Abstract
Background
Considerable interobserver variability exists in diagnosis of ovarian high-grade endometrioid carcinoma (HGEC) and high-grade serous carcinoma (HGSC) due to histopathological similarities. While homologous recombination deficiency (HRD) correlates with drug sensitivity in HGSC, the molecular features of HGEC are unclear.
Methods
Fresh-frozen samples from 15 ovarian HGECs and 274 ovarian HGSCs in the JGOG-TR2 cohort were submitted to targeted DNA sequencing, RNA sequencing, DNA methylation array, and SNP array. We additionally analyzed 555 ovarian HGSCs from TCGA-OV and 287 endometrial high-grade carcinomas from TCGA-UCEC.
Results
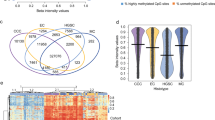
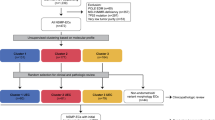
Unsupervised clustering using copy number signatures identified four distinct tumor groups (C1, C2, C3 and C4). C1 (n = 41) showed CCNE1 amplification and poor survival. C2 (n = 160) and C3 (n = 59) showed high BRCA1/2 alteration frequency with low and moderate ploidy, respectively. C4 (n = 22) was characterized by favorable outcome, higher HGEC proportion, no BRCA1/2 alteration or CCNE1 amplification, and low levels of HRD score, ploidy, intra-tumoral heterogeneity, cell proliferation rate, and WT1 gene expression. Notably, C4 exhibited a normal endometrium-like DNA methylation profile, thus, defined as “HGEC-type” tumors, which were also identified in TCGA-OV and TCGA-UCEC.
Conclusions
Ovarian “HGEC-type” tumors present a non-HRD status, favorable prognosis, and endometrial differentiation, possibly constituting a subset of clinically diagnosed HGSCs.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 24 print issues and online access
$259.00 per year
only $10.79 per issue
Buy this article
- Purchase on SpringerLink
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
Data availability
Processed data and analysis codes to reproduce the results in this study are available in Supplementary table 4 and the GitHub project page (https://github.com/shirotak/JGOG_HGEOC). Genomic data of the JGOG3025-TR2 cohort, including RNA-seq, SNP array, and DNA methylation array are available from SRA (PRJNA1092599) and NCBI-GEO (GSE263455). Targeted sequencing data and clinical information are available upon reasonable request to JGOG (info@jgog.gr.jp, https://jgog.gr.jp/en/index.html). Controlled access data for the TCGA cases were obtained through dbGaP (access permission phs000178). Gene expression data for the ICON7 cases were obtained through the European Genome-Phenome Archive (accession number EGAS00001003487).
References
Zhang Y, Luo G, Li M, Guo P, Xiao Y, Ji H, et al. Global patterns and trends in ovarian cancer incidence: age, period and birth cohort analysis. BMC Cancer. 2019;19:984.
Nakai H, Higashi T, Kakuwa T, Matsumura N. Trends in gynecologic cancer in Japan: incidence from 1980 to 2019 and mortality from 1981 to 2021. Int J Clin Oncol. 2024;29:363–71.
Koshiyama M, Matsumura N, Konishi I. Recent concepts of ovarian carcinogenesis: type I and type II. Biomed Res Int. 2014;2014:934261.
Yamaguchi K, Mandai M, Oura T, Matsumura N, Hamanishi J, Baba T, et al. Identification of an ovarian clear cell carcinoma gene signature that reflects inherent disease biology and the carcinogenic processes. Oncogene. 2010;29:1741–52.
Yamaguchi K, Huang Z, Matsumura N, Mandai M, Okamoto T, Baba T, et al. Epigenetic determinants of ovarian clear cell carcinoma biology. Int J Cancer. 2014;135:585–97.
Huang RY, Chen GB, Matsumura N, Lai HC, Mori S, Li J, et al. Histotype-specific copy-number alterations in ovarian cancer. BMC Med Genomics. 2012;5:47.
Yoshihara K, Baba T, Tokunaga H, Nishino K, Sekine M, Takamatsu S, et al. Homologous recombination inquiry through ovarian malignancy investigations: JGOG3025 Study. Cancer Sci. 2023;114:2515–23.
Nakai H, Matsumura N. The roles and limitations of bevacizumab in the treatment of ovarian cancer. Int J Clin Oncol. 2022;27:1120–6.
Nakai H, Matsumura N. Individualization in the first-line treatment of advanced ovarian cancer based on the mechanism of action of molecularly targeted drugs. Int J Clin Oncol. 2022;27:1001–12.
Jang JYA, Yanaihara N, Pujade-Lauraine E, Mikami Y, Oda K, Bookman M, et al. Update on rare epithelial ovarian cancers: based on the Rare Ovarian Tumors Young Investigator Conference. J Gynecol Oncol. 2017;28:e54.
Mandai M, Yamaguchi K, Matsumura N, Baba T, Konishi I. Ovarian cancer in endometriosis: molecular biology, pathology, and clinical management. Int J Clin Oncol. 2009;14:383–91.
Soyama H, Miyamoto M, Takano M, Iwahashi H, Kato K, Sakamoto T, et al. A Pathological Study Using 2014 WHO Criteria Reveals Poor Prognosis of Grade 3 Ovarian Endometrioid Carcinomas. Vivo. 2018;32:597–602.
Schwartz DR, Kardia SL, Shedden KA, Kuick R, Michailidis G, Taylor JM, et al. Gene expression in ovarian cancer reflects both morphology and biological behavior, distinguishing clear cell from other poor-prognosis ovarian carcinomas. Cancer Res. 2002;62:4722–9.
Tothill RW, Tinker AV, George J, Brown R, Fox SB, Lade S, et al. Novel molecular subtypes of serous and endometrioid ovarian cancer linked to clinical outcome. Clin Cancer Res. 2008;14:5198–208.
Kurman RJ, Carcangiu ML, Herrington CS, Young RH, editors. WHO Classification of Tumours of the Female Reproductive Organs: WHO Classification of Tumours, 4th ed., Volume 6. Lyon: International Agency for Research on Cancer; 2014.
WHO Classification of Tumours Editorial Board. Female genital tumours: WHO Classification of Tumours, 5th ed., Volume 4. Lyon: International Agency for Research on Cancer; 2020.
Lim D, Murali R, Murray MP, Veras E, Park KJ, Soslow RA. Morphological and Immunohistochemical Reevaluation of Tumors Initially Diagnosed as Ovarian Endometrioid Carcinoma With Emphasis on High-grade Tumors. Am J Surg Pathol. 2016;40:302–12.
Mirza MR, Monk BJ, Herrstedt J, Oza AM, Mahner S, Redondo A, et al. Niraparib Maintenance Therapy in Platinum-Sensitive, Recurrent Ovarian Cancer. N Engl J Med. 2016;375:2154–64.
Ledermann J, Harter P, Gourley C, Friedlander M, Vergote I, Rustin G, et al. Olaparib maintenance therapy in platinum-sensitive relapsed ovarian cancer. N Engl J Med. 2012;366:1382–92.
Moore K, Colombo N, Scambia G, Kim BG, Oaknin A, Friedlander M, et al. Maintenance Olaparib in Patients with Newly Diagnosed Advanced Ovarian Cancer. N Engl J Med. 2018;379:2495–505.
Ray-Coquard I, Pautier P, Pignata S, Pérol D, González-Martín A, Berger R, et al. Olaparib plus Bevacizumab as First-Line Maintenance in Ovarian Cancer. N Engl J Med. 2019;381:2416–28.
González-Martín A, Pothuri B, Vergote I, DePont Christensen R, Graybill W, Mirza MR, et al. Niraparib in Patients with Newly Diagnosed Advanced Ovarian Cancer. N Engl J Med. 2019;381:2391–402.
Li N, Zhu J, Yin R, Wang J, Pan L, Kong B, et al. Treatment With Niraparib Maintenance Therapy in Patients With Newly Diagnosed Advanced Ovarian Cancer: A Phase 3 Randomized Clinical Trial. JAMA Oncol. 2023;9:1230–7.
Takamatsu S, Yoshihara K, Baba T, Shimada M, Yoshida H, Kajiyama H, et al. Prognostic relevance of HRDness gene expression signature in ovarian high-grade serous carcinoma; JGOG3025-TR2 study. Br J Cancer. 2023;128:1095–104.
Niu B, Ye K, Zhang Q, Lu C, Xie M, McLellan MD, et al. MSIsensor: microsatellite instability detection using paired tumor-normal sequence data. Bioinformatics. 2014;30:1015–6.
De’ Angelis GL, Bottarelli L, Azzoni C, De’ Angelis N, Leandro G, Di Mario F, et al. Microsatellite instability in colorectal cancer. Acta Biomed. 2018;89:97–101.
Wang K, Li M, Hakonarson H. ANNOVAR: functional annotation of genetic variants from high-throughput sequencing data. Nucleic Acids Res. 2010;38:e164.
Chakravarty D, Gao J, Phillips SM, Kundra R, Zhang H, Wang J, et al. OncoKB: A Precision Oncology Knowledge Base. JCO Precis Oncol. 2017;2017:PO.17.00011.
Cline MS, Liao RG, Parsons MT, Paten B, Alquaddoomi F, Antoniou A, et al. BRCA Challenge: BRCA Exchange as a global resource for variants in BRCA1 and BRCA2. PLoS Genet. 2018;14:e1007752.
Van Loo P, Nordgard SH, Lingjærde OC, Russnes HG, Rye IH, Sun W, et al. Allele-specific copy number analysis of tumors. Proc Natl Acad Sci USA. 2010;107:16910–5.
Khandekar A, Vangara R, Barnes M, Díaz-Gay M, Abbasi A, Bergstrom EN, et al. Visualizing and exploring patterns of large mutational events with SigProfilerMatrixGenerator. BMC Genomics. 2023;24:469.
Sztupinszki Z, Diossy M, Krzystanek M, Reiniger L, Csabai I, Favero F, et al. Migrating the SNP array-based homologous recombination deficiency measures to next generation sequencing data of breast cancer. NPJ Breast Cancer. 2018;4:16.
Ritchie ME, Phipson B, Wu D, Hu Y, Law CW, Shi W, et al. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 2015;43:e47.
Law CW, Chen Y, Shi W, Smyth GK. voom: Precision weights unlock linear model analysis tools for RNA-seq read counts. Genome Biol. 2014;15:R29.
Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15:550.
Barbie DA, Tamayo P, Boehm JS, Kim SY, Moody SE, Dunn IF, et al. Systematic RNA interference reveals that oncogenic KRAS-driven cancers require TBK1. Nature. 2009;462:108–12.
Loyfer N, Magenheim J, Peretz A, Cann G, Bredno J, Klochendler A, et al. A DNA methylation atlas of normal human cell types. Nature. 2023;613:355–64.
Minegishi R, Gotoh O, Tanaka N, Maruyama R, Chang JT, Mori S. A method of sample-wise region-set enrichment analysis for DNA methylomics. Epigenomics. 2021;13:1081–93.
Cavalcante RG, Sartor MA. annotatr: genomic regions in context. Bioinformatics. 2017;33:2381–3.
Bartlett TE, Jones A, Goode EL, Fridley BL, Cunningham JM, Berns EM, et al. Intra-Gene DNA Methylation Variability Is a Clinically Independent Prognostic Marker in Women’s Cancers. PLoS One. 2015;10:e0143178.
Pisanic TR, Wang Y, Sun H, Considine M, Li L, Wang TH, et al. Methylomic Landscapes of Ovarian Cancer Precursor Lesions. Clin Cancer Res. 2020;26:6310–20.
Chan DW, Lam WY, Chen F, Yung MMH, Chan YS, Chan WS, et al. Genome-wide DNA methylome analysis identifies methylation signatures associated with survival and drug resistance of ovarian cancers. Clin Epigenetics. 2021;13:142.
Beddows I, Fan H, Heinze K, Johnson BK, Leonova A, Senz J, et al. Cell State of Origin Impacts Development of Distinct Endometriosis-Related Ovarian Carcinoma Histotypes. Cancer Res. 2024;84:26–38.
Wu R, Zhai Y, Kuick R, Karnezis AN, Garcia P, Naseem A, et al. Impact of oviductal versus ovarian epithelial cell of origin on ovarian endometrioid carcinoma phenotype in the mouse. J Pathol. 2016;240:341–51.
Ramakrishna M, Williams LH, Boyle SE, Bearfoot JL, Sridhar A, Speed TP, et al. Identification of candidate growth promoting genes in ovarian cancer through integrated copy number and expression analysis. PLoS One. 2010;5:e9983.
Wu YH, Chang TH, Huang YF, Huang HD, Chou CY. COL11A1 promotes tumor progression and predicts poor clinical outcome in ovarian cancer. Oncogene. 2014;33:3432–40.
Uehara Y, Oda K, Ikeda Y, Koso T, Tsuji S, Yamamoto S, et al. Correction: Integrated Copy Number and Expression Analysis Identifies Profiles of Whole-Arm Chromosomal Alterations and Subgroups with Favorable Outcome in Ovarian Clear Cell Carcinomas. PLoS One. 2015;10:e0132751.
Desbois M, Udyavar AR, Ryner L, Kozlowski C, Guan Y, Dürrbaum M, et al. Integrated digital pathology and transcriptome analysis identifies molecular mediators of T-cell exclusion in ovarian cancer. Nat Commun. 2020;11:5583.
Bell D, Berchuck A, Birrer M, Chien J, Cramer D, Dao F, et al. Integrated genomic analyses of ovarian carcinoma. Nature. 2011;474:609–15.
Kandoth C, Schultz N, Cherniack AD, Akbani R, Liu Y, Shen H, et al. Integrated genomic characterization of endometrial carcinoma. Nature. 2013;497:67–73.
Grossman RL, Heath AP, Ferretti V, Varmus HE, Lowy DR, Kibbe WA, et al. Toward a Shared Vision for Cancer Genomic Data. N Engl J Med. 2016;375:1109–12.
Wu P, Heins ZJ, Muller JT, Katsnelson L, de Bruijn I, Abeshouse AA, et al. Integration and Analysis of CPTAC Proteomics Data in the Context of Cancer Genomics in the cBioPortal. Mol Cell Proteom. 2019;18:1893–8.
Gao J, Aksoy BA, Dogrusoz U, Dresdner G, Gross B, Sumer SO, et al. Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sci Signal. 2013;6:pl1.
Lin Q, Wagner W, Zenke M. Analysis of genome-wide DNA methylation profiles by BeadChip technology. Methods Mol Biol. 2013;1049:21–33.
Zhang Y, Parmigiani G, Johnson WE. ComBat-seq: batch effect adjustment for RNA-seq count data. NAR Genom Bioinform. 2020;2:lqaa078.
Cybulska P, Paula ADC, Tseng J, Leitao MM, Bashashati A, Huntsman DG, et al. Molecular profiling and molecular classification of endometrioid ovarian carcinomas. Gynecol Oncol. 2019;154:516–23.
Hollis RL, Thomson JP, Stanley B, Churchman M, Meynert AM, Rye T, et al. Molecular stratification of endometrioid ovarian carcinoma predicts clinical outcome. Nat Commun. 2020;11:4995.
Assem H, Rambau PF, Lee S, Ogilvie T, Sienko A, Kelemen LE, et al. High-grade Endometrioid Carcinoma of the Ovary: A Clinicopathologic Study of 30 Cases. Am J Surg Pathol. 2018;42:534–44.
Konishi I, Abiko K, Hayashi T, Yamanoi K, Murakami R, Yamaguchi K, et al. Peritoneal dissemination of high-grade serous ovarian cancer: pivotal roles of chromosomal instability and epigenetic dynamics. J Gynecol Oncol. 2022;33:e83.
Tao Z, Wang S, Wu C, Wu T, Zhao X, Ning W, et al. The repertoire of copy number alteration signatures in human cancer. Brief Bioinform. 2023;24:bbad053.
Kuhn E, Wang TL, Doberstein K, Bahadirli-Talbott A, Ayhan A, Sehdev AS, et al. CCNE1 amplification and centrosome number abnormality in serous tubal intraepithelial carcinoma: further evidence supporting its role as a precursor of ovarian high-grade serous carcinoma. Mod Pathol. 2016;29:1254–61.
Otsuka I. Mechanisms of High-Grade Serous Carcinogenesis in the Fallopian Tube and Ovary: Current Hypotheses, Etiologic Factors, and Molecular Alterations. Int J Mol Sci. 2021;22:4409.
Cochrane DR, Tessier-Cloutier B, Lawrence KM, Nazeran T, Karnezis AN, Salamanca C, et al. Clear cell and endometrioid carcinomas: are their differences attributable to distinct cells of origin. J Pathol. 2017;243:26–36.
Madore J, Ren F, Filali-Mouhim A, Sanchez L, Köbel M, Tonin PN, et al. Characterization of the molecular differences between ovarian endometrioid carcinoma and ovarian serous carcinoma. J Pathol. 2010;220:392–400.
Liu Z, Yamanouchi K, Ohtao T, Matsumura S, Seino M, Shridhar V, et al. High levels of Wilms’ tumor 1 (WT1) expression were associated with aggressive clinical features in ovarian cancer. Anticancer Res. 2014;34:2331–40.
Köbel M, Kalloger SE, Boyd N, McKinney S, Mehl E, Palmer C, et al. Ovarian carcinoma subtypes are different diseases: implications for biomarker studies. PLoS Med. 2008;5:e232.
Gilks CB, Ionescu DN, Kalloger SE, Köbel M, Irving J, Clarke B, et al. Tumor cell type can be reproducibly diagnosed and is of independent prognostic significance in patients with maximally debulked ovarian carcinoma. Hum Pathol. 2008;39:1239–51.
Pierson WE, Peters PN, Chang MT, Chen LM, Quigley DA, Ashworth A, et al. An integrated molecular profile of endometrioid ovarian cancer. Gynecol Oncol. 2020;157:55–61.
Kramer C, Lanjouw L, Ruano D, Ter Elst A, Santandrea G, Solleveld-Westerink N, et al. Causality and functional relevance of BRCA1 and BRCA2 pathogenic variants in non-high-grade serous ovarian carcinomas. J Pathol. 2024;262:137–46.
Takaya H, Nakai H, Takamatsu S, Mandai M, Matsumura N. Homologous recombination deficiency status-based classification of high-grade serous ovarian carcinoma. Sci Rep. 2020;10:2757.
Khoo SK, Hurst T, Kearsley J, Dickie G, Free K, Parsons PG, et al. Prognostic significance of tumor ploidy in patients with advanced ovarian carcinoma. Gynecol Oncol. 1990;39:284–8.
Takaya H, Nakai H, Sakai K, Nishio K, Murakami K, Mandai M, et al. Intratumor heterogeneity and homologous recombination deficiency of high-grade serous ovarian cancer are associated with prognosis and molecular subtype and change in treatment course. Gynecol Oncol. 2020;156:415–22.
D’Andrilli G, Kumar C, Scambia G, Giordano A. Cell cycle genes in ovarian cancer: steps toward earlier diagnosis and novel therapies. Clin Cancer Res. 2004;10:8132–41.
D’Andrilli G, Giordano A, Bovicelli A. Epithelial ovarian cancer: the role of cell cycle genes in the different histotypes. Open Clin Cancer J. 2008;2:7–12.
Wu R, Hendrix-Lucas N, Kuick R, Zhai Y, Schwartz DR, Akyol A, et al. Mouse model of human ovarian endometrioid adenocarcinoma based on somatic defects in the Wnt/beta-catenin and PI3K/Pten signaling pathways. Cancer Cell. 2007;11:321–33.
Singhal A, Li BT, O’Reilly EM. Targeting KRAS in cancer. Nat Med. 2024;30:969–83.
Hein KZ, Stephen B, Fu S. Therapeutic Role of Synthetic Lethality in ARID1A-Deficient Malignancies. J Immunother Precis Oncol. 2024;7:41–52.
Glaviano A, Foo ASC, Lam HY, Yap KCH, Jacot W, Jones RH, et al. PI3K/AKT/mTOR signaling transduction pathway and targeted therapies in cancer. Mol Cancer. 2023;22:138.
Murakami K, Kotani Y, Nakai H, Matsumura N. Endometriosis-Associated Ovarian Cancer: The Origin and Targeted Therapy. Cancers. 2020;12:1676.
Reade CJ, McVey RM, Tone AA, Finlayson SJ, McAlpine JN, Fung-Kee-Fung M, et al. The fallopian tube as the origin of high grade serous ovarian cancer: review of a paradigm shift. J Obstet Gynaecol Can. 2014;36:133–40.
Garavaglia E, Sigismondi C, Ferrari S, Candiani M. The origin of endometriosis-associated ovarian cancer from uterine neoplastic lesions. Med Hypotheses. 2018;110:80–2.
Moran S, Martínez-Cardús A, Sayols S, Musulén E, Balañá C, Estival-Gonzalez A, et al. Epigenetic profiling to classify cancer of unknown primary: a multicentre, retrospective analysis. Lancet Oncol. 2016;17:1386–95.
Papanicolau-Sengos A, Aldape K. DNA Methylation Profiling: An Emerging Paradigm for Cancer Diagnosis. Annu Rev Pathol. 2022;17:295–321.
Paracchini L, Mannarino L, Romualdi C, Zadro R, Beltrame L, Fuso Nerini I, et al. Genomic instability analysis in DNA from Papanicolaou test provides proof-of-principle early diagnosis of high-grade serous ovarian cancer. Sci Transl Med. 2023;15:eadi2556.
Agarwal A, Yadav S, Dusane R, Menon S, Rekhi B, Deodhar KK. Endometrial serous carcinoma: A retrospective review of histological features & their clinicopathological association with disease-free survival & overall survival. Indian J Med Res. 2022;156:83–93.
Kommoss F, Faruqi A, Gilks CB, Lamshang Leen S, Singh N, Wilkinson N, et al. Uterine Serous Carcinomas Frequently Metastasize to the Fallopian Tube and Can Mimic Serous Tubal Intraepithelial Carcinoma. Am J Surg Pathol. 2017;41:161–70.
Jarboe EA, Miron A, Carlson JW, Hirsch MS, Kindelberger D, Mutter GL, et al. Coexisting intraepithelial serous carcinomas of the endometrium and fallopian tube: frequency and potential significance. Int J Gynecol Pathol. 2009;28:308–15.
de Jonge MM, de Kroon CD, Jenner DJ, Oosting J, de Hullu JA, Mourits MJE, et al. Endometrial Cancer Risk in Women With Germline BRCA1 or BRCA2 Mutations: Multicenter Cohort Study. J Natl Cancer Inst. 2021;113:1203–11.
Cortés-Ciriano I, Gulhan DC, Lee JJ, Melloni GEM, Park PJ. Computational analysis of cancer genome sequencing data. Nat Rev Genet. 2022;23:298–314.
Acknowledgements
This work was supported by AstraZeneca K.K. and Merck Sharp & Dohme Corp as the programme of Externally Sponsored Research (ESR-19-14550) and in part by the Japan Agency for Medical Research and development, AMED under Grant Number JP21tm0124005. We would like to thank all the JGOG members who participated in the JGOG3025-TR2 study; Drs. Makio Shozu, Hiroyuki Shigeta, Kazuhiro Takehara, Akira Kikuchi, Toyomi Sato, Akinori Oki, Shinya Yoshioka, Shinya Sato, Ryuji Kawaguchi, Hisafumi Okura, Takeshi Iwasa, Shoji Kamiura, Masato Kamitomo, Yoichi Aoki, Nao Suzuki, Yoshio Yoshida, Tadashi Kimura, Daisuke Aoki, Kazuyoshi Kato, Hiroaki Kobayashi, Hidemichi Watari, Etsuko Miyagi, Tsuyoshi Saito, Yoshihito Yokoyama, Tsunekazu Kita, Takashi Matsumoto, Satoshi Nagase, Toshiya Yamamoto, Yukio Hirano, Tomoaki Ikeda, Shiro Suzuki, Keiya Fujimori, Nagamasa Maeda, Naohiko Umesaki, Masatoshi Sugita, and Akira Kouyama.
Author information
Authors and Affiliations
Contributions
ST: data analysis and writing the manuscript; RTH: data analysis and review of the manuscript ; KY: sample collection and data analysis; TB: sample collection and review of the manuscript; MS: sample collection and review of the manuscript; HY: sample collection and review of the manuscript; AO: sample collection and review of the manuscript; HK: sample collection and review of the manuscript; KO: sample collection and data analysis; MM: data analysis and review of the manuscript; TE: sample collection and review of the manuscript; NM: design of this study, data analysis and writing the manuscript.
Corresponding author
Ethics declarations
Competing interests
NM received a research grant from AstraZeneca. NM received lecture fees from AstraZeneca, Takeda Pharmaceutical, Eisai, MSD and Chugai Pharmaceutical, and is an outside director of Takara Bio. TB received lecture fees from AstraZeneca. KY received lecture fees and a research grant from AstraZeneca. There are no other competing interests related to this paper.
Ethics
For the JGOG3025 study, i.e., clinical data analysis, frozen tumor tissue collection, target sequencing, and future analyses of tumor tissue, written informed consent was obtained from all patients with approval from the Institutional Review Board at each JGOG participating site prior to the start of the study [7]. The JGOG3025-TR2 study was then conducted with the approval of the Ethics Committee of JGOG and the Institutional Ethics Committee of Kindai University (approval number; 29-167), with opt-out patient consent. This study was performed in accordance with the Declaration of Helsinki.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Takamatsu, S., Hillman, R.T., Yoshihara, K. et al. Molecular classification of ovarian high-grade serous/endometrioid carcinomas through multi-omics analysis: JGOG3025-TR2 study. Br J Cancer (2024). https://doi.org/10.1038/s41416-024-02837-x
Received:
Revised:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41416-024-02837-x