post-surgery in patients with gastric cancer, indicating the potential application of H19 in the early diagnosis and postoperative monitoring settings [28]. Furthermore, many strategies for developing ncRNA-targeting small-molecule compounds have emerged, such as small-molecule microarray, structure-based designing, phenotypic screening, and pharmacological validation approaches [8]. As a result, several small-molecule compounds, including bisphenol-A, mitoxantrone, and enoxacin have been shown to modulate or selectively target ncRNAs in different types of cancer [8]. To date, some ncRNA-regulating small-molecule compounds have been pharmacologically confirmed. Bisphenol-A (BPA) and diethylstilbestrol (DES) are typical examples. LncRNA HOTAIR was oncogenic in a variety of cancers. HOTAIR, which prompted the proliferation and migration of breast cancer cells, is transcriptionally activated by estradiol [29]. The same team also found that low concentrations of BPA and DES could significantly induce HOTAIR expression in breast cancer cells [30], suggesting that small-molecule compounds could regulate the levels of lncRNAs.
Other teams and we have shown that lncRNA HAR1A is a tumor suppressor in several cancers, including HCC [31], oral cancer [10], and NSCLC [9]. This study aimed to discover the molecular mechanisms underpinning HAR1A’s tumor-suppressing functions in NSCLC. We found that overexpression of HAR1A could reduce the proliferation, migration, invasion, and EMT of NSCLC cells. HAR1A strengthened the interaction between the oncogenic protein ANXA2 and its E3 ligase TRIM65, thereby promoting ubiquitination and degradation of ANXA2 to inactivate the NF-κB signaling pathway. Similarly, Lee et al. demonstrated that si-HAR1A treatment augmented proliferation, migration, and EMT in oral squamous cell carcinoma (OSCC) cells [10].
Shi et al. reported that loss of HAR1A in HBV-Induced HCC in Chinese Patients was related to inferior clinical outcomes [31]. Tumor-suppressive lncRNAs are very common. LncRNA DRAIC was downregulated in castration-resistant advanced PC. This lncRNA tumor suppressor inhibited tumor growth and invasion by inactivating nuclear factor-κB (NF-κB). lncRNA DRAIC binds to the IκB kinase complex (IKK) complex to inhibit the phosphorylation and proteasomal degradation of NF-κB inhibitor-α (IκBα), thereby suppressing the NF-κB signaling pathway [5]. LINC00261 was epigenetically silenced in LUAD, and its deficiency was validated in liver, breast, and gastric cancer. Enforced expression of LINC00261 inhibited migration and proliferation of LUAD cell lines through induction of a G2/M cell cycle arrest and activation of the DNA damage response [32].
The regulatory mechanisms of lncRNAs have been studied extensively, which interact with DNA, RNA, or proteins to regulate various cellular processes, including transcriptional regulation, chromatin structure, RNA stability, and cell proliferation. Wang and colleagues categorize the numerous functions of lncRNA into four archetypes of molecular mechanisms: signals, guides, scaffolds, and decoys; however, these mechanisms are not mutually exclusive [33]. First, lncRNAs can transduce signals because the transcription of single lncRNAs is tightly regulated in response to diverse stimuli. Moreover, lncRNAs do not require translation and respond more quickly than proteins regarding signal transduction. Second, lncRNAs can serve as molecular decoys. For instance, lncRNAs bind and sequester a protein target without exerting extra functions. In other words, lncRNA trapped RBPs (e.g., transcription factors, enzymes, or other regulatory factors) to not fulfill their tasks. LINC00941 promoted the progress of pancreatic cancer (PC) through the decoying mechanism. LINC00941 bound to ANXA2 as a decoy to competitively inhibit the interaction of ANXA2 with NEDD4L, an E3 ligase, consequently preventing ANXA2 from NEDD4L-mediated ubiquitination and proteasomal degradation. The upregulated ANXA2, in turn, activated FAK/AKT signaling to fuel PC cell proliferation and metastasis [34]. The third molecular mechanism of lncRNA is the guide-lncRNA interacts with and conveys protein to a specific location, especially in the case of regulating gene expression in trans. Finally, lncRNAs can provide a platform to facilitate intermolecular interactions since lncRNA contains different domains that bind more than one effector molecule. In the current study, HAR1A might act as a scaffold to aid the interaction between TRIM65 and ANXA2, destabilizing the latter to inactivate the NF-κB signaling pathway. In other words, HAR1A bonds TRIM65 and ANXA2 simultaneously to bring them together in both time and space. By doing so, HAR1A greatly enhanced the enzymatic activity of TRIM65 to attach ubiquitins to ANXA2 for subsequent proteasomal degradation. The future exhaustive understanding of how these molecules are assembled and regulated may enable therapeutic strategies to target signaling components and reshape cellular behavior precisely.
ANXA2 is a multi-functional protein and participates in various cellular processes involving signal transduction, endocytosis, cytokinesis, actin remodeling, mRNA translocation, and DNA repair. Accumulating evidence has shown the contribution of Annexin 2 in the development of lung cancer. Coculture with cancer-associated fibroblasts (CAFs) led to acquired resistance of NSCLC cells to tyrosine kinase inhibitors (TKIs). The underpinning mechanism was that growth factors HGF and IGF-1 released by CAFs upregulated ANXA2 and increased its phosphorylation, thereby inducing EMT of NSCLC cells [35]. ANXA2 interacted with T-cell immunoglobulin and mucin domain-containing molecule 4 (TIM-4) and mediated TIM-4-induced lung cancer progression by activating PI3K/AKT/OPA1 axis [36]. Ubiquitination is a critical post-translational modification to control ANXA2 activity in carcinogenesis, and it is subjected to ubiquitination-mediated degradation [16]. E3 ubiquitin ligase FBXW7 facilitated the ubiquitination and subsequent disintegration of ANXA2. Loss of FBXW7 promotes esophageal squamous cell carcinoma (ESCC) progression by enhancing ANXA2 to activate the MAPK pathway [16]. Moreover, Wei et al. showed that another E3 enzyme, TRIM65, installed ubiquitin in amino acid residues of ANXA2 in urothelial carcinoma of the bladder [25]. Therefore, decreased HAR1A in NSCLC potentiates cancer growth and metastasis by upregulating the ANXA2.
We also investigated the effector signaling pathway downstream of ANXA2. Several studies disclosed the direct interaction of ANXA2 with p65 [17, 18, 37]. Trim33 promoted ANXA2’s interaction with p50/p65 subunits of NF-κB by catalyzing lysine 63 (K63)-linked ubiquitination of Annexin A2 (Anxa2) to stir inflammation response in psoriasis [18]. In the LUAD, LINC01614 formed a trimeric complex with ANXA2 and p65 to facilitate the latter two molecules’ interaction and the activation of NF-κB, eventually elevating cancer cell growth [17]. Consistently, we demonstrated that HAR1A overexpression lowered the activity of the NF-κB pathway by downregulating ANXA2. Therefore, decreased HAR1A in NSCLC potentiates cancer growth and metastasis by upregulating the ANXA2/NF-κB axis.
Finally, our results demonstrated that METTL3 suppressed ANXA2 levels and reduced stability. It turned out that YTHDF2 recognized the m6A modification in HAR1A to accelerate its degradation. Dysregulation of RNA m6A modification has been well known to be implicated in carcinogenesis. Methyltransferase Like 3 (METTL3), installing m6A in RNAs, is highly expressed in NSCLC and promotes cancer progress [19,20,21]. Apart from mRNAs, lncRNAs can also be regulated by m6A modification. Many studies demonstrated that YTHDF2 mediated the stability of m6A-modified RNAs in a cotext-dependent manner [38,39,40,41]. LINC02038, subjected to METTL3-mediated m6A modification, was a tumor suppressor and decreased in CRC. YTHDF2 was shown to recognize m6A modification in LINC02038 and trigger its degradation [38]. Consistently, Ye et al. reported that YTHDF2 sensed m6A modifications in the tumor-suppressive lncRNA CARMN and enabled m6A-caused breakdown of this lncRNA in cervical cancer [41]. Another study reported that METTL3-mediated m6A modification could stabilize LINC00839, an oncogenic lncRNA, in a YTHDF2-dependent manner. The upregulated LINC00839 was associated with glioblastoma progression and radiation resistance by activating Wnt/β-catenin activation [39].
In conclusion, our results indicated that HAR1A deficiency disables the ubiquitination and proteasomal degradation of ANXA2 to activate the NF-κB pathway, leading to lung cancer progress. The METTL3- YTHDF2 axis partially interprets the loss of HAR1A in NSCLC.
Materials and methods
Clinical samples
The NSCLC and matched adjacent normal tissues were collected from August 2022 to October 2022 in the Harbin Medical Cancer Hospital. Fresh tissue samples were snap-frozen in liquid nitrogen and stored in a deep freezer (−80 °C) until usage. The Ethical Committee of the Harbin Medical Cancer Hospital approved the study. Written informed consent was obtained by all participants in the study.
Cell culture
Normal human lung bronchial epithelial cell lines (HBE) and NSCLC cell lines (H1299, A549, H1650, and H1993) were purchased from the Cell Bank of Type Culture Collection of Chinese Academy of Sciences (CBTCCCAS, Shanghai, China). All the cells were cultured in DMEM (Corning, USA) with the addition of 10% fetal calf serum and 10% penicillin/streptomycin (Gibco, USA). The cultures were maintained in an incubator preset at 37 °C in a 5% CO2 humid environment.
Transfection
Both lentiviruses expressing HAR1A (LV-HAR1A) and control vectors (LV-vector) were purchased from the Genechem company (Shanghai, China). The full length of human HAR1A cDNA was directly synthesized to construct the HAR1A overexpression vector. HAR1A overexpression lentiviral vector GV717 (CMV enhancer-MCS-sv40-puromycin) and control lentiviral vector were constructed and packed using 293T cells. Stable cell lines overexpressing HAR1A or vector were established by lentiviral infection followed by puromycin (Calbiochem, USA) selection. Then, HAR1A expression was evaluated by quantitative PCR (qPCR). The knockdown of HAR1A was achieved by transfection of siRNA, specifically targeting the lncRNA (General Biol, China).
Short interference RNAs (siRNAs) against human METTL3 (siMETTL3), YTHDF1 (siYTHDF1), YTHDF2 (siYTHDF2), YTHDF3 (siYTHDF3) and matched negative controls (siNC) were provided by Genechem (Shanghai, China). Plasmids overexpressing METTL3, ANXA2, and respective controls (Ctrl) were cloned into the pcDNA3.1 vector (Genechem, China). The transfections were performed with INTERFERin® (Polyplus-transfection® SA) according to the manufacturer’s instructions. The target sequences used for the siRNAs are listed in Table S1.
Reverse transcription and quantitative (RT-qPCR)
TRIZOL reagent (Invitrogen) was used to extract total RNA. First-strand cDNA was prepared with the Transcriptor First-Strand cDNA Synthesis Kit (Roche, USA). Real-time PCR was performed using FastStart Universal SYBR Green Master (ROX) (Roche) on a 7500 Fast Real-Time PCR system (ABI, USA). The relative expression of HAR1A, Vimentin, N-cadherin, E-cadherin, ANXA2, U6, and GAPDH was detected at least in triplicate with indicated primers (Table S1). The melting-curve analyses were used to confirm PCR product specificity. Relative expression of genes of interest was determined using the 2–ΔΔCt method and normalized to GAPDH.
Western blot
The harvested cells were pelleted and lysed in ice-cold RIPA buffer with 0.1 M PMSF. Protein concentrations of samples were quantified by using the BCA Protein Assay Kit. Protein samples (30 µg) were separated by polyacrylamide electrophoresis, transferred onto polyvinylidene fluoride (PVDF) membranes and incubated with specific antibodies. The primary detection antibodies that were used in the study are listed in Table S1.
Subcellular fractionation
Cytoplasmic & Nuclear RNA Purification Kit (NORGEN BIOTEK CORP. Thorold, ON, Canada) was used to extract RNA from nuclear and cytoplasmic fractions of A549 and H1299 cells according to the manufacturer’s instructions. GAPDH and U6 exclusively expressed in nuclear and cytoplasm, respectively, were used as positive controls for HAR1A in the qRT-PCR assay.
Cell counting kit (CCK8) assay
Cell proliferation was assessed using a CCK8 kit (Dojindo, Japan) according to the manufacturer’s instructions. A total of 4 × 103 cells were seeded into 96-well plates and cultured at 37 °C. Medium containing 10% CCK8 replaced the original medium and incubated at 37 °C for 2 h, and the absorbance was finally determined at 450 nm using a microplate reader. For paclitaxel sensitivity, the cells were treated with different concentrations of paclitaxel for 24 h. After treatment, the media were removed, and absorbance was then measured at 450 nm.
Colony formation assay
In total, 600 cells were seeded in 6-well plates and cultured in a complete medium supplemented with 10% FBS. After 14 days, the cells were washed with PBS, fixed in methanol for 30 min, and stained with 0.1% crystal violet dye. Finally, the number of colonies was counted by three different individuals.
EdU assay
Cells were incubated with 100 μM EdU (Ribobio, Guangzhou, China) for 3 h at 37 °C and then fixed in 4% paraformaldehyde for 30 min. After that, the cells were permeabilized with 0.4% Triton X-100 for 10 min and incubated with Apollo® reagent (100 μl) for 30 min. Finally, the cells were stained with Hoechst for 30 min. The images were observed under an inverted fluorescence microscope. The cell proliferation rate was calculated using the ratio of EdU-positive cells (red) to Hoechst-positive cells (blue).
TUNEL assay
Apoptotic cells were identified by terminal deoxynucleotidyl transferase-mediated dUTP nick end-labeling (TUNEL) staining using the In Situ Cell Death Detection Kit (11684817910, Roche). The sections were analyzed by fluorescence microscopy (Olympus, Japan). The average number of TUNEL-positive cells in three images from each treatment group was calculated.
Tumor cell migration and invasion assays
Both cell migration and invasion experiments were conducted with Transwell polycarbonate membrane cell culture inserts (BD Biosciences, CA, USA). A total of 3 × 104 cells were added to the upper chamber, and the cells were allowed to migrate to the lower chamber for 24 h. (Matrigel was applied for the filter for invasion assay (BD Biosciences, CA, USA). The migrated/invaded cells were fixed with 4% paraformaldehyde and stained with 0.1% crystal violet staining for 30 min. The cells were counted in three independent fields under an optical microscope to calculate the transwell capacity.
Wound healing assay
Cells were seeded to 6‐well plate and incubated until approximately 90% confluence. A plastic tip was used to create a wound at the cell surface. Then, PBS buffer was used to remove dissociated cellular fragments. The speed of wound healing was observed and imaged using a microscope, and the closure rate was estimated. Experiments were repeated in triplicates.
Immunofluorescence staining
The cells were fixed with 4% paraformaldehyde for 30 min at room temperature and then permeabilized with 0.1% Triton X-100 for 15 min. After blocking with 5% BSA, the cells were incubated with the primary antibodies E-cadherin (ab40772, Abcam, 1:500), N-cadherin (ab18203, Abcam, 1:200), and Vimentin (ab92547, Abcam, 1:250) at 4 °C overnight. The next day, the cells were washed and incubated with secondary antibodies for 1 h at room temperature. Subsequently, cells were washed, and nuclei were counterstained with DAPI (Beyotime Company, China). All images were acquired using an inverted fluorescence microscope.
Animal models
For in vivo experiments, H1299 cell lines stably transfected with negative control (NC) and HAR1A expression (HAR1A) were first established. BALB/c nude mice (5 weeks old) were obtained from Beijing Vital River Laboratory Animal Technology Co, Ltd (Beijing, China). For the proliferation model, 20 nude mice were randomly assigned to four groups, and mice were injected subcutaneously with 1 ml of cell suspension (2 × 106 cells/mouse). After the tumors grew to approximately 100 mm3 in size, 15 mg/kg paclitaxel was administered once every four days for a total of three courses. Tumor volume was measured once every two days by using calipers at the indicated time points. The tumor volume was estimated by the following formula: volume (mm3) = 0.5 × length (mm) × width2 (mm2). The whole body weight of mice was measured once every two days as indicated. For the metastasis model, 1 × 106 cells were injected intravenously. Five weeks later, mice were sacrificed, and lungs were collected and fixed in 4% formaldehyde for H&E and immunofluorescent (IF) staining, respectively. The number of metastatic nodes in lung tissues was counted in five mice in each group. The Institutional Animal Care and Use Committee of the Center of Harbin Medical University approved the protocol.
RNA pull-down assay
HAR1A RAN pull-down assay was conducted using the Pierce™ Magnetic RNA-Protein Pull-Down (Thermo Scientific, USA) and MAXIscript® (Ambion, USA) kits based on protocols provided by manufacturers. Cells lysed in RIPA buffer were incubated with in vitro transcribed full-length HAR1A probes and antisense probes labeled by biotins (Gene Create, China). Briefly, NSCLC cell extracts were incubated with biotinylated HAR1A or antisense-HAR1A (control), affinity precipitated using streptavidin-conjugated beads. The RNA-binding protein complexes were collected, washed, eluted, and analyzed by Liquid Chromatography and Mass Spectrometry (LC/MS) and western blot.
Shotgun analysis
RNA pull-down samples were resolved in SDS-PAGE and stained with Bio-Safe Coomassie (Bio-Rad). Gels with protein bands were excised and minced into small pieces, followed by overnight digestion at 37OC. Peptides retrieved from the gel were dried and resuspended in a loading buffer. The Thermo Scientific UltiMate 3000 RSLCnano system combined with Thermo Scientific Q Exactive HF-X hybrid quadrupole-Orbitrap mass spectrometer was adopted to analyze peptides. Data-dependent (DDA) acquisition mode was applied to obtain the tandem mass spectrometry (MS/MS) data. We processed raw MS data using MaxQuant software (V1.6.6) using Andromeda, a peptide search engine based on a probabilistic scoring model. Protein sequences retrieved from the complete human proteome of Uniprot (Dec 2021, 20381 protein sequences) were utilized to determine the protein identities. The relative quantitative analysis was performed by calculating iBAQ values for identified proteins.
RNA immunoprecipitation (RIP)
Cells were lysed with lysis buffer containing protease and RNase inhibitors. The lysates were centrifuged at 15,000 × g for 10 min, and the resulting supernatants were used to perform RIP assay with the Magna RIP™ RNA-Binding Protein Immunoprecipitation Kit (Millipore, USA). Antibodies used for immunoprecipitation include primary antibodies against ANXA2, TRIM65, YTHDF2, m6A, and IgG. The precipitated complexes were then washed, and the RNA was isolated using RNAiso Plus (TAKARA) for subsequent qRT-PCR analysis.
RNA fluorescence in situ hybridization (FISH)
For FISH experiments, cells were grown on coverslips and fixed with 4% paraformaldehyde (Sigma-Aldrich, Germany) for 15 min. Subsequently, permeabilization was performed using 0.1% Triton X-100 (BioFroxx, Guangzhou, China) for 10 min. Following that, sections were treated with 20 μg/mL protease K (Servicebio, China) at 37 °C for 5 min. The samples were then hybridized overnight with 8 ng/μL m-HAR1A probe (5′-CY3-CGGACCGGAGGGAGAGCCGGGCGCAGAGACCGAGGCACA-CY3-3′) (Gene Pharma, China) at 37 °C, and subsequently incubated overnight at 4 °C with anti-ANXA2 (#8235, 1:100; CST, USA). After three PBS washes, cells were incubated with the secondary antibody (BosterBio, BA1105, China) for 1 h at room temperature in the dark. Nuclei were stained with DAPI (Servicebio) for 5 min. Finally, the slices were sealed with anti-fluorescence quenching tablets. Confocal microscopy (LSM880, Carl Zeiss) was used to capture all microscopic images.
Protein degradation assay
Cycloheximide (CHX) chase assay was used to determine ANXA2 protein stability. A549 and H1299 cells overexpressing HAR1A or control vector cells were seeded in the cell culture plates. After 24 h, CHX was added to the culture media at a final concentration of 200 μg/mL to halt de novo protein synthesis. Cells were collected at specified time points after CHX treatment, and the protein levels of ANXA2 and β-Actin were assessed using western blotting.
Ubiquitination assay
Ubiquitination assays were performed in LUAD cells with either control vectors or HAR1A overexpression. These cells were transfected with pcDNA3.1-HA-ubiquitin (HA-Ub) and Flag-ANXA2 plasmids. After 24 h of transfection, cells were treated with 50 μg/mL MG132 for 8 h, followed by lysis with ice-cold RIPA buffer (P0013C, Beyotime, China). The proteins in the cell lysate were immunoprecipitated using an anti-Flag antibody to enrich ubiquitinated ANXA2. The resulting immunoprecipitates were subjected to western blot analysis to determine ANXA2 protein and its ubiquitination levels using anti-HA and anti-Flag antibodies.
Co-immunoprecipitation (Co-IP)
Whole-cell lysates prepared with IP lysis buffer were used for endogenous IP. The supernatants were incubated with primary antibodies against TRIM65, ANXA2, or normal mouse/rabbit IgG. The antibody-protein complexes were then incubated with Protein A/G PLUS-Agarose (Santa Cruz Biotechnology). The agarose-antibody-protein complexes were collected using a magnetic separator and analyzed by western blot with corresponding antibodies.
RNA stability assay
The RNA stability assay was conducted following the previously described method (PMID: 31230592). A549 and H1299 cells were cultured in 6-well plates overnight. Actinomycin D (Med-ChemExpress) was added to inhibit gene transcription at a concentration of 5 μg/mL. The total RNA was subsequently extracted from harvested cells at different time points, and levels were quantified using RT-qPCR. The RNA levels in the indicated groups at different time points were calculated and normalized to GAPDH.
Statistical methods
Data are presented as the mean ± standard deviation of at least three independent experiments for each cellular experimental group and at least five independent experiments for each animal group. Between-group differences concerning continuous variables were assessed using Student’s t-test or one-way/two-way ANOVA whenever appropriate. P values < 0.05 were indicative of statistical significance. Statistical analyses were performed using SPSS 17.0 software and GraphPad Prism 8 software.
Data availability
The data sets in this study are available from the corresponding author upon reasonable request.
References
Statello L, Guo CJ, Chen LL, Huarte M. Gene regulation by long non-coding RNAs and its biological functions. Nat Rev Mol Cell Biol. 2021;22:96–118.
Goodall GJ, Wickramasinghe VO. RNA in cancer. Nat Rev Cancer. 2021;21:22–36.
Zhu Y, Zhou B, Hu X, Ying S, Zhou Q, Xu W, et al. LncRNA LINC00942 promotes chemoresistance in gastric cancer by suppressing MSI2 degradation to enhance c-Myc mRNA stability. Clin Transl Med. 2022;12:e703.
Chen Z, Chen X, Chen P, Yu S, Nie F, Lu B, et al. Long non-coding RNA SNHG20 promotes non-small cell lung cancer cell proliferation and migration by epigenetically silencing of P21 expression. Cell Death Dis. 2017;8:e3092.
Saha S, Kiran M, Kuscu C, Chatrath A, Wotton D, Mayo MW, et al. Long non-coding RNA DRAIC inhibits prostate cancer progression by interacting with IKK to Inhibit NF-kappaB activation. Cancer Res. 2020;80:950–63.
Wang H, Meng Q, Qian J, Li M, Gu C, Yang Y. Review: RNA-based diagnostic markers discovery and therapeutic targets development in cancer. Pharmacol Ther. 2022;234:108123.
Nagasaka M, Uddin MH, Al-Hallak MN, Rahman S, Balasubramanian S, Sukari A, et al. Liquid biopsy for therapy monitoring in early-stage non-small cell lung cancer. Mol. Cancer. 2021;20:82.
Zhao R, Fu J, Zhu L, Chen Y, Liu B. Designing strategies of small-molecule compounds for modulating non-coding RNAs in cancer therapy. J Hematol Oncol. 2022;15:14.
Ma J, Cao K, Ling X, Zhang P, Zhu J. LncRNA HAR1A suppresses the development of non-small cell lung cancer by inactivating the STAT3 pathway. Cancers. 2022;14:2845.
Lee CP, Ko AM, Nithiyanantham S, Lai CH, Ko YC. Long non-coding RNA HAR1A regulates oral cancer progression through the alpha-kinase 1, bromodomain 7, and myosin IIA axis. J. Mol. Med. 2021;99:1323–34.
Herrera-Lopez EE, Guerrero-Escalera D, Aguirre-Maldonado I, Lopez-Hernandez A, Montero H, Gutierrez-Nava MA, et al. Annexins A2 and A5 are potential early biomarkers of hepatocarcinogenesis. Sci. Rep. 2023;13:6948.
Yuan J, Yang Y, Gao Z, Wang Z, Ji W, Song W, et al. Tyr23 phosphorylation of Anxa2 enhances STAT3 activation and promotes proliferation and invasion of breast cancer cells. Breast Cancer Res Treat. 2017;164:327–40.
Luo CH, Liu QQ, Zhang PF, Li MY, Chen ZC, Liu YF. Prognostic significance of annexin II expression in non-small cell lung cancer. Clin Transl Oncol. 2013;15:938–46.
Liu ZY, Lin XT, Zhang YJ, Gu YP, Yu HQ, Fang L, et al. FBXW10-S6K1 promotes ANXA2 polyubiquitination and KRAS activation to drive hepatocellular carcinoma development in males. Cancer Lett. 2023;566:216257.
Sarkar S, Swiercz R, Kantara C, Hajjar KA, Singh P. Annexin A2 mediates up-regulation of NF-kappaB, beta-catenin, and stem cell in response to progastrin in mice and HEK-293 cells. Gastroenterology. 2011;140:583–95.e584.
Li Z, Pan Y, Yao J, Gao Y, Qian Y, Zheng M, et al. ANXA2 as a novel substrate of FBXW7 promoting esophageal squamous cell carcinoma via ERK phosphorylation. Biochem Biophys Res Commun. 2023;649:93–100.
Liu T, Han C, Fang P, Ma Z, Wang X, Chen H, et al. Cancer-associated fibroblast-specific lncRNA LINC01614 enhances glutamine uptake in lung adenocarcinoma. J Hematol Oncol. 2022;15:141.
Zhang J, Zhu J, Chen X, Xia H, Yang L. E3 ubiquitin ligase Trim33 ubiquitylates Annexin A2 to promote NF-kappaB induced skin inflammation in psoriasis. J Dermatol Sci. 2022;107:160–8.
Xiao H, Zhao R, Meng W, Liao Y. Effects and translatomics characteristics of a small-molecule inhibitor of METTL3 against non-small cell lung cancer. J Pharm Anal. 2023;13:625–39.
Xu X, Zhang P, Huang Y, Shi W, Mao J, Ma N, et al. METTL3-mediated m6A mRNA contributes to the resistance of carbon-ion radiotherapy in non-small-cell lung cancer. Cancer Sci. 2023;114:105–14.
Zhao S, Song P, Zhou G, Zhang D, Hu Y. METTL3 promotes the malignancy of non-small cell lung cancer by N6-methyladenosine modifying SFRP2. Cancer Gene Ther. 2023;30:1094–104.
Cheng L, Tong Q. Interaction of FLNA and ANXA2 promotes gefitinib resistance by activating the Wnt pathway in non-small-cell lung cancer. Mol Cell Biochem. 2021;476:3563–75.
Hwang SK, Han SY, Jeong YJ, Magae J, Bae YS, Chang YC. 4-O-methylascochlorin activates autophagy by activating AMPK and suppressing c-Myc in glioblastoma. J Biochem Mol. Toxicol. 2020;34:e22552.
Saidova AA, Potashnikova DM, Tvorogova AV, Paklina OV, Veliev EI, Knyshinsky GV, et al. Myosin 1C isoform A is a novel candidate diagnostic marker for prostate cancer. PLoS ONE. 2021;16:e0251961.
Wei WS, Chen X, Guo LY, Li XD, Deng MH, Yuan GJ, et al. TRIM65 supports bladder urothelial carcinoma cell aggressiveness by promoting ANXA2 ubiquitination and degradation. Cancer Lett. 2018;435:10–22.
Zhao S, Li B, Zhao R, Pan Z, Zhang S, Qiu W, et al. Hypoxia-induced circADAMTS6 in a TDP43-dependent manner accelerates glioblastoma progression via ANXA2/ NF-kappaB pathway. Oncogene. 2023;42:138–53.
Tang G, Yu C, Xiang K, Gao M, Liu Z, Yang B, et al. Inhibition of ANXA2 regulated by SRF attenuates the development of severe acute pancreatitis by inhibiting the NF-kappaB signaling pathway. Inflamm Res. 2022;71:1067–78.
Zhang L, Zhou Y, Huang T, Cheng AS, Yu J, Kang W, et al. The interplay of LncRNA-H19 and its binding partners in physiological process and gastric carcinogenesis. Int J Mol Sci. 2017;18:450.
Bhan A, Hussain I, Ansari KI, Kasiri S, Bashyal A, Mandal SS. Antisense transcript long non-coding RNA (lncRNA) HOTAIR is transcriptionally induced by estradiol. J Mol Biol. 2013;425:3707–22.
Bhan A, Hussain I, Ansari KI, Bobzean SA, Perrotti LI, Mandal SS. Bisphenol-A and diethylstilbestrol exposure induces the expression of breast cancer associated long non-coding RNA HOTAIR in vitro and in vivo. J Steroid Biochem Mol Biol. 2014;141:160–70.
Shi Z, Luo Y, Zhu M, Zhou Y, Zheng B, Wu D, et al. Expression analysis of long non-coding RNA HAR1A and HAR1B in HBV-induced hepatocullular carcinoma in Chinese patients. Lab Med. 2019;50:150–7.
Shahabi S, Kumaran V, Castillo J, Cong Z, Nandagopal G, Mullen DJ, et al. LINC00261 is an epigenetically regulated tumor suppressor essential for activation of the DNA damage response. Cancer Res. 2019;79:3050–62.
Wang KC, Chang HY. Molecular mechanisms of long non-coding RNAs. Mol. Cell. 2011;43:904–14.
Wang J, He Z, Liu X, Xu J, Jiang X, Quan G, et al. LINC00941 promotes pancreatic cancer malignancy by interacting with ANXA2 and suppressing NEDD4L-mediated degradation of ANXA2. Cell Death Dis. 2022;13:718.
Yi Y, Zeng S, Wang Z, Wu M, Ma Y, Ye X, et al. Cancer-associated fibroblasts promote epithelial-mesenchymal transition and EGFR-TKI resistance of non-small cell lung cancers via HGF/IGF-1/ANXA2 signaling. Biochim Biophys Acta Mol Basis Dis. 2018;1864:793–803.
Wang Y, Wang Y, Liu W, Ding L, Zhang X, Wang B, et al. TIM-4 orchestrates mitochondrial homeostasis to promote lung cancer progression via ANXA2/PI3K/AKT/OPA1 axis. Cell Death Dis. 2023;14:141.
Cheng L, Wang H, Maboh R, Mao G, Wu X, Chen H. LncRNA LINC00281/Annexin A2 regulates vascular smooth muscle cell phenotype switching via the nuclear factor-kappa B signaling pathway. J. Cardiovasc Transl Res. 2022;15:971–84.
Liu W, Zhang Z, Luo X, Qian K, Huang B, Liang J, et al. m(6)A‑mediated LINC02038 inhibits colorectal cancer progression via regulation of the FAM172A/PI3K/AKT pathway via competitive binding with miR‑552‑5p. Int J Oncol. 2023;63:81.
Yin J, Ding F, Cheng Z, Ge X, Li Y, Zeng A, et al. METTL3-mediated m6A modification of LINC00839 maintains glioma stem cells and radiation resistance by activating Wnt/beta-catenin signaling. Cell Death Dis. 2023;14:417.
Tu B, Song K, Zhou Y, Sun H, Liu ZY, Lin LC, et al. METTL3 boosts mitochondrial fission and induces cardiac fibrosis by enhancing LncRNA GAS5 methylation. Pharm Res. 2023;194:106840.
Yu B, Li X, Yan W, Ding B, Zhang X, Shen S, et al. Post-transcriptional regulation of tumor suppressor gene lncRNA CARMN via m(6)A modification and miRNA regulation in cervical cancer. J Cancer Res Clin Oncol. 2023;149:10307–18.
Funding
This study was supported by the National Natural Science Foundation of China (No. 82172786 to JM), the Haiyan Foundation of Harbin Medical University Cancer Hospital (No. JJMS2023-04 to XL) and Beijing Medical Award Foundation (No. YXJL-2023-0091-0054 to XL).
Author information
Authors and Affiliations
Contributions
JM, JZ, and XL designed the experiments and wrote the manuscript. XL, CQ, KC, and ML performed experiments and analyzed the data. LZ sorted out the pictures and drew them into figures. YY and JZ collected the human clinical samples. ML drew a schematic diagram of molecular mechanisms. All authors reviewed and approved the manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Ling, X., Qi, C., Cao, K. et al. METTL3-mediated deficiency of lncRNA HAR1A drives non-small cell lung cancer growth and metastasis by promoting ANXA2 stabilization. Cell Death Discov. 10, 203 (2024). https://doi.org/10.1038/s41420-024-01965-w
Received:
Revised:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41420-024-01965-w