Dear Editor,
Approximately 25–35% of adult patients with acute myeloid leukemia (AML) carries NPM1 mutation, which generally indicated a favorable outcome in the absence of FLT3-ITD mutation [1]. NPM1 mutations are absent in clonal hematopoiesis, and have been considered as AML initiating lesions [2]. Research on co-mutation characteristics of NPM1-mutated patients concentrated on FLT3-ITD, which has been suggested to hold a negative prognostic impact on NPM1-mutated patients by several large retrospective clinical studies [3, 4]. Besides FLT3-ITD, although there remains controversy, other high-frequency co-mutations such as DNMT3A, IDH1, IDH2, FLT3-TKD, NRAS, and WT1 mutations have also been pointed out to affect the prognosis of NPM1-mutated patients [3, 5,6,7,8,9]. Indeed, identification of specific co-mutation combinations other than FLT3-ITD mutation is essential for precise risk stratification and treatment strategy optimization for NPM1-mutated AML patients. Since allogeneic hematopoietic stem cell transplantation (allo-HSCT) is generally considered to improve the long-term outcome of most adverse-risk and suitable intermediate-risk AML patients, for NPM1-mutated AML patients, it is imperative to revisit the co-mutation profiles to determine the optimal population who may benefit from allo-HSCT.
In this study, we conducted a retrospective analysis of newly diagnosed adult AML patients with NPM1 mutations (acute promyelocytic leukemia excluded) in our center diagnosed from October 2018 to December 2022, focusing on exploring the therapeutic and prognostic significance of co-mutation characteristics in AML patients with NPM1 mutations. Patients who received at least one complete course of induction therapy were included in the further outcome analysis. Table S1 provided details of induction chemotherapy. We evaluated efficacy after two induction cycles, unless patients achieved CR/CRi after receiving only one induction cycle or discontinued treatment. Response evaluation was performed according to the NCCN guidelines for AML (version 3. 2023) and was categorized as CR/CRi or non-CR/CRi (including PR and NR) cohort [10]. Overall survival (OS) was defined as the time interval from treatment initiation until death due to any reason. Event-free survival (EFS) was defined as the time interval from treatment initiation to the occurrence of induction failure, relapse, or death, whichever came first. Disease-free survival (DFS) was defined as the time interval from disease remission to the occurrence of relapse or death, whichever came first. The study was conducted in accordance to the Declaration of Helsinki and was approved by the Ethics Committee of the First Affiliated Hospital of Zhejiang University College of Medicine (Hangzhou, China, Ethics Approval Number: IIT20240304A). All statistical analyses were performed using GraphPad Prism 7.0 software (GraphPad Software, CA, USA) and SPSS 23.0 (SPSS Inc., Chicago, IL).
One hundred ninety-two newly diagnosed NPM1-mutated AML patients detected through next-generation sequencing (NGS) were analyzed (Tables S2–S4). Twenty NPM1 mutants were identified, most of which were located in exon 12 and manifested as 4 base pair duplication/insertion alteration. Seven non-exon 12 mutants were located in exon 5, 8, 9 and exon 11, respectively (Fig. 1A and Table S5). A total of 56 co-mutated genes were detected in the cohort (Fig. 1B). Co-mutated genes with a detection rate of ≥10% included FLT3 (56.77%), DNMT3A (48.44%), TET2 (29.69%), IDH2 (23.96%), IDH1 (14.58%), PTPN11 (11.46%), and NRAS (11.46%). Co-mutated genes related to epigenetics and signal transduction were the most common by functional classification (Table S6).
A Protein domain structure and location of amino acids affected by mutations in NPM1. Several nuclear import and export signals of NPM1 assist its nucleocytoplasmic shuttling and cytological localization. The conserved N-terminal domain of NPM1 contains a leucine-rich nuclear export signal (NES). The middle domain contains two nuclear localization signals (NLS) that drive NPM1 to move from the cytoplasm to the nucleus. The C-terminus contains a nucleolar localization signal (NoLS), in which two highly conserved tryptophan residues (W288 and W290) are responsible for the correct folding of the helix to stabilize the hydrophobic core of NoLS. Most of the insertion mutations in exon 12 led to the loss of the original NoLS signal and generated a new NES signal, leading to aberrant cytoplasmic dislocation of NPM1 protein. B Co-mutation distribution map of NPM1-mutated AML patients.
One hundred seventy-eight patients (92.71%) received at least one complete course of intensive induction chemotherapy and underwent efficacy assessment, of which 133 patients (74.72%) achieved CR/CRi within two courses of induction chemotherapy. The median follow-up of the 178 patients was 26.23 months (95% confidence interval [CI], 23.31–29.16). The median OS and DFS have not been reached, with the median EFS of 15.03 months (95% CI, 8.25–21.82). The 3-year expected OS, EFS, and DFS were 51.5%, 40.3%, and 53.7%, respectively.
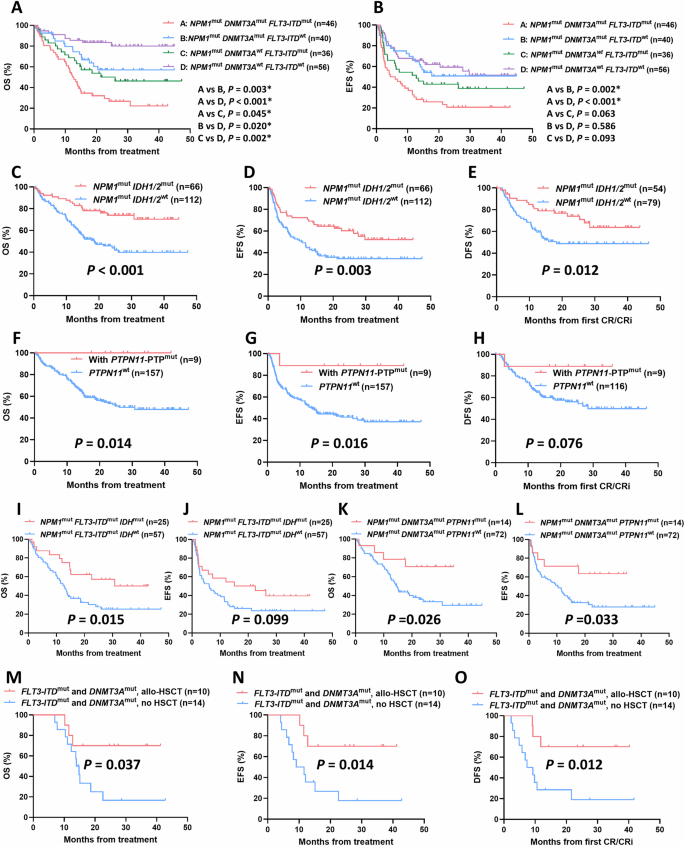
Regardless of the cut-off value of variant allele frequency (VAF) levels, there was no significant difference in OS, EFS, and DFS between NPM1low VAF group and NPM1high VAF group (Fig. S1). Then we focused on impact of co-mutations on response and outcome of AML patients with NPM1 mutations. Among the 178 NPM1-mutated patients included in the follow-up, we noticed that patients with either FLT3-ITD or DNMT3A mutations showed significantly worse CR/CRi rates and prognosis trends than wild type group (FLT3-ITD, CR/CRi rates, 63.41% vs. 84.38%, p = 0.001; median OS, 14.3 months vs. NR, p < 0.001; median EFS, 7.3 months vs. NR, p < 0.001; median DFS, 21.6 months vs. NR, p = 0.044; DNMT3A, CR/CRi rates, 67.44% vs. 81.53%, p = 0.013; median OS, 15.3 months vs. NR, p < 0.001; Median EFS, 11.6 months vs 27.7 months, p = 0.031; Median DFS, p = 0.337) (Table S7 and Fig. S2). We further divided patients into four subgroups according to the FLT3-ITD and DNMT3A mutation status. NPM1/FLT3-ITD/DNMT3A triple mutants showed extremely poor OS and EFS trends among four groups (Fig. 2A, B). Besides, we noticed that when combined with DNMT3A mutations, FLT3-ITD mutated patients exhibited significantly worse OS than that of FLT3-ITD wild-type patients (p = 0.003), while similar results were found in DNMT3A wild-type patients (p = 0.002); We also noticed that when combined with FLT3-ITD mutations, DNMT3A mutated patients exhibited significantly worse OS than that of DNMT3A wild-type patients (p = 0.045), with similar results occurred in FLT3-ITD wild-type patients (p = 0.020) (Fig. 2A).
A OS and B EFS of NPM1-mutated AML patients with different combination patterns of FLT3-ITD and DNMT3A mutations. C OS, D EFS, and E DFS of NPM1-mutated AML patients with IDH1/2 mutation. F OS, G EFS, and H DFS of NPM1-mutated AML patients with PTPN11-PTP mutation. I OS and J EFS of NPM1mutFLT3-ITDmut AML patients with IDH mutations. K OS and L EFS of NPM1mutDNMT3Amut AML patients with PTPN11 mutations. M OS, N EFS, and O DFS of allo-HSCT on NPM1-mutated AML patients harbored both FLT3-ITD and DNMT3A mutations.
For patients combined with IDH1/2 mutations, we observed that the IDH1/2 mutant group significantly improved OS, EFS, and DFS compared with wild-type group (Median OS, NR vs. 18.6 months, p < 0.001; Median EFS, NR vs 10.2 months, p = 0.003; Median DFS, NR vs 18.3 months, p = 0.012) (Figs. 2C–E and S3). Although patients combined with PTPN11 mutations showed a trend toward improved outcome compared with PTPN11 wild-type, the difference was not significant (Fig. S4). PTPN11 mutations have been reported to be mainly clustered in the N-terminal Src homology region 2 (N-SH2) and phosphatase (PTP) domains. Since mutations in both two domains involved in attenuating the autoinhibition of the protein, SHP2, encoded by PTPN11 [11], we further investigated whether mutations in different domains of PTPN11 led to comparable outcome. The OS and EFS of patients with PTPN11-PTP domain mutations were significantly improved compared to those with PTPN11 wild-type (Median OS, NR vs 26.0 months, p = 0.014; Median EFS, NR vs 13.5 months, p = 0.016). Similar trends were found in DFS, whereas patients with PTPN11-N-SH2 domain mutations showed no significant improvement in outcome (Figs. 2F–H and S4). In addition, Fig. S5 showed the prognostic impact of other co-mutation genes with a detection rate of ≥10% in the follow-up patients, including TET2, FLT3-TKD, NRAS, and WT1, with trends all non-significant.
Further, we took into account the presence of IDH or PTPN11 mutations in NPM1-mutated patients combined with FLT3-ITD or DNMT3A mutations to explore the prognostic impact of the specific co-mutation interaction patterns. Separately, carrying IDH mutations significantly improved OS and exhibited an improved EFS trend in patients with NPM1/FLT3-ITD dual mutations (Median OS, 30.8 vs 12.8 months, p = 0.015; Median EFS, 22.6 vs 6.1 months, p = 0.099), but has no significant impact on the outcome of patients with NPM1/DNMT3A mutations (Figs. 2I, J and S6). Similarly, carrying PTPN11 mutations significantly improved OS and EFS in patients with NPM1/DNMT3A dual mutations (Median OS, NR vs. 14.6 months, p = 0.026; Median EFS, NR vs. 10.2 months, p = 0.033), but has no significant impact on the outcome of patients with NPM1/FLT3-ITD mutations (Figs. 2K, L and S6).
Previous research generally acknowledged that allo-HSCT is beneficial for FLT3-ITD mutated AML patients without NPM1 mutations. To identify the subgroup of NPM1-mutated AML patients likely to benefit from allo-HSCT, we explored the prognosis of patients who underwent allo-HSCT during post-remission after achieving CR/CRi within two courses of induction. A total of 32 patients received allo-HSCT, with another four patients relapsed and received salvage-HSCT during post-remission. For patients with NPM1 mutation, receiving allo-HSCT or salvage-HSCT did not significantly improve the outcome compared with non-transplanted patients (Fig. S7). For patients with NPM1 mutations combined with either FLT3-ITD or DNMT3A mutation, allo-HSCT showed a trend toward improved outcome, but the difference was not significant. When further focused on patients with NPM1/FLT3-ITD/DNMT3A triple mutations characterized by poor prognosis, we observed that allo-HSCT significantly improved the OS, EFS, and DFS of these subgroup (Median OS, NR vs. 14.0 months, p = 0.037; Median EFS, NR vs. 9.1 months, p = 0.014; Median DFS, NR vs. 7.4 months, p = 0.012) (Fig. 2M–O). Nevertheless, for NPM1-mutated patients with wild type FLT3-ITD and DNMT3A, administration of allo-HSCT showed no improved outcome (Fig. S7).
Our results indicated that in NPM1-mutated AML, co-mutations of IDH1/2 and PTPN11-PTP domain were correlated with favorable prognosis, whereas FLT3-ITD and DNMT3A co-mutations were indicative of poor prognosis. Notably, the presence of NPM1/FLT3-ITD/DNMT3A triple mutations is associated with exceptionally adverse OS and EFS trends. Several studies have reported NPM1/FLT3-ITD/DNMT3A, the most common triple mutation pattern in NPM1-mutated patients, defined an AML subgroup with extremely poor prognosis [7, 12], which aligned with our findings. Further, our results on specific co-mutation combinations indicated that IDH and PTPN11 co-mutations, respectively, ameliorated the adverse prognosis of patients with NPM1/FLT3-ITD or NPM1/DNMT3A dual mutations, thus two subsets with improved prognosis were redefined from the original adverse-prognosis subset of NPM1-mutated AML. Besides, for patients with NPM1/FLT3-ITD dual mutations, allo-HSCT post-first remission has demonstrated a significant enhancement in both OS and DFS juxtaposed with the continued administration of chemotherapy alone [13, 14]. However, another large cohort study on pediatric AML reported opposite results [15]. Our research endeavored to identify the optimal population who may benefit from allo-HSCT. The findings underscored the therapeutic potential of allo-HSCT, particularly for AML patients with NPM1/FLT3-ITD/DNMT3A triple mutations during post-remission.
In summary, these findings underscored the importance of co-mutation analysis in NPM1-mutated AML for risk stratification and therapeutic decision-making, suggesting that allo-HSCT may be a recommended strategy for NPM1-mutated patients with specific adverse co-mutation profiles. Nevertheless, further research is needed to confirm these findings and explore how these co-mutations interact to diversify the outcome of NPM1-mutated AML patients.
Data availability
The data are not publicly available, owing to ethics considerations and privacy restriction, but can be requested from the corresponding author if necessary.
References
Grimwade D, Ivey A, Huntly BJ. Molecular landscape of acute myeloid leukemia in younger adults and its clinical relevance. Blood. 2016;127:29–41. https://doi.org/10.1182/blood-2015-07-604496.
McKerrell T, Park N, Moreno T, Grove CS, Ponstingl H, Stephens J, et al. Leukemia-associated somatic mutations drive distinct patterns of age-related clonal hemopoiesis. Cell Rep. 2015;10:1239–45. https://doi.org/10.1016/j.celrep.2015.02.005.
Papaemmanuil E, Gerstung M, Bullinger L, Gaidzik VI, Paschka P, Roberts ND, et al. Genomic classification and prognosis in acute myeloid leukemia. N Engl J Med. 2016;374:2209–21. https://doi.org/10.1056/NEJMoa1516192.
Boddu PC, Kadia TM, Garcia-Manero G, Cortes J, Alfayez M, Borthakur G, et al. Validation of the 2017 European LeukemiaNet classification for acute myeloid leukemia with NPM1 and FLT3-internal tandem duplication genotypes. Cancer. 2019;125:1091–100. https://doi.org/10.1002/cncr.31885.
Gaidzik VI, Weber D, Paschka P, Kaumanns A, Krieger S, Corbacioglu A, et al. DNMT3A mutant transcript levels persist in remission and do not predict outcome in patients with acute myeloid leukemia. Leukemia. 2018;32:30–7. https://doi.org/10.1038/leu.2017.200.
Boddu P, Kantarjian H, Borthakur G, Kadia T, Daver N, Pierce S, et al. Co-occurrence of FLT3-TKD and NPM1 mutations defines a highly favorable prognostic AML group. Blood Adv. 2017;1:1546–50. https://doi.org/10.1182/bloodadvances.2017009019.
Bezerra MF, Lima AS, Pique-Borras MR, Silveira DR, Coelho-Silva JL, Pereira-Martins DA, et al. Co-occurrence of DNMT3A, NPM1, FLT3 mutations identifies a subset of acute myeloid leukemia with adverse prognosis. Blood. 2020;135:870–5. https://doi.org/10.1182/blood.2019003339.
Eisfeld AK, Kohlschmidt J, Mims A, Nicolet D, Walker CJ, Blachly JS, et al. Additional gene mutations may refine the 2017 European LeukemiaNet classification in adult patients with de novo acute myeloid leukemia aged <60 years. Leukemia. 2020;34:3215–27. https://doi.org/10.1038/s41375-020-0872-3.
Patel JP, Gonen M, Figueroa ME, Fernandez H, Sun Z, Racevskis J, et al. Prognostic relevance of integrated genetic profiling in acute myeloid leukemia. N Engl J Med. 2012;366:1079–89. https://doi.org/10.1056/NEJMoa1112304.
Benson AB, Venook AP, Al-Hawary MM, Arain MA, Chen YJ, Ciombor KK, et al. Colon Cancer, Version 2.2021, NCCN clinical practice guidelines in oncology. J Natl Compr Canc Netw. 2021;19:329–59. https://doi.org/10.6004/jnccn.2021.0012.
Alfayez M, Issa GC, Patel KP, Wang F, Wang X, Short NJ, et al. The Clinical impact of PTPN11 mutations in adults with acute myeloid leukemia. Leukemia. 2021;35:691–700. https://doi.org/10.1038/s41375-020-0920-z.
Heiblig M, Duployez N, Marceau A, Lebon D, Goursaud L, Plantier I, et al. The impact of DNMT3A status on NPM1 MRD predictive value and survival in elderly AML patients treated intensively. Cancers. 2021;13. https://doi.org/10.3390/cancers13092156.
Pratcorona M, Brunet S, Nomdedeu J, Ribera JM, Tormo M, Duarte R, et al. Favorable outcome of patients with acute myeloid leukemia harboring a low-allelic burden FLT3-ITD mutation and concomitant NPM1 mutation: relevance to post-remission therapy. Blood. 2013;121:2734–8. https://doi.org/10.1182/blood-2012-06-431122.
Sakaguchi M, Yamaguchi H, Najima Y, Usuki K, Ueki T, Oh I, et al. Prognostic impact of low allelic ratio FLT3-ITD and NPM1 mutation in acute myeloid leukemia. Blood Adv. 2018;2:2744–54. https://doi.org/10.1182/bloodadvances.2018020305.
Xu LH, Fang JP, Liu YC, Jones AI, Chai L. Nucleophosmin mutations confer an independent favorable prognostic impact in 869 pediatric patients with acute myeloid leukemia. Blood Cancer J. 2020;10:1. https://doi.org/10.1038/s41408-019-0268-7.
Funding
This work was supported in part by National Natural Science Foundation of China (82370162); Natural Science Foundation of Zhejiang Province, China (LY23H080005); Key R&D Program of Zhejiang (2024C03162) and the Fundamental Research Funds for the Central Universities (226-2022-00003).
Author information
Authors and Affiliations
Contributions
YY and HW designed the study, collected and analyzed the data, and wrote the first draft of the manuscript. YZ, NZ, WX, HM, YL, LM, HT, JQ, MY, WY, and DZ collected and analyzed the data, and reviewed the manuscript. JJ and HW read and reviewed the manuscript. HW accessed and verified the data, and provided administrative support. All authors had full access to all the data in the study and had final responsibility for the decision to submit for publication.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Ethics approval and consent to participate
This study was approved by local ethics committees and was conducted in accordance with the Declaration of Helsinki. All patients signed written informed consent.
Consent for publication
All patients signed informed consent and also consented to the publication of these data.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Yao, Y., Zhou, Y., Zhuo, N. et al. Co-mutation landscape and its prognostic impact on newly diagnosed adult patients with NPM1-mutated de novo acute myeloid leukemia. Blood Cancer J. 14, 118 (2024). https://doi.org/10.1038/s41408-024-01103-w
Received:
Revised:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41408-024-01103-w