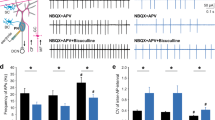
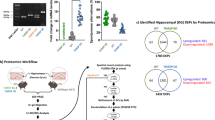
Abstract
Although circadian and sleep disorders are frequently associated with autism spectrum disorders (ASD), it remains elusive whether clock gene disruption can lead to autistic-like phenotypes in animals. The essential clock gene Bmal1 has been associated with human sociability and its missense mutations are identified in ASD. Here we report that global Bmal1 deletion led to significant social impairments, excessive stereotyped and repetitive behaviors, as well as motor learning disabilities in mice, all of which resemble core behavioral deficits in ASD. Furthermore, aberrant cell density and immature morphology of dendritic spines were identified in the cerebellar Purkinje cells (PCs) of Bmal1 knockout (KO) mice. Electrophysiological recordings uncovered enhanced excitatory and inhibitory synaptic transmission and reduced firing rates in the PCs of Bmal1 KO mice. Differential expression of ASD- and ataxia-associated genes (Ntng2, Mfrp, Nr4a2, Thbs1, Atxn1, and Atxn3) and dysregulated pathways of translational control, including hyperactivated mammalian target of rapamycin complex 1 (mTORC1) signaling, were identified in the cerebellum of Bmal1 KO mice. Interestingly, the antidiabetic drug metformin reversed mTORC1 hyperactivation and alleviated major behavioral and PC deficits in Bmal1 KO mice. Importantly, conditional Bmal1 deletion only in cerebellar PCs was sufficient to recapitulate autistic-like behavioral and cellular changes akin to those identified in Bmal1 KO mice. Together, these results unveil a previously unidentified role for Bmal1 disruption in cerebellar dysfunction and autistic-like behaviors. Our findings provide experimental evidence supporting a putative role for dysregulation of circadian clock gene expression in the pathogenesis of ASD.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout






Similar content being viewed by others
References
Herzog ED, Hermanstyne T, Smyllie NJ, Hastings MH. Regulating the Suprachiasmatic Nucleus (SCN) Circadian Clockwork: Interplay between Cell-Autonomous and Circuit-Level Mechanisms. Cold Spring Harb Perspect Biol. 2017;9:a027706.
Takahashi JS, Hong HK, Ko CH, McDearmon EL. The genetics of mammalian circadian order and disorder: implications for physiology and disease. Nat Rev Genet. 2008;9:764–75.
Rosbash M. The implications of multiple circadian clock origins. PLoS Biol. 2009;7:e62.
Bunger MK, Wilsbacher LD, Moran SM, Clendenin C, Radcliffe LA, Hogenesch JB, et al. Mop3 is an essential component of the master circadian pacemaker in mammals. Cell. 2000;103:1009–17.
Lai MC, Lombardo MV, Baron-Cohen S. Autism. Lancet. 2014;383:896–910.
Lorsung E, Karthikeyan R, Cao R. Biological timing and neurodevelopmental disorders: a role for circadian dysfunction in autism spectrum disorders. Front Neurosci. 2021;15:642745.
Nicholas B, Rudrasingham V, Nash S, Kirov G, Owen MJ, Wimpory DC. Association of Per1 and Npas2 with autistic disorder: support for the clock genes/social timing hypothesis. Mol Psychiatry. 2007;12:581–92.
Iossifov I, O’Roak BJ, Sanders SJ, Ronemus M, Krumm N, Levy D, et al. The contribution of de novo coding mutations to autism spectrum disorder. Nature. 2014;515:216–21.
Yang Z, Matsumoto A, Nakayama K, Jimbo EF, Kojima K, Nagata K, et al. Circadian-relevant genes are highly polymorphic in autism spectrum disorder patients. Brain Dev. 2016;38:91–99.
Abrahams BS, Arking DE, Campbell DB, Mefford HC, Morrow EM, Weiss LA, et al. SFARI Gene 2.0: a community-driven knowledge base for the autism spectrum disorders (ASDs). Mol Autism. 2013;4:36.
Bralten J, Mota NR, Klemann C, De Witte W, Laing E, Collier DA, et al. Genetic underpinnings of sociability in the general population. Neuropsychopharmacology. 2021;46:1627–34.
Lipton JO, Yuan ED, Boyle LM, Ebrahimi-Fakhari D, Kwiatkowski E, Nathan A, et al. The Circadian protein BMAL1 regulates translation in response to S6K1-mediated phosphorylation. Cell. 2015;161:1138–51.
Kelleher RJ 3rd, Bear MF. The autistic neuron: troubled translation? Cell. 2008;135:401–6.
Gkogkas CG, Khoutorsky A, Ran I, Rampakakis E, Nevarko T, Weatherill DB, et al. Autism-related deficits via dysregulated eIF4E-dependent translational control. Nature. 2013;493:371–7.
Santini E, Huynh TN, MacAskill AF, Carter AG, Pierre P, Ruggero D, et al. Exaggerated translation causes synaptic and behavioural aberrations associated with autism. Nature. 2013;493:411–5.
Hering H, Sheng M. Dendritic spines: structure, dynamics and regulation. Nat Rev Neurosci. 2001;2:880–8.
Cao R, Gkogkas CG, de Zavalia N, Blum ID, Yanagiya A, Tsukumo Y, et al. Light-regulated translational control of circadian behavior by eIF4E phosphorylation. Nat Neurosci. 2015;18:855–62.
Amorim IS, Kedia S, Kouloulia S, Simbriger K, Gantois I, Jafarnejad SM, et al. Loss of eIF4E phosphorylation engenders depression-like behaviors via selective mRNA translation. J Neurosci. 2018;38:2118–33.
Gantois I, Khoutorsky A, Popic J, Aguilar-Valles A, Freemantle E, Cao R, et al. Metformin ameliorates core deficits in a mouse model of fragile X syndrome. Nat Med. 2017;23:674–7.
Dziuk MA, Gidley Larson JC, Apostu A, Mahone EM, Denckla MB, Mostofsky SH. Dyspraxia in autism: association with motor, social, and communicative deficits. Dev Med Child Neurol. 2007;49:734–9.
Whitney ER, Kemper TL, Rosene DL, Bauman ML, Blatt GJ. Calbindin-D28k is a more reliable marker of human Purkinje cells than standard Nissl stains: a stereological experiment. J Neurosci Methods. 2008;168:42–47.
De Zeeuw CI, Hoebeek FE, Bosman LW, Schonewille M, Witter L, Koekkoek SK. Spatiotemporal firing patterns in the cerebellum. Nat Rev Neurosci. 2011;12:327–44.
Kawamura Y, Nakayama H, Hashimoto K, Sakimura K, Kitamura K, Kano M. Spike timing-dependent selective strengthening of single climbing fibre inputs to Purkinje cells during cerebellar development. Nat Commun. 2013;4:2732.
Wardlaw SM, Phan TX, Saraf A, Chen X, Storm DR. Genetic disruption of the core circadian clock impairs hippocampus-dependent memory. Learn Mem. 2014;21:417–23.
Snider KH, Dziema H, Aten S, Loeser J, Norona FE, Hoyt K, et al. Modulation of learning and memory by the targeted deletion of the circadian clock gene Bmal1 in forebrain circuits. Behav Brain Res. 2016;308:222–35.
Musiek ES, Lim MM, Yang G, Bauer AQ, Qi L, Lee Y, et al. Circadian clock proteins regulate neuronal redox homeostasis and neurodegeneration. J Clin Invest. 2013;123:5389–5400.
Crawford JD, Chandley MJ, Szebeni K, Szebeni A, Waters B, Ordway GA. Elevated GFAP protein in anterior cingulate cortical white matter in males with autism spectrum disorder. Autism Res. 2015;8:649–57.
Laurence JA, Fatemi SH. Glial fibrillary acidic protein is elevated in superior frontal, parietal and cerebellar cortices of autistic subjects. Cerebellum. 2005;4:206–10.
Khan S, Gramfort A, Shetty NR, Kitzbichler MG, Ganesan S, Moran JM, et al. Local and long-range functional connectivity is reduced in concert in autism spectrum disorders. Proc Natl Acad Sci USA. 2013;110:3107–12.
Hickman RA, O’Shea SA, Mehler MF, Chung WK. Neurogenetic disorders across the lifespan: from aberrant development to degeneration. Nat Rev Neurol. 2022;18:117–24.
Wang SS, Kloth AD, Badura A. The cerebellum, sensitive periods, and autism. Neuron. 2014;83:518–32.
Bauman ML, Kemper TL. The neuropathology of the autism spectrum disorders: what have we learned? Novartis Found Symp. 2003;251:112–22.
Zhou H, Lin Z, Voges K, Ju C, Gao Z, Bosman LW, et al. Cerebellar modules operate at different frequencies. Elife. 2014;3:e02536.
Yang YM, Arsenault J, Bah A, Krzeminski M, Fekete A, Chao OY, et al. Identification of a molecular locus for normalizing dysregulated GABA release from interneurons in the Fragile X brain. Mol Psychiatry. 2020;25:2017–35.
Kondratov RV, Kondratova AA, Gorbacheva VY, Vykhovanets OV, Antoch MP. Early aging and age-related pathologies in mice deficient in BMAL1, the core component of the circadian clock. Genes Dev. 2006;20:1868–73.
Leliavski A, Shostak A, Husse J, Oster H. Impaired glucocorticoid production and response to stress in Arntl-deficient male mice. Endocrinology. 2014;155:133–42.
Monyak RE, Emerson D, Schoenfeld BP, Zheng X, Chambers DB, Rosenfelt C, et al. Insulin signaling misregulation underlies circadian and cognitive deficits in a Drosophila fragile X model. Mol Psychiatry. 2017;22:1140–8.
Dy ABC, Tassone F, Eldeeb M, Salcedo-Arellano MJ, Tartaglia N, Hagerman R. Metformin as targeted treatment in fragile X syndrome. Clin Genet. 2018;93:216–22.
Biag HMB, Potter LA, Wilkins V, Afzal S, Rosvall A, Salcedo-Arellano MJ, et al. Metformin treatment in young children with fragile X syndrome. Mol Genet Genom Med. 2019;7:e956.
Ramanathan C, Kathale ND, Liu D, Lee C, Freeman DA, Hogenesch JB, et al. mTOR signaling regulates central and peripheral circadian clock function. PLoS Genet. 2018;14:e1007369.
Cao R, Robinson B, Xu H, Gkogkas C, Khoutorsky A, Alain T, et al. Translational control of entrainment and synchrony of the suprachiasmatic circadian clock by mTOR/4E-BP1 signaling. Neuron. 2013;79:712–24.
Cao R, Anderson FE, Jung YJ, Dziema H, Obrietan K. Circadian regulation of mammalian target of rapamycin signaling in the mouse suprachiasmatic nucleus. Neuroscience. 2011;181:79–88.
Lipton JO, Sahin M. The neurology of mTOR. Neuron. 2014;84:275–91.
Tsai PT, Hull C, Chu Y, Greene-Colozzi E, Sadowski AR, Leech JM, et al. Autistic-like behaviour and cerebellar dysfunction in Purkinje cell Tsc1 mutant mice. Nature. 2012;488:647–51.
Bhattacharya A, Kaphzan H, Alvarez-Dieppa AC, Murphy JP, Pierre P, Klann E. Genetic removal of p70 S6 kinase 1 corrects molecular, synaptic, and behavioral phenotypes in fragile X syndrome mice. Neuron. 2012;76:325–37.
Gkogkas CG, Khoutorsky A, Cao R, Jafarnejad SM, Prager-Khoutorsky M, Giannakas N, et al. Pharmacogenetic inhibition of eIF4E-dependent Mmp9 mRNA translation reverses fragile X syndrome-like phenotypes. Cell Rep. 2014;9:1742–55.
Acknowledgements
We thank S. Fatemi and I. Gantois for critical reading of the manuscript. The work was supported by grants from NIH (NS118026 and DK109714 to RC; MH119355 to AA and PK.; NS097312, DA048822 to AA; NS022920 and NS045667 to HTO; GM114142 and CA217297 to VXJ), from the Winston and Maxine Wallin Neuroscience Discovery Fund, from the University of Minnesota Medical School Foundation and from Whiteside Institute for Clinical Research to RC.
Author information
Authors and Affiliations
Contributions
DL, CN, PK, AA, HTO, CGG, and RC designed research; DL, CN, KS, EL, ISA, KC, SP, and RC performed research; DL, CN, JL, KF, CGG, and RC analyzed data; DL, CN, EL, JCG, VXJ, PK, AA, HTO, CGG and RC wrote the manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
About this article
Cite this article
Liu, D., Nanclares, C., Simbriger, K. et al. Autistic-like behavior and cerebellar dysfunction in Bmal1 mutant mice ameliorated by mTORC1 inhibition. Mol Psychiatry 28, 3727–3738 (2023). https://doi.org/10.1038/s41380-022-01499-6
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41380-022-01499-6
This article is cited by
-
Neural function of Bmal1: an overview
Cell & Bioscience (2023)
-
The comparative effectiveness of metformin and risperidone in a rat model of valproic acid-induced autism, Potential role for enhanced autophagy
Psychopharmacology (2023)
-
Krüppel-like factor 7 deficiency causes autistic-like behavior in mice via regulating Clock gene
Cell & Bioscience (2022)
-
The trilateral interactions between mammalian target of rapamycin (mTOR) signaling, the circadian clock, and psychiatric disorders: an emerging model
Translational Psychiatry (2022)
-
Signalling pathways in autism spectrum disorder: mechanisms and therapeutic implications
Signal Transduction and Targeted Therapy (2022)