Abstract
Sinonasal papillomas are benign epithelial tumors of the sinonasal tract that are associated with a synchronous or metachronous sinonasal carcinoma in a subset of cases. Our group recently identified mutually exclusive EGFR mutations and human papillomavirus (HPV) infection in inverted sinonasal papillomas and frequent KRAS mutations in oncocytic sinonasal papillomas. We also demonstrated concordant mutational and HPV infection status in sinonasal papilloma-associated sinonasal carcinomas, confirming a clonal relationship between these tumors. Despite our emerging understanding of the oncogenic mechanisms driving formation of sinonasal papillomas, little is currently known about the molecular mechanisms of malignant progression to sinonasal carcinoma. In the present study, we utilized targeted next-generation DNA sequencing to characterize the molecular landscape of a large cohort of sinonasal papilloma-associated sinonasal carcinomas. As expected, EGFR or KRAS mutations were present in the vast majority of tumors. In addition, highly recurrent TP53 mutations, CDKN2A mutations, and/or CDKN2A copy-number losses were detected; overall, nearly all tumors (n = 28/29; 96.6%) harbored at least one TP53 or CDKN2A alteration. TERT copy-number gains also occurred frequently (27.6%); however, no TERT promoter mutations were identified. Other recurrent molecular alterations included NFE2L2 and PIK3CA mutations and SOX2, CCND1, MYC, FGFR1, and EGFR copy-number gains. Importantly, TP53 mutations and CDKN2A alterations were not detected in matched sinonasal papillomas, suggesting that these molecular events are associated with malignant transformation. Compared to aerodigestive tract squamous cell carcinomas from The Cancer Genome Atlas (TCGA) project, sinonasal papilloma-associated sinonasal carcinomas have a distinct molecular phenotype, including more frequent EGFR, KRAS, and CDKN2A mutations, TERT copy-number gains, and low-risk human papillomavirus (HPV) infection. These findings shed light on the molecular mechanisms of malignant progression of sinonasal papillomas and may have important diagnostic and therapeutic implications for patients with advanced sinonasal cancer.
Similar content being viewed by others
Introduction
Sinonasal papillomas are uncommon benign epithelial tumors of the sinonasal tract; however, a small subset of cases are associated with a synchronous or metachronous sinonasal carcinoma—most commonly squamous cell carcinoma or one of its variants (i.e., adenosquamous carcinoma, etc.) [1, 2]. Over the past several years, our group has utilized a variety of conventional and next-generation sequencing (NGS) approaches to define the oncogenic events that occur in specific sinonasal papilloma subtypes, including mutually exclusive EGFR mutations and Human papillomavirus (HPV) infection in inverted sinonasal papilloma and KRAS mutations in oncocytic sinonasal papillomas [3,4,5]. We have also demonstrated that matched pairs of sinonasal papillomas and associated sinonasal carcinomas have concordant EGFR, KRAS, and HPV genotypes, indicating a clonal molecular relationship between these tumors.
Despite our emerging understanding of sinonasal papilloma oncogenesis, the molecular mechanisms underlying malignant progression to sinonasal carcinoma are relatively understudied. While several studies have reported a high incidence of TP53 mutations in sinonasal papilloma-associated sinonasal carcinomas [6, 7], a comprehensive assessment of the molecular landscape of these tumors is lacking. Thus, in this study, we sought to characterize mutations and copy-number alterations in sinonasal papilloma-associated sinonasal carcinomas utilizing targeted next-generation DNA sequencing (DNAseq) of frequently altered pan-cancer genes. We also evaluated molecular alterations within matched sinonasal papilloma–carcinoma pairs and compared the molecular landscape of sinonasal papilloma-associated sinonasal carcinomas to available large cohorts of aerodigestive tract squamous cell carcinomas.
Materials and methods
Case selection and DNA extraction
With Institutional Review Board approval, sinonasal papilloma-associated sinonasal carcinomas were retrospectively identified from surgical pathology records databases at Michigan Medicine. Sinonasal papilloma-associated sinonasal carcinomas were defined as a sinonasal carcinoma with either a concurrent or previously diagnosed sinonasal papilloma. (A majority of the cases in the current study were included in previous studies; see Supplementary Table 1 for details [3,4,5].) For a subset of the cases (n = 11), matched sinonasal papilloma material was available for comparison. Available hematoxylin and eosin slides from each case were reviewed by experienced head and neck pathologists (AMU and JBM) to confirm the diagnosis and select areas of tumor for sequencing; corresponding formalin-fixed paraffin-embedded (FFPE) tissue was macrodissected from glass slides, and DNA was isolated using the Pinpoint Slide DNA Isolation System Kit (D3001; Zymo Research, Irvine, CA).
Targeted next-generation sequencing (NGS)
Targeted next-generation DNAseq was performed essentially as described previously [8]. Briefly, FFPE-extracted DNA was quantitated using the QubitTM dsDNA HS Assay Kit (Q32851; Thermo Fisher Scientific, Waltham, MA), and for each sample, amplicon-based NGS libraries were generated from up to 20 ng of DNA by multiplex PCR using the Ion AmpliSeq Library Kit 2.0 (4475345; Thermo Fisher Scientific) and a custom pan-cancer DNA AmpliSeq panel (Oncomine Comprehensive Panel, version 1c; Thermo Fisher Scientific). NGS libraries were quantitated using qPCR, and sequencing templates were generated using the Ion PI™ Hi-Q™ OT2 200 Kit (A26434; Thermo Fisher Scientific). Templated libraries were pooled and sequenced on an Ion Torrent Proton machine using the Ion PI™ Chip Kit v3 (A26771; Thermo Fisher Scientific). NGS quality control metrics for all samples are provided in Supplementary Table 2. Sequence alignment and analysis was performed using Ion Torrent Suite Software (version 5.0.4; Thermo Fisher Scientific) and established in-house bioinformatics pipelines. Prioritized mutations and copy-number alterations were manually curated by an experienced molecular pathologist (AMU) using previously established criteria [9].
HPV infection status
For a subset of samples, the presence and subtype of HPV genomic DNA was assessed using GP5+/GP6+ consensus primers for L1 as described previously [5]. This method detects both low-risk and high-risk HPV subtypes.
TERT promoter mutation testing
TERT promoter mutation testing was performed as described previously [10]. Briefly, an allele-specific PCR assay was performed, targeting the most common TERT promoter mutations including c.-146C>T (Chr.5:1295250C>T), c.-124C>T (Chr.5:1295228C>T), c.-138_139CC>TT (Chr.5:1295242_1295243CC>TT), and c.-124_125CC>TT (Chr.5:1295228_1295229CC>TT).
The cancer genome atlas (TCGA) analysis
TCGA data for aerodigestive tract squamous cell carcinomas, including lung (LUSC) and head and neck (HNSC), were visualized using cBioPortal (www.cbioportal.org) [11,12,13,14]. Genes with one or more prioritized alterations in the sinonasal papilloma-associated sinonasal carcinoma cohort were selected for comparison to the TCGA cohorts, and only mutations classified as putative drivers or significant copy-number alterations (i.e., amplifications and deep deletions) in the TCGA datasets were retained for subsequent analysis. HPV infection data for head and neck squamous cell carcinomas were obtained from the HNSC TCGA dataset [12], while HPV infection data for lung squamous cell carcinomas were obtained from the TCGA Pan-Cancer Atlas [15]. The relative frequency of specific molecular alterations across these cohorts was examined using Chi-squared or Fisher’s Exact tests, as indicated.
Results
Recurrent molecular alterations in sinonasal papilloma-associated sinonasal carcinomas
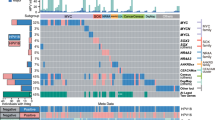
A total of 29 sinonasal papilloma-associated sinonasal carcinomas were available for the purposes of this study. All tumors were squamous cell carcinomas (either keratinizing or non-keratinizing) or squamous cell carcinoma variants (i.e., adenosquamous carcinoma, etc.). To explore the molecular landscape of these tumors, we utilized targeted DNAseq using a custom pan-cancer 133-gene panel that detects mutations and copy-number changes in recurrently altered oncogenes and tumor suppressor genes [8]. Overall, a total of 76 mutations (median per tumor = 3; range = 0–6) and 38 copy-number alterations (median per tumor = 1; range = 0–4) were identified by this panel (see Fig. 1 and Supplementary Table 1 for details). As expected, targeted DNAseq confirmed the presence of recurrent mutually exclusive EGFR (n = 21) and KRAS (n = 5) mutations in sinonasal papilloma-associated sinonasal carcinomas; three tumors lacked both EGFR and KRAS mutations—two of which harbored low-risk HPV subtype 11, and one for which HPV PCR analysis failed. As described previously, nearly all EGFR mutations occurred in exons 19 or 20; however, a nonsynonymous exon 6 mutation (p.R222C) was detected in one case. Aside from EGFR and KRAS, recurrent mutations included TP53 (n = 22), CDKN2A (n = 12), NFE2L2 (n = 4), PIK3CA (n = 4), ATM (n = 2), FBXW7 (n = 2), NOTCH1 (n = 2), and PIK3R1 (n = 2); recurrent copy-number gains included TERT (n = 8), SOX2 (n = 6), CCND1 (n = 5), MYC (n = 4), FGFR1 (n = 3), MYCL (n = 2), and PIK3CA (n = 2), while recurrent copy-number losses included two-copy loss (“deep deletion”) of CDKN2A (n = 10). Integration of mutation and copy-number data revealed that nearly all tumors (n = 28; 96.6%) harbored at least one TP53 or CDKN2A alteration; in addition, three tumors with EGFR mutations harbored concurrent EGFR copy-number gain (two of the mutant allele and one of the wild-type allele), while one tumor with a KRAS mutation showed copy-number gain of the mutant allele.
Heatmap of prioritized somatic variants and copy-number alterations highlights recurrent molecular alterations identified in 29 sinonasal papilloma-associated sinonasal carcinomas, including: 24 inverted sinonasal papilloma-associated sinonasal carcinomas (I); and, 5 oncocytic sinonasal papilloma-associated sinonasal carcinomas (O). Recurrent somatic variants include EGFR, KRAS, TP53, CDKN2A, NFE2L2, PIK3CA, ATM, FBXW7, NOTCH1, and PIK3R1; recurrent copy-number gains include EGFR, KRAS, TERT, SOX2, CCND1, MYC, FGFR1, MYCL, and PIK3CA, while recurrent copy-number losses include CDKN2A. Integration of somatic variant and copy-number data reveals that nearly all tumors (n = 28; 96.6%) harbor at least one TP53 or CDKN2A alteration. Tumor samples are ordered from top to bottom by type (I or O) and then increasing NGS ID number. Human papillomavirus infection status is indicated as follows: positive (P), negative (N), or unknown (U). Molecular alterations are ordered from left to right by decreasing frequency and then alphabetical order. Somatic variants are annotated by type: nonsynonymous = yellow; frame-preserving indel = green; stopgain (nonsense) mutation = pink; frameshift indel = orange; and splicing variant = purple. Copy-number alterations are annotated by type: amplification = red; and deep deletion (two-copy loss) = blue.
Non-EGFR/non-KRAS molecular alterations are uncommon in sinonasal papillomas associated with sinonasal carcinomas
Our previous studies highlighted the clonal molecular relationship between sinonasal papillomas and associated sinonasal carcinomas [3,4,5], however, the molecular events underlying malignant progression of sinonasal papillomas remain incompletely explored. A matched sinonasal papilloma sample was available from 11 of the sequenced sinonasal carcinoma cases. As expected, EGFR and KRAS genotypes were concordant for all matched papilloma–carcinoma pairs; however, no copy-number alterations were identified in any of the matched sinonasal papillomas, and aside from EGFR and KRAS, only one other mutation (an inactivating PIK3R1 frameshift mutation present in one matched papilloma–carcinoma pair) was detected (see Fig. 2 and Table 1 for details). Overall, these results indicate that sinonasal papillomas have a low mutational burden and genomic complexity and only infrequently harbor mutations in genes commonly altered in associated sinonasal carcinomas. Strikingly, despite the high prevalence of TP53 and CDKN2A alterations in associated sinonasal carcinomas, these alterations were not identified in any of the 11 matched sinonasal papillomas, suggesting that TP53 and/or CDKN2A alterations are early molecular events in the progression to sinonasal carcinoma.
Annotated copy-number plots for four matched sinonasal papilloma–carcinoma pairs (patients #2, 4, 9, and 10) depicting prioritized somatic variants and copy-number alterations (see Table 1 for additional details). The presence of concordant EGFR (patient #2, 4, and 9) or KRAS (patient #10) genotypes confirms clonality of these matched sample pairs. In contrast to carcinoma samples—which demonstrate frequent prioritized TP53 mutations, CDKN2A mutations, and/or copy-number alterations (i.e., TERT amplification, CCND1 amplification, EGFR amplification, KRAS amplification, etc.)—papilloma samples do not frequently harbor additional prioritized molecular alterations (beyond EGFR or KRAS mutations). Each circle represents a different targeted gene on the next-generation sequencing (NGS) panel; gray = no somatic variant or copy-number alteration, yellow = somatic variant without copy-number alteration, red = amplification, and blue = deep deletion (two-copy loss). Log2 copy-number ratio is depicted on the y-axis, and genes are ordered in ascending genomic position from left to right. Error bars indicate 95% confidence intervals for prioritized copy-number alterations.
Sinonasal papilloma-associated sinonasal carcinomas are molecularly distinct from other squamous cell carcinomas of the aerodigestive tract
Squamous cell carcinomas account for the vast majority of sinonasal papilloma-associated sinonasal carcinomas; however, the molecular landscape of these tumors relative to other aerodigestive tract squamous cell carcinomas has never been directly explored. Thus, we sought to compare the results of targeted DNAseq in our cohort of sinonasal papilloma-associated sinonasal carcinomas to available large cohorts of sequenced lung and head and neck squamous cell carcinomas from the TCGA project [11, 12, 15]. As expected, EGFR and KRAS mutations are significantly enriched in sinonasal papilloma-associated sinonasal carcinomas relative to other aerodigestive tract squamous cell carcinomas (P < 0.001); CDKN2A mutations, TERT copy-number gains, and low-risk HPV infection also occur more frequently in these tumors (P < 0.05) (see Fig. 3 and Table 2). Overall, these data indicate that sinonasal papilloma-associated sinonasal carcinomas are molecularly distinct from squamous cell carcinomas of the aerodigestive tract.
Bar graph showing the relative frequency of specific molecular alterations in sinonasal papilloma-associated sinonasal carcinomas compared to aerodigestive tract squamous cell carcinomas (SCC) in The Cancer Genome Atlas (TCGA) cohort, including head and neck (HNSCC) and lung SCC. Sinonasal papilloma-associated sinonasal carcinomas show a number of distinct molecular features, including increased proportion of tumors with EGFR mutations, KRAS mutations, low-risk human papillomavirus (HPV) infection, CDKN2A mutations, and/or TERT amplifications (see Table 2 for details). Asterisks indicate statistical significance (P < 0.05) for pairwise comparisons between sinonasal papilloma-associated sinonasal carcinomas with TCGA HNSCC and TCGA lung SCC.
TERT promoter mutations are uncommon in sinonasal papillomas and associated sinonasal carcinomas
Finally, given the relatively high frequency of TERT copy-number gains in sinonasal papilloma-associated sinonasal carcinomas (27.6% in our cohort), as well as the comparatively low frequency of TERT copy-number gains in aerodigestive tract squamous cell carcinomas from the TCGA cohorts (see above for details), we wondered whether TERT dysregulation may be a more general feature of malignant progression of sinonasal papillomas. Thus, we performed TERT promoter mutation testing on a subset of sinonasal carcinoma and papilloma cases from our cohort using an allele-specific PCR-based approach that detects the majority of such mutations in cancer and has been previously validated in a large cohort of urothelial carcinoma specimens [10]. Surprisingly, we found no evidence of TERT promoter mutations in sinonasal papillomas or carcinomas, suggesting that such mutations are uncommon in these tumors.
Discussion
In this manuscript, we report the first comprehensive assessment of mutations and copy-number alterations in sinonasal papilloma-associated sinonasal carcinomas. As expected, based on our previous work (which constitutes most of the specimens profiled in this study), the majority of tumors harbored either activating EGFR or KRAS mutations, while a small subset of tumors demonstrated low-risk HPV infection. Inactivating TP53 and/or CDKN2A mutations with loss of heterozygosity or two-copy loss (“deep deletion”) of CDKN2A was frequently observed in sinonasal carcinomas but was not present in matched sinonasal papillomas, indicating that these alterations may be early molecular events in malignant progression to sinonasal carcinoma. In addition, sinonasal papilloma-associated sinonasal carcinomas harbor a number of recurrent molecular alterations that are commonly found in other aerodigestive tract squamous cell carcinomas, although subsets of these alterations are relatively enriched in sinonasal carcinomas—suggesting potential novel carcinogenic pathways in these tumors.
Previous studies have implicated TP53 mutations and HPV infection (particularly high-risk subtypes) in the malignant progression of sinonasal papillomas [1, 6, 7]. While our results clearly support the hypothesis that TP53 mutations are early molecular events in malignant progression to sinonasal carcinoma, there appears to be an emerging consensus against a role for high-risk HPV infection in these tumors. Indeed, in addition to our current and prior data [5], a recent large tissue microarray-based study by Rooper et al. failed to show evidence of transcriptionally active high-risk HPV infection in sinonasal papilloma-associated sinonasal carcinomas [16]. In contrast, prior work from our group and others has indicated that low-risk HPV infection in inverted sinonasal papilloma may be associated with an increased risk of malignant progression [5, 17]. Whether low-risk HPV infection is truly a risk factor for malignant progression needs additional study in larger multi-institutional and/or prospective cohorts; however, given the fact that low-risk E6 and E7 oncoproteins exert only weak effects on p53 and Rb, respectively, if low-risk HPV infection is a risk factor for malignant progression, nontraditional oncogenic mechanisms may be involved [18]. For example, our group previously demonstrated that genomic integration of the low-risk HPV subtype 11 occurs in subsets of sinonasal papilloma-associated sinonasal carcinoma but not the associated sinonasal papilloma [19]. Overall, these data indicate a need for additional study of the molecular mechanisms underlying malignant progression of sinonasal papillomas with low-risk HPV infection.
In addition to TP53 mutations, our study highlights a central role for CDKN2A inactivation—either through mutation and subsequent loss of heterozygosity or focal “deep deletion” of the gene locus—in sinonasal papilloma-associated sinonasal carcinoma. Indeed, the vast majority of tumors (72.4%) showed evidence of at least one CDKN2A alteration, and all except one (96.6%) harbored at least one TP53 or CDKN2A alteration. Importantly, none of these alterations were detected in the subset of matched sinonasal papillomas from our cohort, indicating that TP53 and CDKN2A are likely to be early molecular events in the malignant progression to sinonasal carcinoma. The frequency of these alterations also suggests that tobacco exposure may play an etiologic role in many sinonasal papilloma-associated sinonasal carcinomas, as the concurrent presence of TP53 and CDKN2A alterations is strongly associated with tobacco exposure in the TCGA head and neck squamous cell carcinoma cohort [12]. Indeed, we have previously reported that the majority of patients with sinonasal papilloma-associated sinonasal carcinoma have prior or ongoing tobacco exposure [3]. Future studies should examine the molecular changes accompanying epithelial dysplasia in sinonasal papillomas and assess the potential diagnostic utility of common ancillary tools (i.e., p53 and/or p16 immunohistochemistry, etc.) for detecting dysplastic lesions.
Aside from TP53 and CDKN2A alterations, sinonasal papilloma-associated sinonasal carcinomas demonstrate a number of recurrently altered genes, which are likely to be secondary events in the malignant progression to sinonasal carcinoma. These secondary alterations include NFE2L2 mutations and SOX2, CCND1, MYC, and FGFR1 copy-number gains, which are frequently observed in other aerodigestive tract squamous cell carcinomas. Interestingly, the most common secondary alteration is TERT copy-number gain (occurring in 27.6% of tumors), which is relatively infrequently observed in lung and head and neck squamous cell carcinomas (occurring in 9.0% and 6.7% of tumors, respectively). TERT copy-number gain drives aberrant TERT overexpression, which facilitates carcinogenesis via inappropriate telomere maintenance in otherwise rapidly dividing cells [20]. Given that inappropriate telomere maintenance can occur via a variety of different mechanisms, including TERT promoter mutations, we examined a subset of sinonasal papillomas and associated carcinomas without TERT copy-number gains for TERT promoter mutations but did not identify any such mutations. Future studies should investigate other potential mechanisms of inappropriate telomere maintenance in sinonasal papilloma-associated sinonasal carcinomas, as well as possible additional transcriptional and/or epigenomic mechanisms of malignant progression to sinonasal carcinoma.
The aerodigestive tract—a contiguous luminal structure comprising the respiratory tract and upper portion of the digestive system—is a hotspot for human malignancy, including lung and head and neck cancers. However, the types and etiologies of tumors in this region vary dramatically by anatomic site: squamous cell carcinoma of the lung typically involves the bronchial tree and shows a strong association with tobacco exposure; laryngeal and oral cavity squamous cell carcinoma similarly shows a strong association with tobacco exposure; and, oropharyngeal squamous cell carcinoma is typically associated with infection by high-risk HPV subtypes (although tobacco exposure is still a risk factor) [11, 12]. The sinonasal tract is a unique anatomic subset of the aerodigestive tract, and although the majority of its malignant tumors are squamous cell carcinomas associated with tobacco exposure or high-risk HPV infection, recent morphologic and molecular profiling studies have delineated a number of characteristic tumors with specific molecular alterations (i.e., NUT carcinoma, SMARCB1 (INI-1)-deficient sinonasal carcinoma, IDH2-mutant sinonasal undifferentiated carcinoma, etc.) [2]. The results of this study (and others) clearly indicate that sinonasal papilloma-associated sinonasal carcinomas are similarly molecularly distinct from other aerodigestive tract squamous cell carcinomas, with frequent EGFR and KRAS mutations that are only exceptionally seen at other anatomic sites [11, 12]. In addition, a small (but significant) subset of tumors appears to be related to low-risk HPV infection, which is not commonly observed in other aerodigestive tract squamous cell carcinomas. As we have shown, the presence of these distinctive oncogenic alterations in sinonasal carcinomas is due to their clonal relationship with associated sinonasal papillomas, which highlights the need for additional molecular investigation of both sinonasal papillomas and associated sinonasal carcinomas.
Finally, although sinonasal papilloma-associated sinonasal carcinomas are relatively uncommon tumors, they are associated with the potential for significant morbidity and mortality. As we have shown previously, the presence of activating EGFR mutations in a large subset of these tumors indicates the potential for targeted therapeutic approaches with EGFR inhibitors, including next-generation molecules that have increased efficacy against tumors with exon 20 insertions (e.g., poziotinib) [3, 21, 22]. In contrast, based on collective experience in lung and colon cancers, the presence of KRAS mutations in a subset of sinonasal papilloma-associated sinonasal carcinomas likely predicts primary resistance to anti-EGFR-based therapies. Novel therapeutics targeting the downstream components of the RAF/RAS pathway (i.e., MEK and ERK inhibitors) are in development and clinical trials for patients with KRAS-mutant tumors; however, no FDA-approved drugs that target these alterations are currently available. Importantly, the results of our current study suggest that subsets of sinonasal papilloma-associated sinonasal carcinomas harbor potentially therapeutically targetable alterations, including PI3K/AKT pathway alterations (i.e., PIK3CA or PIK3R1 mutations, PTEN deletion, etc.) and other rare molecular alterations (e.g., CDK6, MYC, or FGFR1 amplification).
In conclusion, our results shed light on the molecular mechanisms underlying malignant progression of sinonasal papillomas. It is likely that as our understanding and awareness of the unique molecular landscape of sinonasal papilloma-associated sinonasal carcinoma improves, there may potentially be important diagnostic and therapeutic implications for patients with sinonasal cancer.
References
Barnes L. Schneiderian papillomas and nonsalivary glandular neoplasms of the head and neck. Mod Pathol. 2002;15:279–97.
Weindorf SC, Brown NA, McHugh JB, Udager AM. Sinonasal papillomas and carcinomas a contemporary update with review of an emerging molecular classification. Arch Pathol Lab Med. 2019;143:1304–16.
Udager AM, Rolland DCM, McHugh JB, Betz BL, Murga-Zamalloa C, Carey TE, et al. High-frequency targetable EGFR mutations in sinonasal squamous cell carcinomas arising from inverted sinonasal papilloma. Cancer Res. 2015;75:2600–6.
Udager AM, McHugh JB, Betz BL, Montone KT, Livolsi VA, Seethala RR, et al. Activating KRAS mutations are characteristic of oncocytic sinonasal papilloma and associated sinonasal squamous cell carcinoma. J Pathol. 2016;239:394–8.
Udager AM, McHugh JB, Goudsmit CM, Weigelin HC, Lim MS, Elenitoba-Johnson KSJ, et al. Human papillomavirus (HPV) and somatic EGFR mutations are essential, mutually exclusive oncogenic mechanisms for inverted sinonasal papillomas and associated sinonasal squamous cell carcinomas. Ann Oncol. 2018;29:466–71.
Caruana SM, Zwiebel N, Cocker R, McCormick SA, Eberle RC, Lazarus P. p53 alteration and human papilloma virus infection in paranasal sinus cancer. Cancer. 1997;79:1320–8.
Finkelstein SD, Tiffee JC, Bakker A, Swalsky P, Barnes L. Malignant transformation in sinonasal papillomas is closely associated with aberrant p53 expression. Mol Diagn. 1998;3:37–41.
Hovelson DH, McDaniel AS, Cani AK, Johnson B, Rhodes K, Williams PD, et al. Development and validation of a scalable next-generation sequencing system for assessing relevant somatic variants in solid tumors. Neoplasia. 2015;17:385–99.
Lazo de la Vega L, McHugh JB, Cani AK, Kunder K, Walocko FM, Liu CJ, et al. Comprehensive molecular profiling of olfactory neuroblastoma identifies potentially targetable FGFR3 amplifications. Mol Cancer Res. 2017;15:1551–7.
Brown NA, Lew M, Weigelin HC, Weizer AZ, Montgomery JS, Betz BL, et al. Comparative study of TERT promoter mutation status within spatially, temporally and morphologically distinct components of urothelial carcinoma. Histopathology. 2018;72:354–6.
Hammerman PS, Voet D, Lawrence MS, Voet D, Jing R, Cibulskis K, et al. Comprehensive genomic characterization of squamous cell lung cancers. Nature. 2012;489:519–25.
Lawrence MS, Sougnez C, Lichtenstein L, Cibulskis K, Lander E, Gabriel SB, et al. Comprehensive genomic characterization of head and neck squamous cell carcinomas. Nature. 2015;517:576–82.
Cerami E, Gao J, Dogrusoz U, Gross BE, Sumer SO, Aksoy BA, et al. The cBio cancer genomics portal: an open platform for exploring multidimensional cancer genomics data. Cancer Discov. 2012;2:401–4.
Gao J, Aksoy BA, Dogrusoz U, Dresdner G, Gross B, Sumer SO, et al. Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sci Signal. 2013;6:pl1.
Campbell JD, Yau C, Bowlby R, Liu Y, Brennan K, Fan H, et al. Genomic, pathway network, and immunologic features distinguishing squamous carcinomas. Cell Rep. 2018;23:194–212.e6.
Rooper LM, Bishop JA, Westra WH. Transcriptionally active high-risk human papillomavirus is not a common etiologic agent in the malignant transformation of inverted schneiderian papillomas. Head Neck Pathol. 2017;11:346–53.
Mehrad M, Stelow EB, Bishop JA, Wang X, Haynes W, Oliver D, et al. Transcriptionally active HPV and targetable EGFR mutations in sinonasal inverted papilloma: an association between low-risk HPV, condylomatous morphology, and cancer risk? Am J Surg Pathol. 2020;44:340–6.
Egawa N, Doorbar J. The low-risk papillomaviruses. Virus Res. 2017;231:119–27.
Scheel A, Lin GC, Mchugh JB, Komarck CM, Walline HM, Prince ME, et al. Human papillomavirus infection and biomarkers in sinonasal inverted papillomas: clinical significance and molecular mechanisms. Int Forum Allergy Rhinol. 2015;5:701–7.
Yuan X, Larsson C, Xu D. Mechanisms underlying the activation of TERT transcription and telomerase activity in human cancer: old actors and new players. Oncogene. 2019;38:6172–83.
Robichaux JP, Elamin YY, Tan Z, Carter BW, Zhang S, Liu S, et al. Mechanisms and clinical activity of an EGFR and HER2 exon 20-selective kinase inhibitor in non-small cell lung cancer. Nat Med. 2018;24:638–46.
Vyse S, Huang PH. Targeting EGFR exon 20 insertion mutations in non-small cell lung cancer. Signal Transduct Target Ther. 2019;4:5.
Acknowledgements
Funding for this study was provided by the Anatomic Pathology Projects Committee, Department of Pathology, Division of Anatomic Pathology, University of Michigan Medical School (AP Project #94) and the University of Michigan Head and Neck Specialized Program of Research Excellence (SPORE; NCI P50 CA97248).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
SAT is co-founder and chief medical officer of Strata Oncology. KRP is a current employee of Strata Oncology. The other authors declare that they have no conflict of interest.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
About this article
Cite this article
Brown, N.A., Plouffe, K.R., Yilmaz, O. et al. TP53 mutations and CDKN2A mutations/deletions are highly recurrent molecular alterations in the malignant progression of sinonasal papillomas. Mod Pathol 34, 1133–1142 (2021). https://doi.org/10.1038/s41379-020-00716-3
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41379-020-00716-3
This article is cited by
-
Low-grade papillary Schneiderian carcinoma with TP53 mutation: a case report and review of the literature
Diagnostic Pathology (2023)
-
An in vitro model and the underlying pathways of sinonasal inverted papilloma development
Scientific Reports (2023)
-
Histopathologic Diagnosis of Sinonasal Tumors: Challenges and the Importance of Establishing the Correct Diagnosis
Current Otorhinolaryngology Reports (2023)
-
Evaluation of high-risk human papillomavirus in sinonasal papillomas and squamous cell carcinomas
Virchows Archiv (2023)
-
Experimental Models of Sinonasal Tumors for Preclinical Testing of Candidate Targeted Therapies
Current Otorhinolaryngology Reports (2023)