Abstract
Constitutively active AR-V7, one of the major androgen receptor (AR) splice variants lacking the ligand-binding domain, plays a key role in the development of castration-resistant prostate cancer (CRPC) and anti-androgen resistance. However, our understanding of the regulatory mechanisms of AR-V7-driven transcription is limited. Here we report DBC1 as a key regulator of AR-V7 transcriptional activity and stability in CRPC cells. DBC1 functions as a coactivator for AR-V7 and is required for the expression of AR-V7 target genes including CDH2, a mesenchymal marker linked to CRPC progression. DBC1 is required for recruitment of AR-V7 to its target enhancers and for long-range chromatin looping between the CDH2 enhancer and promoter. Mechanistically, DBC1 enhances DNA-binding activity of AR-V7 by direct interaction and inhibits CHIP E3 ligase-mediated ubiquitination and degradation of AR-V7 by competing with CHIP for AR-V7 binding, thereby stabilizing and activating AR-V7. Importantly, DBC1 depletion suppresses the tumorigenic and metastatic properties of CRPC cells. Our results firmly establish DBC1 as a critical AR-V7 coactivator that plays a key role in the regulation of DNA binding and stability of AR-V7 and has an important physiological role in CRPC progression.
Similar content being viewed by others
Introduction
As a hormone-dependent transcription factor, androgen receptor (AR) mediates diverse biological functions in normal prostate and also plays an important role during all stages of prostate cancer (PCa) [1,2,3]. Upon androgen binding, AR binds to the androgen response element (ARE) in the regulatory regions (enhancers and promoters) of target genes and then regulates gene expression via the recruitment of coactivators that mediate the communication with the basal transcription machinery, as well as local chromatin remodeling [1, 3, 4]. Full-length AR (AR-FL) comprises the N-terminal activation domain (NTD) harboring constitutively active activation function 1 (AF-1), DNA-binding domain (DBD), and the ligand-binding domain (LBD) harboring the hormone-binding pocket and AF-2 [1,2,3].
All current anti-hormone therapies target the AR LBD either directly with anti-androgens (e.g., enzalutamide) or indirectly with androgen biosynthesis inhibitors (e.g., abiraterone) [1, 3]. PCa initially regresses in response to anti-androgens and androgen deprivation therapy (ADT) but eventually relapses and progresses to castration-resistant prostate cancer (CRPC). CRPC is characterized by an aberrant activation or reactivation of AR signaling through various mechanisms including overexpression of coactivators and AR gene mutation or amplification [1, 3]. A newly emerging mechanism of CRPC progression is the expression or upregulation of constitutively active AR variants (AR-Vs) [1,2,3, 5]. These alternatively spliced AR-Vs encode a truncated AR protein that retains the NTD and DBD but lacks the C-terminal LBD, resulting in ligand-independent constitutive activation and resistance to ADT and anti-androgens. Among AR-Vs identified to date, AR-V7 is the most frequently expressed in PCa cell lines and CRPC tissues and contains intact AR NTD and DBD followed by a unique C-terminal sequence of 16 amino acids encoded by a cryptic exon 3b [1,2,3, 5]. AR-V7 has been implicated in promoting both primary and acquired resistance to ADT and anti-androgens [5,6,7]. However, how the activity and expression of AR-V7 are regulated in CRPC cells remains elusive. Although numerous AR-FL coactivators have been identified and shown to activate AR-FL activity by binding to the LBD, little is known about their roles in the regulation of AR-V7 activity in CRPC progression. In addition, previous studies have identified upregulation of CDH2 (N-cadherin/cadherin-2), a mesenchymal marker associated with the epithelial–mesenchymal transition (EMT), as another mechanism for CRPC progression [8, 9]. CDH2 is expressed in metastatic CRPC tissues and multiple CRPC cell lines but not in androgen-sensitive PCa tissues and cell lines. Moreover, monoclonal antibodies targeting CDH2 could delay the emergence of CRPC and suppress CRPC xenograft growth [8]. Although CDH2 has been reported as a promising therapeutic target for CRPC [8, 9] and as a target of AR-V7 [10, 11], molecular mechanisms by which CDH2 expression is regulated in CRPC remains unclear.
Deleted in breast cancer (DBC1/CCAR2) acts as a coactivator for various transcription factors and also functions as an inhibitor of epigenetic regulators (e.g., methyltransferase SUV39H1, deacetylases HDAC3 and SIRT1, and E3 ligase MDM2) [12,13,14,15,16]. We have previously shown that DBC1 inhibits SIRT1-mediated repression and deacetylation of estrogen receptor, PEA3/ETV4, and β-catenin and therefore positively regulates their transcriptional activities and/or DNA-binding activities [12, 17, 18]. Additionally, a previous study identified DBC1 as an AR-FL coactivator and suggested that DBC1 has the potential to participate in androgen-driven PCa [19]. However, if DBC1 has a direct role in CRPC remains unknown. In this study, we report a role of DBC1 as a key regulator of AR-V7 function and AR-V7-CDH2 signaling-mediated CRPC progression.
Results
DBC1 interacts with and enhances the transcriptional activity of AR-V7
As DBC1 has been shown to bind to the LBD of AR-FL and serve as a coactivator for AR-FL [19], we first examined if DBC1 also interacts with AR-V7 lacking the LBD. In coimmunoprecipitation experiments, endogenous interaction between AR-V7 and DBC1 was detected in 22RV1 and VCaP cells, which express both AR-FL and AR-V7 (Fig. 1a). Similarly, DBC1 was coimmunoprecipitated with AR-V7 from lysates of transfected 293T cells (Fig. 1b). GST pull-down experiments confirmed this interaction and demonstrated that DBC1 binds directly to the C-terminal domain (CTD) of AR-V7 (Fig. 1c, d). We next performed reporter gene assays with AR-FL and AR-V7 to test the coactivator activity of DBC1. Consistent with previous reports [12, 19, 20], DBC1 enhanced the androgen-induced AR-FL activity (Fig. 1e). DBC1 also stimulated AR-V7 activity in a dose-dependent fashion (Fig. 1f). These results suggest that DBC1 interacts directly with AR-V7 and functions as an AR-V7 coactivator.
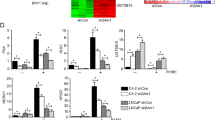
Coactivator activity of DBC1 for AR-V7 and identification of AR-V7 target genes. a Endogenous interaction between AR-V7 and DBC1. Whole cell lysates of 22RV1 and VCaP were immunoprecipitated with anti-AR-V7 antibody or normal IgG. The immunoprecipitates were analyzed by immunoblot using the indicated antibodies. b 293T cell lysates transfected with FLAG-AR-V7 and DBC1-V5 expression vectors were immunoprecipitated and immunoblotted with the indicated antibodies. c Schematic representation of AR-V7 and deletion mutants tested in GST pull-down assays. AF-1 activation function 1, DBD DNA-binding domain, CE cryptic exon, NTD N-terminal domain, CTD C-terminal domain. d In vitro-translated FLAG-tagged AR-V7 and its fragments were incubated with recombinant GST-DBC1. Bound proteins were analyzed by immunoblot with anti-FLAG antibody. e, f CV-1 cells were transfected with MMTV-LUC reporter and pSG5.HA-AR e or pSG5.HA-AR-V7 f in combination with various amounts of pSG5.HA-DBC1, and luciferase assays were performed. Data are means ± s.d. (n = 3). g, h Identification of AR-V7 target genes. LNCaP/Tet-V7 cells were treated with Dox for 48 h and DHT for 24 h, as indicated. Protein levels were determined by immunoblot using the indicated antibodies. For detection of both AR-FL and AR-V7, anti-AR antibody (N-20) was used g. Total RNA was subjected to qRT–PCR analysis for the indicated mRNAs, and results were visualized by heatmap h. i Total RNA from 22RV1 cells infected with lentivirus expressing shNS or shAR-V7 was analyzed by qRT–PCR using primers specific for the indicated mRNAs. Data are expressed as fold change relative to control shNS (set at 1) and are means ± s.d. (n = 3)
DBC1 is required for the efficient expression of AR-FL and AR-V7 target genes
AR-V7 regulates some canonical AR-FL target genes, as well as its own unique set of genes [10, 11, 21,22,23]. To confirm and identify AR-V7-specific target genes and to study the regulatory mechanisms of AR-V7 activity, we chose LNCaP cells, which express AR-FL but not a detectable level of AR-V7, and generated a cell line with doxycycline (Dox) inducible AR-V7 expression (LNCaP/Tet-V7). The expression of AR-V7 was induced in the presence, but not in the absence, of Dox in LNCaP/Tet-V7 cells (Fig. 1g), thus allowing to compare the activities of AR-V7 and endogenous AR-FL by Dox or dihydrotestosterone (DHT) treatment, respectively. DHT treatment stimulated the expression of well-characterized AR-FL target genes, such as KLK3, TMPRSS2, FKBP5, PMEPA1, KLK2, and SNAI2, and Dox treatment also increased their expression (Fig. 1h), suggesting that AR-V7 activates canonical AR-FL target genes in the absence of DHT in LNCaP cells. Consistent with these results, AR-V7 depletion reduced the expression of its target genes in 22RV1 cells, a CRPC cell line expressing high levels of AR-V7 (Fig. 1i). As reported previously [10, 11, 21], the expression of EDN2 and CDH2 was increased by AR-V7, but not by AR-FL, in LNCaP/Tet-V7 and C4-2B/Tet-V7 cells (Figs. 1h and 2a and Supplementary Fig. 1), suggesting that they are preferentially regulated by AR-V7.
Requirement of DBC1 for AR-V7 activity. a, b Identification of DBC1-regulated AR-V7 target genes. LNCaP/Tet-V7 cells were infected with lentivirus expressing shNS or shDBC1#8. Protein levels were determined by immunoblot with the indicated antibodies a. Total RNA was analyzed by qRT–PCR for the indicated mRNAs b. mRNA levels are shown relative to the control shNS, vehicle-treated cells (set at 1). Data are means ± s.d. (n = 3). c, d LNCaP/Tet-V7 cells transfected with MMTV-LUC and indicated siRNAs against DBC1 were treated with Dox for 48 h or DHT for 24 h and harvested for luciferase assays d. Data are means ± s.d. (n = 3). Protein levels were monitored by immunoblot using the indicated antibodies c. e VCaP cells infected with lentivirus expressing shNS or shDBC1#8 were treated with DHT for 24 h. Total RNA was analyzed by qRT–PCR with primers specific for the indicated mRNAs. mRNA levels are shown relative to the control shNS, vehicle-treated cells (set at 1). Data are means ± s.d. (n = 3). f 22RV1 cells were infected with lentiviruses expressing shNS, shDBC1#5, or shDBC1#8. Total RNA was examined by qRT–PCR with primers specific for the indicated mRNAs. mRNA levels are shown relative to control shNS-treated cells (set at 1). Data are means ± s.d. (n = 3)
Next, we assessed the functional involvement of DBC1 in AR-FL and AR-V7 activities. Depletion of DBC1 reduced the DHT-induced expression of KLK3, TMPRSS2, FKBP5, and PMEPA1 (Fig. 2a, b). Similarly, Dox-induced expression of AR-FL target genes and the AR-V7-specific target CDH2, but not EDN2, was also suppressed by DBC1 depletion. Consistent with these results, DBC1 depletion inhibited both DHT-induced and Dox-induced MMTV-LUC reporter activity (Fig. 2c, d). Similar effects of DBC1 depletion on the expression of DHT-induced AR-FL target genes and AR-V7-specific target CDH2 were also observed in VCaP cells (Fig. 2e). DBC1 depletion also reduced the expression of AR-V7 target genes in castration-resistant 22RV1 cells (Fig. 2f). Together, these results suggest that DBC1 is required for the transcriptional activities of both AR-FL and AR-V7 in CRPC cells.
DBC1 functions as a coactivator for AR-V7 to drive CDH2 expression
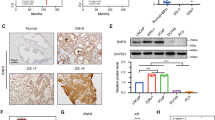
Given that CDH2 has been shown to be frequently upregulated in CRPC [8, 9] and our results showing AR-V7-specific regulation of CDH2 expression, we focused on CDH2 for further analysis. CRPC 22RV1 cells express higher mRNA and protein levels of CDH2 compared to those in AR-V7-negative, androgen-sensitive LNCaP cells (Fig. 3a, b). In addition, AR-V7 depletion reduced both mRNA and protein levels of CDH2 in 22RV1 cells (Figs. 1i and 3c), and conversely, AR-V7 overexpression increased CDH2 expression (Fig. 3d), confirming that CDH2 is a direct target of AR-V7.
DBC1 functions as a coactivator to stimulate AR-V7-mediated CDH2 gene expression. a, b Comparison of CDH2 expression levels in LNCaP and 22V1 cells. CDH2 mRNA and protein levels were determined by qRT-PCR (data are means ± s.d. (n = 3)) a and immunoblot b, respectively. c Protein levels from 22RV1 cells infected with lentivirus expressing shNS or shAR-V7 were monitored by immunoblot with the indicated antibodies. d 22RV1 cell lysates transfected with empty or FLAG-AR-V7 expression vectors were immunoblotted with the indicated antibodies. e, f CV-1 cells were transfected with CDH2.Enh-TA-LUC reporter and either pSG5.HA-AR-V7 alone e or in combination with various amounts of pSG5.HA-DBC1 f, and luciferase assays were performed. Data are means ± s.d. (n = 3). g LNCaP/Tet-V7 cells transfected with CDH2.Enh-TA-LUC were treated with DHT for 24 h or Dox for 48 h and harvested for luciferase assays. Data are means ± s.d. (n = 3). Protein levels were monitored by immunoblot using the indicated antibodies. h 22RV1 cells infected with lentiviruses encoding a NS or AR-V7 shRNA were transfected with CDH2.Enh-TA-LUC, and luciferase assays were performed. Data are means ± s.d. (n = 3). Protein levels were determined by immunoblot using the indicated antibodies. i CDH2.Enh-TA-LUC reporter assays in LNCaP/Tet-V7 cells transfected with indicated siRNAs against DBC1 were performed as described in g. DBC1 knockdown efficiency was monitored by immunoblot. j CDH2.Enh-TA-LUC reporter assays in 22RV1 cells infected with lentivirus expressing shNS or shDBC1#8 were performed as described in h. DBC1 knockdown efficiency was monitored by immunoblot. k Total RNA from 22RV1 cells infected with lentiviruses expressing the indicated shRNAs was examined by qRT-PCR for CDH2 expression. Data are means ± s.d. (n = 3). Protein levels were determined by immunoblot using the indicated antibodies
A 15-repeat of half ARE sequences (hereafter referred to as CDH2 enhancer) is located in CDH2 gene intron 1, ~20 kb downstream of the transcription start site (Supplementary Fig. 2) [24]. In luciferase assays using a reporter containing the CDH2 enhancer, AR-V7 activated the reporter activity in a dose-dependent manner, and this activity was further enhanced by increasing levels of DBC1 (Fig. 3e, f). Consistent with the quantitative real-time PCR (qRT-PCR) data, AR-FL was much less effective than AR-V7 in stimulating the CDH2 enhancer activity in LNCaP/Tet-V7 and 293T cells (Fig. 3g and Supplementary Fig. 3). Furthermore, AR-V7 depletion decreased the reporter activity in 22RV1 cells (Fig. 3h), suggesting that the CDH2 enhancer activity is preferentially regulated by AR-V7 rather than AR-FL. In addition, DBC1 depletion decreased AR-V7-dependent reporter activity in LNCaP/Tet-V7 and 22RV1 cells (Fig. 3i, j) and also inhibited the endogenous expression of CDH2 in both cell lines and VCaP cells (Fig. 2a, b, e, f, and 3k), suggesting that DBC1 functions as a coactivator to stimulate AR-V7-mediated CDH2 gene expression.
DBC1 is required for efficient occupancy of AR-V7 on target enhancers and for chromatin loop formation between the promoter and enhancer of the CDH2 gene
To test the direct involvement of AR-V7 and DBC1 in CDH2 gene transcription, we performed chromatin immunoprecipitation (ChIP) experiments in 22RV1 cells. AR-V7 and DBC1 were recruited to the CDH2 enhancer, and similar recruitment patterns were observed on the PMEPA1 and FKBP5 enhancers [25, 26] (Fig. 4a), suggesting that DBC1 directly regulates AR-V7 target gene transcription. Reciprocal sequential ChIP analyses showed that AR-V7 associates with DBC1 on the CDH2 enhancer, but not on a region lacking AR-V7-binding sequences (Fig. 4b and Supplementary Fig. 4). Moreover, DBC1 depletion greatly reduced the occupancy of AR-V7 at the CDH2, PMEPA1, and FKBP5 enhancers (Fig. 4c), suggesting requirement of DBC1 for efficient binding of AR-V7 to target enhancers.
DBC1 is required for AR-V7 occupancy on its target enhancers and a long range chromatin interaction between the CDH2 enhancer and promoter. a ChIP-qPCR analysis of AR-V7 and DBC1 occupancy at the indicated enhancers in 22RV1 cells. Data are means ± s.d. (n = 3). b Re-ChIP assays were performed by sequential IP with the indicated antibodies. qPCR was performed with primers specific for the indicated regions of the CDH2 gene. Data are means ± s.d. (n = 3). c ChIP assays using soluble chromatin from 22RV1 cells infected with lentiviruses expressing shNS or shDBC1#8 were performed as in a. d 3C analysis was performed using ApoI-digested, cross-linked chromatin from 22RV1 cells. After ligation and reverse cross-linking, 3C-PCR was performed with one primer in the CDH2 promoter and another in the enhancer (primer P+/E−) using equal amounts of 3C DNA. Control PCR (primer E+/E−) for the input was also performed. e 3C assays were performed using chromatin from LNCaP/Tet-V7 cells treated with or without Dox for 24 h as described in d. Protein levels were determined by immunoblot using the indicated antibodies (right). f 3C assays were performed using chromatin from 22RV1 cells expressing shNS or shDBC1#8 as described in d. g FAIRE was performed with 22RV1 cells expressing shNS or shDBC1#8, and FAIRE DNA was analyzed by qPCR using primers specific for the CDH2 enhancer. Data are means ± s.d. (n = 3)
To test whether the 20 kb downstream enhancer could physically communicate with the CDH2 promoter via chromatin looping, we performed chromatin conformation capture (3C) experiments in 22RV1 cells. As shown in Fig. 4d, 3C-PCR product was obtained in a ligase-dependent manner, indicating that the 20 kb downstream enhancer interacts specifically with the CDH2 promoter through chromatin looping. The 458 bp 3C-PCR product was sequenced to verify the interaction of the enhancer with the CDH2 promoter (Supplementary Fig. 5). In addition, 3C assays using LNCaP/Tet-V7 cells demonstrated that the long-range chromatin interaction is dependent upon the presence of AR-V7 (Fig. 4e). Interestingly, DBC1 depletion in 22RV1 cells almost completely eliminated the CDH2 enhancer–promoter interaction (Fig. 4f), indicating the involvement of AR-V7 and DBC1 in bridging the enhancer and promoter of the CDH2 gene. Moreover, formaldehyde-assisted isolation of regulatory elements (FAIRE) analysis showed that DBC1 depletion reduced chromatin accessibility at the CDH2 enhancer region (Fig. 4g), suggesting that DBC1 contributes to regulating chromatin architecture of the CDH2 gene locus by maintaining open chromatin and mediating long-range chromatin interactions.
DBC1 positively regulates DNA binding and stability of AR-V7
To further elucidate mechanisms by which DBC1 regulates AR-V7 activity, we performed DNA affinity precipitation assays (DAPA). As shown in Fig. 5a, AR-V7 was precipitated with CDH2 enhancer probe, indicating that AR-V7 binds directly to the CDH2 enhancer. Interestingly, the enhancer binding activity of AR-V7 was enhanced in the presence of DBC1, and DBC1 was precipitated only in the presence of AR-V7 (Fig. 5b). DAPA using extracts from AR-V7-depeleted 22RV1 cells confirmed AR-V7-dependent recruitment of DBC1 to the CDH2 enhancer (Fig. 5c). These results suggest that DBC1 is recruited to the CDH2 enhancer through the interaction with AR-V7 and that the interaction enhances the DNA binding of AR-V7.
DBC1 enhances DNA binding and stability of AR-V7. a, b DAPA was performed with 293T cell extracts transfected with the indicated expression vectors and biotinylated CDH2 enhancer fragments. Bound proteins were detected by immunoblot. c DAPA using 22RV1 cell lysates infected with lentivirus expressing shNS or shAR-V7 was performed as described above. d 22RV1 cell lysates infected with lentiviruses expressing the indicated shRNA were examined by immunoblot with the indicated antibodies. e 22RV1 cell lysates transfected with empty or FLAG-DBC1 expression vector were immunoblotted with the indicated antibodies. f 293T cell lysates transfected with FLAG-AR-V7 and HA-ubiquitin expression vectors were immunoprecipitated with anti-FLAG antibody or normal IgG and immunoblotted with the indicated antibodies. g 293T cells transfected with His-AR-V7 and HA-ubiquitin expression vectors were lysed by 6 M Guanidine–HCl, and AR-V7 ubiquitination was examined by Ni-NTA Agarose or control agarose beads (BDs) pull-down (PD) and immunoblots. h 22RV1 cell lysates treated with or without 20 µM MG132 for 4 h were analyzed by immunoblot with the indicated antibodies. i 22RV1 cells infected with lentiviruses expressing the indicated shRNA were treated with 50 µg/ml cycloheximide (CHX) and harvested at the indicated time. Cell lysates were analyzed by immunoblot with the indicated antibodies, and representative immunoblot (n = 3) is shown (left). The band intensities were quantified by ImageJ software, and AR-V7 levels are shown relative to controls (time point 0 h in each shRNA group, set at 100) (right). j 22RV1 cell lysates transfected with empty or FLAG-DBC1 expression vector were immunoprecipitated with anti-AR-V7 antibody and immunoblotted with the indicated antibodies. k 293T cell lysates transfected with the indicated expression vectors were immunoprecipitated with anti-FLAG antibody and immunoblotted with the indicated antibodies. l 22RV1 cell lysates infected with lentiviruses expressing the indicated shRNA were immunoprecipitated with anti-AR-V7 antibody and immunoblotted with the indicated antibodies
In our investigation of DBC1 knockdown effect on AR-V7 function, we interestingly found that DBC1 depletion decreased AR-V7 protein levels in 22RV1 and VCaP cells (Fig. 5d and Supplementary Fig. 6c). AR-FL levels were also decreased, but to a lesser extent, by DBC1 depletion. In contrast, DBC1 overexpression increased AR-V7 protein levels in 22RV1 cells (Fig. 5e). DBC1 depletion did not affect mRNA levels of either AR-V7 or AR-FL in 22RV1 and VCaP cells (Supplementary Fig. 6a and b), suggesting that, in addition to enhancing DNA binding, DBC1 has an additional mechanism to control AR-V7 function, which may regulate AR-V7 expression at the post-translational level.
Whereas AR-FL levels are known to be regulated by ubiquitin-proteasome degradation pathway [27,28,29,30,31], little is known about how the stability and turnover of AR-V7 are regulated. To investigate whether AR-V7 can be poly-ubiquitinated, we performed ubiquitination assays in 293T cells transfected with plasmids expressing AR-V7 and HA-ubiquitin. After immunoprecipitation of AR-V7, ubiquitinated levels of AR-V7 were determined by immunoblots. A series of higher molecular weight bands of AR-V7 were readily detected when HA-ubiquitin was coexpressed (Fig. 5f). Similar results were obtained under denaturing conditions (Fig. 5g), indicating that AR-V7 is directly poly-ubiquitinated. In addition, treatment of 22RV1 cells with MG132 increased AR-V7 protein levels (Fig. 5h), suggesting that AR-V7 is targeted for degradation through the ubiquitination-proteasome pathway. When 22RV1 cells were treated with cycloheximide, AR-V7 was degraded much faster in DBC1-depleted cell (Fig. 5i). In addition, DBC1 overexpression decreased the ubiquitination levels of both endogenous and ectopically expressed AR-V7 (Fig. 5j, k). Conversely, DBC1 depletion increased the ubiquitination level of endogenous AR-V7 (Fig. 5l), suggesting that DBC1 contributes to AR-V7 stability by blocking its ubiquitination.
DBC1 inhibits CHIP-mediated ubiquitination of AR-V7
Although AR-FL has been shown to be ubiquitinated by several E3 ligases including MDM2, SPOP, and CHIP [27,28,29,30,31], E3 ligases for AR-V7 have not been elucidated. Given previous studies showing that AR-Vs lacking LBD are not ubiquitinated by MDM2 and SPOP [30, 31], we tested whether CHIP can associate with and function as an E3 ligase for AR-V7. In coimmunoprecipitation experiments, endogenous CHIP bound to AR-V7 in 22RV1 cells (Fig. 6a). Coimmunoprecipitation assays with exogenously expressed proteins showed similar results (Fig. 6b). GST pull-down experiments confirmed this interaction and showed further that CHIP binds to the CTD of AR-V7 (Fig. 6c). In ubiquitination assays, overexpression of CHIP, but not a dominant-negative, catalytically inactive mutant (CHIP-P269A), led to a dramatic increase in the ubiquitination levels of endogenous and ectopically expressed AR-V7 (Fig. 6d and Supplementary Fig. 7), suggesting that CHIP interacts directly with and functions as an E3 ligase for AR-V7. Consistent with these results, CHIP overexpression reduced protein levels of AR-V7 and CDH2 in 22RV1 cells (Fig. 6e). In reporter gene assays, CHIP repressed AR-V7 activity, and coexpression of CHIP–P269A reversed CHIP-mediated repression of AR-V7 (Fig. 6f), indicating that the repression of AR-V7 by CHIP involves its E3 ligase activity.
DBC1 inhibits CHIP-mediated ubiquitination and repression of AR-V7. a 22RV1 cell lysates were immunoprecipitated with anti-AR-V7 antibody or normal IgG. The immunoprecipitates were examined by immunoblot with the indicated antibodies. b 293T cell lysates transfected with FLAG-AR-V7 and HA-CHIP expression vectors were immunoprecipitated and immunoblotted with the indicated antibodies. c In vitro-translated FLAG-tagged AR-V7 and its fragments were incubated with GST-CHIP. Bound proteins were analyzed by immunoblot with anti-FLAG antibody. d 22RV1 cell lysates transfected with wild type or catalytically inactive (P269A) CHIP expression vector were immunoprecipitated with anti-AR-V7 antibody and immunoblotted with the indicated antibodies. e 22RV1 cell lysates transfected with empty or CHIP-V5 expression vectors were immunoblotted with the indicated antibodies. f 293T cells were transfected with CDH2.Enh-TA-LUC reporter in combination with the indicated expression vectors, and luciferase assays were performed. Data are means ± s.d. (n = 3). g 293T cells lysates transfected with the indicated expression vectors were immunoprecipitated with anti-FLAG antibody and immunoblotted with the indicated antibodies. CHIP and DBC1 coimmunoprecipitated with FLAG-AR-V7 were also detected with the indicated antibodies. h Reporter gene assays were performed using CDH2.Enh-TA-LUC reporter and the indicated expression vectors. i In vitro-translated DBC1 and recombinant CHIP were incubated with GST-AR-V7 as indicated. Bound proteins were analyzed by immunoblot with the indicated antibodies. j The role of DBC1 as an AR-V7 coactivator. DBC1 inhibits CHIP-mediated AR-V7 ubiquitination, thereby increasing DNA binding and stability of AR-V7
We next examined the effect of DBC1 on CHIP-mediated ubiquitination and repression of AR-V7. CHIP-mediated AR-V7 ubiquitination was inhibited by DBC1 expression (Fig. 6g), and DBC1 rescued AR-V7 activity from CHIP-mediated repression (Fig. 6h). As both DBC1 and CHIP bind to the CTD of AR-V7, we tested whether DBC1 and CHIP compete for binding to AR-V7. Indeed, CHIP binding to AR-V7 was blocked by DBC1 in in vivo and in vitro competition binding assays (Fig. 6g, i). Collectively, these results suggest that DBC1 inhibits CHIP-mediated ubiquitination and repression of AR-V7 by blocking the interaction between AR-V7 and CHIP (Fig. 6j).
DBC1 is required for metastatic and tumorigenic properties of CRPC cells
We next examined the effect of depletion of AR-V7 or DBC1 on CRPC cell proliferation. AR-V7 depletion attenuated the proliferation and clonogenic survival of 22RV1 cells (Fig. 7a, b and Supplementary Fig. 8a). Similar results were obtained by DBC1 depletion in 22RV1 cells, but not in AR-negative DU145 and PC3 cells (Fig. 7c, d and Supplementary Fig. 8b–d), suggesting a critical role of AR-V7-DBC1 signaling axis in CRPC cell growth and survival. DBC1 knockdown also inhibited the migration and invasion of 22RV1 cells (Fig. 7e, f and Supplementary Fig. 8e, f), indicating that DBC1 is required for metastatic potential of CRPC cells. CRPC cells possess sphere-forming ability, and these prostaspheres are enriched in cells with functional properties of cancer stem cells [32]. In sphere formation assays using 22RV1 cells, DBC1 depletion reduced the size and number of prostaspheres (Fig. 7g), indicating that DBC1 contributes to the self-renewal of stem-like CRPC cells.
DBC1 is required for metastatic and tumorigenic properties of CRPC cells. a, b Cell proliferation a and colony formation b assays using 22RV1 cells infected with lentivirus expressing shNS or shAR-V7. Cell proliferation data are means ± s.d. (n = 5). For the colony formation assay, viable colonies were stained with crystal violet and quantified by spectrophotometry. **P < 0.01. c, d Cell proliferation c and colony formation d assays using 22RV1 cells infected with lentiviruses expressing shNS or the indicated shRNAs targeting DBC1 were performed as described above. **P < 0.01. e, f Quantitative analysis of migration e and invasion f assays using 22RV1 cells infected with lentivirus expressing shNS or the indicated shRNAs against DBC1. Data are means ± s.d. (n = 3). **P < 0.01, *P < 0.05. g Sphere formation analysis of 22RV1 cells infected with lentivirus expressing shNS or shDBC1#8. *P < 0.05. h, i Effect of DBC1 depletion on the growth of 22RV1 xenograft tumors. 22RV1-LUC cells expressing shNS or shDBC1#8 were subcutaneously injected in male nude mice. Representative luminescence images of tumor-bearing mice and their tumors are shown (left panel), and the average signal intensity (n = 5, ± SD) of regions of interest (ROIs) is quantitated (right panel) h. Tumor growth curves are shown i. **P < 0.01. j qRT-PCR analysis of DBC1 and CDH2 expression in 22RV1 xenograft tumors infected with lentivirus encoding shNS or shDBC1#8. Data are means ± s.d. (n = 3). **P < 0.01
To further elucidate the role of DBC1 in promoting prostate tumorigenesis, we tested the DBC1 knockdown effect on the growth of 22RV1 xenograft tumors in male nude mice injected with control or DBC1-depleted cells engineered to constitutively express luciferase (22RV1-LUC). The expression and activity of luciferase in 22RV1-LUC cells were not affected by DBC1 knockdown (Supplementary Fig. 9). The tumor volumes of DBC1-depleted xenografts were significantly reduced compared with those of control shNS group (Fig. 7h, i). In addition, DBC1 depletion significantly inhibited the expression of CDH2 at the transcriptional level and reduced AR-V7 protein levels in xenograft tumors (Fig. 7j and Supplementary Fig. 10). These results suggest that DBC1 plays an important role in tumorigenic growth of CRPC cells and in vivo expression of CDH2 by positively regulating AR-V7 activity and stability.
Discussion
Accumulating evidence strongly suggests that AR-V7 is one of key mechanisms contributing to CRPC progression and resistance to anti-androgens [1,2,3, 5, 6]. Thus, defining the mechanisms that regulate AR-V7 activity is essential for our understanding of CRPC and for the development of therapeutic strategies. In this study, we show DBC1 as a key factor in the regulation of AR-V7 activity and stability in CRPC cells. DBC1 enhanced the transcriptional and DNA-binding activities of AR-V7. DBC1 depletion caused reduction in the expression of AR-V7 target genes, abolished recruitment of AR-V7 to target enhancers, and attenuated metastatic and tumorigenic potential of CRPC cells. These results clearly establish DBC1 as a key coactivator that plays a critical physiological role in AR-V7-mediated CRPC progression.
DBC1 has emerged as an important transcriptional coactivator and a critical regulator of epigenetic modifiers including MDM2 [12,13,14,15,16]. As a MDM2 inhibitor, DBC1 stabilizes p53 by inhibiting MDM2-mediated p53 ubiquitination [15]. Here we identified CHIP as an E3 ligase for AR-V7 and as a novel target of DBC1. We provide several lines of evidence that DBC1 acts as a positive regulator for AR-V7 stability and function by inhibiting the E3 ligase activity of CHIP: DBC1 blocks the binding of CHIP to AR-V7; and CHIP-mediated ubiquitination and repression of AR-V7 was reversed by DBC1 expression. Thus, DBC1 contributes to AR-V7 function, probably not only through enhancing the DNA-binding activity of AR-V7 but also by inhibiting CHIP-mediated AR-V7 ubiquitination and degradation (Fig. 6j). DBC1 was previously found to interact with and enhance AR-FL-mediated transcription by increasing its DNA-binding affinity [19]. In addition, a recent study demonstrated that DBC1 affects the stability of AR-FL in osteosarcoma cells [33]. We also observed that DBC1 positively regulates the stability of AR-FL by inhibiting CHIP-mediated AR-FL ubiquitination (Supplementary Fig. 11). Several studies have also shown a positive correlation between DBC1 and AR-FL protein levels in osteosarcoma, B cell lymphoma, and renal cell carcinoma [33,34,35]. Thus, DBC1 may be a key factor in regulating the stability and function of not only AR-FL but also AR-Vs in various cancers. Although a number of proteins have been reported to act as AR-FL coactivators by binding to the LBD [4], only a few coactivators have been shown to interact with both AR-FL and the LBD-truncated AR-Vs through binding to different domains. Among AR-FL coactivators which bind to the LBD in an androgen-dependent manner, FHL2 and MED1 can also bind to and act as coactivators for AR-Vs [36, 37]. Because AR-FL plays a crucial role in all phases of PCa, and considerable evidence indicates a significant role of AR-Vs, particularly AR-V7, in CRPC development and resistance to anti-androgen therapies [1,2,3, 5], further investigation of coactivators shared by both AR-FL and AR-V7 may provide promising new therapeutic targets and effective strategies to treat advanced CRPC.
We have previously reported that DBC1 promotes acetylation of ER and PEA3 through inhibiting SIRT1 [12, 17]. AR-FL is also acetylated at three lysine residues, K630/K632/K633, in the hinge domain by several acetyltransferases including p300 and PCAF, and the acetylation enhances androgen-mediated AR-FL activity [27, 38]. Conversely, SIRT1 deacetylates and represses the activity of AR-FL [27, 39]. However, it is unknown whether AR-V7, which lacks the hinge region, is also acetylated and, if so, how AR-V7 activity is regulated by reversible acetylation. Intriguingly, a recent study identified a new acetylation site in the DBD of AR-FL, and the acetylation at K618 by ARD1 enhances AR-FL activity and PCa cell growth [40]. Thus, further study is needed to explore the possibility that ARD1 could also acetylate AR-V7 and to investigate whether SIRT1 and DBC1 reciprocally regulate the activities of AR-FL and AR-V7 by modulating their acetylation status.
CDH2 has been shown to be associated with progression to CRPC and thus has emerged as a promising therapeutic target for CRPC [8, 9]. A recent study found a significant increase in CDH2 level in CRPC xenografts, as well as in metastatic tumors of CRPC patients and suggested that monoclonal antibody targeting CDH2 could suppress metastasis and progression to CRPC [8]. CDH2 induction after neoadjuvant ADT also supports the association of this mesenchymal marker with CRPC. A growing body of evidence has linked EMT to cancer stem cell properties and raises the possibility that CDH2 may be a marker for CRPC stem cell populations [8]. Recent studies suggest that AR-Vs can contribute to CRPC development and progression during ADT through the induction of EMT and acquisition of stemness properties as characterized by upregulation of CDH2 [10, 11, 22, 41]. However, little is known about the regulatory mechanism of CDH2 gene expression in CRPC cells. Here we showed that DBC1 promotes CDH2 expression by facilitating looping of the CDH2 locus and AR-V7 occupancy on the CDH2 enhancer. CDH2 expression is preferentially stimulated by AR-V7 rather than AR-FL in CRPC cells. These results are consistent with previous reports that AR-Vs, but not AR-FL, are involved in the regulation of EMT and stem cell marker expression in CRPC cells [10, 11]. It is noteworthy that we and others have observed the androgen-dependent repression of CDH2 expression in CRPC cells (Fig. 2e and Supplementary Fig. 1) [10, 22]. These results suggest that AR-V7 and AR-FL can act as an activator and repressor, respectively, and might compete with each other for binding to the CDH2 enhancer to reciprocally regulate the CDH2 expression. Consistent with this idea, blocking AR-FL activity by targeting the LBD via ADT or anti-androgens led to upregulation of CDH2 in CRPC cells [9, 10, 22, 41]. Also of note is that DBC1 depletion had no effect on androgen-mediated repression of CDH2 expression (Fig. 2e), suggesting that DBC1 functions as a coactivator of AR-V7-mediated CDH2 transcription, but is not involved in AR-FL-mediated CDH2 repression. These results, together with the fact that AR-V7 and CDH2 signalings are major causes of both PCa metastasis and castration resistance, imply that upregulation of DBC1 could lead to an increased expression of CDH2 by enhancing AR-V7 function, therefore promoting the development and progression of CRPC.
The role of DBC1 in cancer remains controversial because DBC1 has been shown in different studies to either promote or suppress cancer cell growth [12, 15,16,17,18,19]. This controversy may be explained by the p53 status in cancer cells [15]. Gain-of-function p53 mutants promote cancer cell proliferation and migration. DBC1 stabilizes both WT and mutant p53 and acts as a tumor suppressor in cells expressing WT p53 but as a tumor promoter in cancer cells expressing p53 mutants. TP53 mutations are frequently found in advanced PCa and known to contribute to CRPC progression [42]. Consistent with these notions, our results showed that DBC1 functions as a tumor promoter in CRPC 22RV1 cells, which have a heterozygous dominant-negative mutation (Q331R) in the TP53 gene [43]. However, it was also reported that DBC1 effect on p53 stabilization is cell type-specific due to difference in genetic backgrounds of cancer cells and that p53 can directly regulate AR signaling in PCa cells [15, 44]. Thus, future studies will be required to explore the tumor-promoting role of DBC1 in cross-talk between p53s (wild type and mutant p53s) and ARs (AR-FL and AR-Vs) in the context of CRPC.
All current hormonal therapies targeting AR-FL rely on the presence of the LBD and eventually fail by mechanisms involving the expression of AR-V7 and its target genes [1,2,3, 5]. More research focus is needed on regulatory mechanisms of AR-V7 and its target gene expression because targeting them could be potential therapeutic strategies for the treatment of CRPC. Understanding upstream and downstream effectors of AR-V7 will thus provide useful information for developing therapeutic drugs for CRPC. Our findings reinforce the importance to target AR-V7 in CRPC, provide a novel regulatory mechanism of AR-V7 activity and the expression of its downstream target CDH2 by DBC1, and suggest DBC1 as a potential therapeutic target for CRPC.
Materials and methods
Cell culture, lentiviral infection, and transient transfection
CV-1, 293T, VCaP, DU145, and PC3 cells were maintained in DMEM supplemented with 10% fetal bovine serum (FBS). LNCaP, C4-2B, 22RV1 cells were cultured in RPMI 1640 containing 10% FBS. All cell lines used were purchased from the Korean Cell Line Bank and the American Type Culture Collection, tested for mycoplasma infection, and authenticated by STR analysis (DNA profiling). For lentiviral infection, lentiviral particles were generated as described previously [45]. LNCaP and C4-2B cells with inducible AR-V7 expression were established using Lenti-X Tet-Advanced Inducible Expression System (Clontech). The detailed method for generation of stable cell lines is described in Supplementary Information. Reporter gene assays and transient transfections were performed as described previously [18, 20, 45]. For DHT or Dox treatment, cells were grown in RPMI 1640 or DMEM containing 5% dextran/charcoal-stripped FBS for at least 3 days and treated with 10 nM DHT for 16–24 h or 5 ng/ml Dox for 48 h as indicated in figure legends.
RNAi and quantitative real-time reverse transcriptase-PCR (qRT–PCR)
DBC1 depletion by shRNA or siRNA was performed as previously described [12, 17, 18]. Total RNA was extracted using TRIzol Reagent (Invitrogen), and qRT–PCR was carried out with Brilliant II SYBR Green QRT–PCR Master Mix 1-Step (Agilent). Data were normalized to GAPDH or β-actin mRNA levels.
ChIP assays
ChIP assays were performed as previously described [12, 17, 18, 20, 45]. ChIP DNAs were quantified by quantitative real-time PCR (qPCR).
3C assays
3C experiments were carried out as described previously [18, 20]. Cross-linked chromatin was digested with ApoI, and ligated under 50-fold diluted conditions. After reverse cross-linking and DNA purification, the input and 3C DNAs were amplified by conventional PCR.
FAIRE assays
FAIRE assays were performed as described previously [20]. Cross-linked, sonicated chromatin fraction was subjected to three rounds of phenol/chloroform extractions followed by reverse cross-linking, and the FAIRE DNAs were purified. Enriched open chromatin at the CDH2 enhancer region was quantified by qPCR using ChIP PCR primers.
DAPA
DAPA was performed as described previously [20].
Ubiquitination assays
293T or 22RV1 cells were transfected with expression plasmids for HA-Ubiquitin, AR-V7, DBC1, and CHIP as described in figure legends. After 48 h transfection, the cells were treated with 10 μM MG132 for 8 h and then lysed in FLAG lysis buffer or 6 M Guanidine–HCl. AR-V7 was immunoprecipitated, and ubiquitinated levels were determined by immunoblot using antibodies as indicated in figure legends.
Xenografts
Xenograft experiments were conducted as previously described [17, 18, 20]. The detailed procedure is provided in Supplementary Data. Animal experiments were conducted according to protocols approved by the Institutional Animal Care and Use Committee of the Samsung Biomedical Research Institute.
The detailed methods, siRNAs, shRNAs, primers, and antibodies can be found in Supplementary Information.
References
Shafi AA, Yen AE, Weigel NL. Androgen receptors in hormone-dependent and castration-resistant prostate cancer. Pharmacol Ther 2013;140:223–38.
Lu J, Van der Steen T, Tindall DJ, Are androgen receptor variants a substitute for the full-length receptor?. Nat Rev Urol. 2015;12:137–44.
Imamura Y, Sadar MD. Androgen receptor targeted therapies in castration-resistant prostate cancer: bench to clinic. Int J Urol 2016;23:654–65.
van de Wijngaart DJ, Dubbink HJ, van Royen ME, Trapman J, Jenster G. Androgen receptor coregulators: recruitment via the coactivator binding groove. Mol Cell Endocrinol 2012;352:57–69.
Antonarakis ES, Armstrong AJ, Dehm SM, Luo J. Androgen receptor variant-driven prostate cancer: clinical implications and therapeutic targeting. Prostate Cancer Prostatic Dis 2016;19:231–41.
Antonarakis ES, Lu C, Wang H, Luber B, Nakazawa M, Roeser JC, et al. AR-V7 and resistance to enzalutamide and abiraterone in prostate cancer. N Engl J Med 2014;371:1028–38.
Li Y, Chan SC, Brand LJ, Hwang TH, Silverstein KA, Dehm SM. Androgen receptor splice variants mediate enzalutamide resistance in castration-resistant prostate cancer cell lines. Cancer Res 2013;73:483–9.
Tanaka H, Kono E, Tran CP, Miyazaki H, Yamashiro J, Shimomura T, et al. Monoclonal antibody targeting of N-cadherin inhibits prostate cancer growth, metastasis and castration resistance. Nat Med 2010;16:1414–20.
Jennbacken K, Tesan T, Wang W, Gustavsson H, Damber JE, Welen K. N-cadherin increases after androgen deprivation and is associated with metastasis in prostate cancer. Endocr Relat Cancer 2010;17:469–79.
Cottard F, Asmane I, Erdmann E, Bergerat JP, Kurtz JE, Ceraline J. Constitutively active androgen receptor variants upregulate expression of mesenchymal markers in prostate cancer cells. PloS One 2013;8:e63466.
Lu J, Lonergan PE, Nacusi LP, Wang L, Schmidt LJ, Sun Z, et al. The cistrome and gene signature of androgen receptor splice variants in castration resistant prostate cancer cells. J Urol 2015;193:690–8.
Yu EJ, Kim SH, Heo K, Ou CY, Stallcup MR, Kim JH. Reciprocal roles of DBC1 and SIRT1 in regulating estrogen receptor a activity and co-activator synergy. Nucleic Acids Res 2011;39:6932–43.
Chini CC, Escande C, Nin V, Chini EN. HDAC3 is negatively regulated by the nuclear protein DBC1. J Biol Chem 2010;285:40830–7.
Li Z, Chen L, Kabra N, Wang C, Fang J, Chen J. Inhibition of SUV39H1 methyltransferase activity by DBC1. J Biol Chem 2009;284:10361–6.
Qin B, Minter-Dykhouse K, Yu J, Zhang J, Liu T, Zhang H, et al. DBC1 functions as a tumor suppressor by regulating p53 stability. Cell Rep 2015;10:1324–34.
Kim JE, Chen J, Lou Z. DBC1 is a negative regulator of SIRT1. Nature 2008;451:583–6.
Kim HJ, Kim SH, Yu EJ, Seo WY, Kim JH. A positive role of DBC1 in PEA3-mediated progression of estrogen receptor-negative breast cancer. Oncogene 2015;34:4500–8.
Yu EJ, Kim SH, Kim HJ, Heo K, Ou CY, Stallcup MR, et al. Positive regulation of beta-catenin-PROX1 signaling axis by DBC1 in colon cancer progression. Oncogene 2016;35:3410–8.
Fu J, Jiang J, Li J, Wang S, Shi G, Feng Q, et al. Deleted in breast cancer 1, a novel androgen receptor (AR) coactivator that promotes AR DNA-binding activity. J Biol Chem 2009;284:6832–40.
Seo WY, Jeong BC, Yu EJ, Kim HJ, Kim SH, Lim JE, et al. CCAR1 promotes chromatin loading of androgen receptor (AR) transcription complex by stabilizing the association between AR and GATA2. Nucleic Acids Res 2013;41:8526–36.
Krause WC, Shafi AA, Nakka M, Weigel NL. Androgen receptor and its splice variant, AR-V7, differentially regulate FOXA1 sensitive genes in LNCaP prostate cancer cells. Int J Biochem Cell Biol 2014;54:49–59.
Kong D, Sethi S, Li Y, Chen W, Sakr WA, Heath E, et al. Androgen receptor splice variants contribute to prostate cancer aggressiveness through induction of EMT and expression of stem cell marker genes. Prostate 2015;75:161–74.
Hu R, Lu C, Mostaghel EA, Yegnasubramanian S, Gurel M, Tannahill C, et al. Distinct transcriptional programs mediated by the ligand-dependent full-length androgen receptor and its splice variants in castration-resistant prostate cancer. Cancer Res 2012;72:3457–62.
Takayama K, Kaneshiro K, Tsutsumi S, Horie-Inoue K, Ikeda K, Urano T, et al. Identification of novel androgen response genes in prostate cancer cells by coupling chromatin immunoprecipitation and genomic microarray analysis. Oncogene 2007;26:4453–63.
Masuda K, Werner T, Maheshwari S, Frisch M, Oh S, Petrovics G, et al. Androgen receptor binding sites identified by a GREF_GATA model. J Mol Biol 2005;353:763–71.
Makkonen H, Kauhanen M, Paakinaho V, Jaaskelainen T, Palvimo JJ. Long-range activation of FKBP51 transcription by the androgen receptor via distal intronic enhancers. Nucleic Acids Res 2009;37:4135–48.
van der Steen T, Tindall DJ, Huang H. Posttranslational modification of the androgen receptor in prostate cancer. Int J Mol Sci 2013;14:14833–59.
Lin HK, Wang L, Hu YC, Altuwaijri S, Chang C. Phosphorylation-dependent ubiquitylation and degradation of androgen receptor by Akt require Mdm2 E3 ligase. EMBO J 2002;21:4037–48.
Cardozo CP, Michaud C, Ost MC, Fliss AE, Yang E, Patterson C, et al. C-terminal Hsp-interacting protein slows androgen receptor synthesis and reduces its rate of degradation. Arch Biochem Biophys 2003;410:134–40.
An J, Wang C, Deng Y, Yu L, Huang H. Destruction of full-length androgen receptor by wild-type SPOP, but not prostate-cancer-associated mutants. Cell Rep 2014;6:657–69.
McClurg UL, Cork DMW, Darby S, Ryan-Munden CA, Nakjang S, Mendes Cortes L, et al. Identification of a novel K311 ubiquitination site critical for androgen receptor transcriptional activity. Nucleic Acids Res 2017;45:1793–804.
Portillo-Lara R, Alvarez MM. Enrichment of the cancer stem phenotype in sphere cultures of prostate cancer cell lines occurs through activation of developmental pathways mediated by the transcriptional regulator DeltaNp63alpha. PloS One 2015;10:e0130118.
Wagle S, Park SH, Kim KM, Moon YJ, Bae JS, Kwon KS, et al. DBC1/CCAR2 is involved in the stabilization of androgen receptor and the progression of osteosarcoma. Sci Rep 2015;5:13144.
Noh SJ, Kang MJ, Kim KM, Bae JS, Park HS, Moon WS, et al. Acetylation status of P53 and the expression of DBC1, SIRT1, and androgen receptor are associated with survival in clear cell renal cell carcinoma patients. Pathology 2013;45:574–80.
Park HS, Bae JS, Noh SJ, Kim KM, Lee H, Moon WS, et al. Expression of DBC1 and androgen receptor predict poor prognosis in diffuse large B cell lymphoma. Transl Oncol 2013;6:370–81.
McGrath MJ, Binge LC, Sriratana A, Wang H, Robinson PA, Pook D, et al. Regulation of the transcriptional coactivator FHL2 licenses activation of the androgen receptor in castrate-resistant prostate cancer. Cancer Res 2013;73:5066–79.
Liu G, Sprenger C, Wu PJ, Sun S, Uo T, Haugk K, et al. MED1 mediates androgen receptor splice variant induced gene expression in the absence of ligand. Oncotarget 2015;6:288–304.
Fu M, Wang C, Wang J, Zhang X, Sakamaki T, Yeung YG, et al. Androgen receptor acetylation governs trans activation and MEKK1-induced apoptosis without affecting in vitro sumoylation and trans-repression function. Mol Cell Biol 2002;22:3373–88.
Fu M, Liu M, Sauve AA, Jiao X, Zhang X, Wu X, et al. Hormonal control of androgen receptor function through SIRT1. Mol Cell Biol 2006;26:8122–35.
DePaolo JS, Wang Z, Guo J, Zhang G, Qian C, Zhang H, et al. Acetylation of androgen receptor by ARD1 promotes dissociation from HSP90 complex and prostate tumorigenesis. Oncotarget 2016;7:71417–28.
Sun Y, Wang BE, Leong KG, Yue P, Li L, Jhunjhunwala S, et al. Androgen deprivation causes epithelial-mesenchymal transition in the prostate: implications for androgen-deprivation therapy. Cancer Res 2012;72:527–36.
Beltran H, Yelensky R, Frampton GM, Park K, Downing SR, MacDonald TY, et al. Targeted next-generation sequencing of advanced prostate cancer identifies potential therapeutic targets and disease heterogeneity. Eur Urol 2013;63:920–6.
van Bokhoven A, Varella-Garcia M, Korch C, Johannes WU, Smith EE, Miller HL, et al. Molecular characterization of human prostate carcinoma cell lines. Prostate 2003;57:205–25.
Cronauer MV, Schulz WA, Burchardt T, Ackermann R, Burchardt M. Inhibition of p53 function diminishes androgen receptor-mediated signaling in prostate cancer cell lines. Oncogene 2004;23:3541–9.
Kim JH, Yang CK, Heo K, Roeder RG, An W, Stallcup MR. CCAR1, a key regulator of mediator complex recruitment to nuclear receptor transcription complexes. Mol Cell 2008;31:510–9.
Acknowledgements
We thank Dr. Robert Roeder (Rockefeller University) for HA-Ubiquitin expression plasmid. This work was supported by Basic Science Research Program through the National Research Foundation of Korea (NRF) funded by the Ministry of Science, ICT and Future Planning [2016R1A2B4008661]; and the Korea Health Technology R&D Project through the Korea Health Industry Development Institute (KHIDI), funded by the Ministry of Health & Welfare, Republic of Korea [HI17C0117].
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interests
The authors declare that they have no competing interests.
Electronic supplementary material
Rights and permissions
About this article
Cite this article
Moon, S.J., Jeong, B.C., Kim, H.J. et al. DBC1 promotes castration-resistant prostate cancer by positively regulating DNA binding and stability of AR-V7. Oncogene 37, 1326–1339 (2018). https://doi.org/10.1038/s41388-017-0047-5
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41388-017-0047-5
This article is cited by
-
Multifaceted roles of CCAR family proteins in the DNA damage response and cancer
Experimental & Molecular Medicine (2024)
-
Mechanistic insights into the dual role of CCAR2/DBC1 in cancer
Experimental & Molecular Medicine (2023)
-
Ubiquitination Process Mediates Prostate Cancer Development and Metastasis through Multiple Mechanisms
Cell Biochemistry and Biophysics (2023)
-
DBC1 regulates Wnt/β-catenin-mediated expression of MACC1, a key regulator of cancer progression, in colon cancer
Cell Death & Disease (2018)