Abstract
We report 17 cases of sinusoidal large B-cell lymphoma (SLBCL). Clinical, morphologic, immunophenotypic, and molecular features were detected and analyzed. All cases showed an obvious sinusoidal growth pattern, usually associated with residual atrophic lymphoid tissue. All tumors contained large pleomorphic lymphoid cells and one or more prominent nucleoli, with abundant amphophilic cytoplasms; 15/17 cases showed anaplastic morphologic features. The patient age ranged from 43 to 80 years (median 57 years), and 7 males and 10 females were included. Eleven of 15 (73.3%) patients had Ann Arbor stage III or IV disease, and 10/15 (66.6%) patients had an International Prognostic Index (IPI) score ≥3. Immunophenotypically, 16/17 (94.1%) cases displayed a nongerminal center B-cell (non-GCB) immunophenotype. Furthermore, 16/17 (94.1%) cases were positive for CD30, and p53 was expressed in 10/16 (62.5%) cases. In total, 12/14 (85.7%) cases expressed BCL2 and MYC simultaneously (double expression), and 11/14 (78.6%) cases showed PD-L1 positivity (6/11 had a PD-L1 tumor proportion score ≥50%). Cytogenetically, concurrent MYC and BCL2 and/or BCL6 abnormalities (break-apart or extra copy) were detected in 10/15 cases, and 7/13 (53.8%) cases harbored a PD-L1/L2 amplification. TP53 mutation was found in 7/13 (53.8%) cases by Sanger sequencing. Whole-exome and large-panel sequencing results revealed high mutation frequencies of TP53 (4/7), MYD88 (3/7), KMT2D (3/7), CREBBP (3/7), and PIM1 (3/7). Among the 13 patients with SLBCL treated with aggressive chemotherapy regimens, the median overall survival (OS) was 18 months, and the 2-year OS rate was 34.6%. The OS of patients with SLBCL was markedly worse than that of 35 control group patients with common diffuse large B-cell lymphoma (DLBCL) without sinusoidal features (P < 0.001). SLBCL may represent a specific type of DLBCL that has characteristic pathologic features. The cancer is aggressive in most clinical cases, and outcomes are poor. SLBCL and anaplastic DLBCL (A-DLBCL) have many overlapping clinicopathological and molecular features.
Similar content being viewed by others
Introduction
Diffuse large B-cell lymphoma (DLBCL) is a heterogeneous disease containing morphologic variants and immunophenotypic and molecular subgroups [1]. For large B-cell lymphomas involving lymph nodes, the most common growth mode is diffuse infiltration, which destroys part or all of the lymph node structure [2]; however, tumor cells can selectively invade or be limited to lymph node sinuses [3]. Sinusoidal large B-cell lymphoma (SLBCL) is not well characterized, most likely due to its rarity [3].
Coupland et al. [3] presented 11 cases of CD30 + SLBCL, which can closely imitate anaplastic large cell lymphoma (ALCL). Despite morphologic and immunophenotypic similarities, SLBCL is distinguishable from ALCL by many features, such as the B-cell lineage, lack of ALK-1 expression, and lack of cells with eosinophilic paranuclear regions. As a group, SLBCLs seem to behave aggressively and have worse outcomes than common DLBCLs. Some SLBCL tumor cells are positive for Epstein Barr virus (EBV). Recently, Asano and her colleagues [4] found a sinusoidal pattern in 16/18 cases of anaplastic diffuse large B-cell lymphoma (A-DLBCL). All cases were negative for PD-L1 on tumor cells, although PD-L1-positive nontumor cells surrounded the tumor cells, exhibiting a rosette-like pattern with compartmentalization. Their study highlighted the distinctiveness of A-DLBCL with a sinusoidal pattern [4]. However, only limited genetic information has been reported in previous studies describing these tumors [5,6,7,8,9,10].
To understand in depth the biological behavior of SLBCL, we investigated 17 cases using strict criteria to define clinicopathologic and genetic features [11, 12]. Our results show that patients with SLBCL display features that are clearly distinctive from common DLBCL, and most patients with SLBCL have an aggressive clinical course and poor outcomes. Overall, these results emphasize the importance of recognizing SLBCL, as this malignant tumor has a poor prognosis and distinctive biological features.
Materials and methods
Patient selection
We analyzed 17 patients with SLBCL originally diagnosed and treated at different institutions from January 1, 2003 to September 30, 2019. Five pathologists (JX, ML, SG, QY, and ZW) reviewed all cases according to the 2016 World Health Organization classification of criteria [11, 12]. Diagnoses for the 17 patients were based on lymph node biopsy specimens. The corresponding medical records were reviewed to obtain clinical information. A total of 14 well-documented patients were treated with chemotherapy regimens including cyclophosphamide, doxorubicin, vincristine, and prednisone (CHOP) plus rituximab (R-CHOP); the treatment strategies for the other three patients were unknown. We also collected 40 cases of common DLBCL without sinusoidal features from one institution (Xijing Hospital) spanning from April 1, 2009 to September 30, 2014; only DLBCL, not otherwise specified (DLBCL, NOS) cases, were included [11]. Five patients were lost to follow-up in the common DLBCL group. Institutional ethical approval was obtained in compliance with the Helsinki Declaration.
Immunohistochemistry (IHC), in situ hybridization (ISH), and fluorescence in situ hybridization (FISH) analysis
Fully automated protocols were used to perform IHC analysis with a Bond-III Autostainer (Leica Biosystems, Melbourne, Australia). Histological sections were stained with antibodies against the following: CD5, CD20, CD30, PD-L1, PAX5, MUM1, BCL2, BCL6, MYC, p53, and Ki-67. Detailed information about the antibodies (source, clone, dilution, and cutoff values) is shown in Supplementary Table 1. The PD-L1 status for eligibility was assessed using the anti-PD-L1 antibody clone SP263 (Merck & Co. Inc., Kenilworth, NJ, USA) and a clinical trial version of the approved IHC assay (PharmDx assay; Dako, Carpinteria, CA, USA). Expression was categorized by the tumor proportion score (TPS), which was defined as the percentage of tumor cells with membranous PD-L1 staining. PD-L1 positivity was defined as at least 1% membranous staining [13]. Germinal center B-cell (GCB) and non-GCB subtypes of SLBCL were classified using the Hans algorithm [14]. EBV was detected using ISH with EBV-encoded small RNA (EBER) probes (Leica Biosystems, Newcastle Upon Tyne, UK). For EBER ISH, a positive control ISH probe was used for each case.
FISH analysis was performed using LSI dual-color break-apart probes for MYC, BCL2, and BCL6 according to the manufacturer’s instructions (Guangzhou LBP Medicine Science & Technology Co., Ltd., Guangzhou, China). Images were collected using a workstation equipped with software (Imstar Pathfinder Cellsca FluoSpot, France). Areas with a minimum of 70% tumor cells were counted, and signals from 100 nonoverlapping nuclei were analyzed. Positivity was determined above a 15% threshold for split signals and above a 30% threshold for extra copies (defined as a copy number ≥3/cell). LSI dual-color amplification probes for PD-L1/L2 (Guangzhou LBP Medicine Science & Technology Co., Ltd.) were used according to the manufacturer’s instructions. Due to the close location of PD-L1 and PD-L2, this probe cannot separate PD-L1 and PD-L2. Furthermore, 9p24.1 (PD-L1/L2 gene) was labeled with SpectrumRed, and the centromere was labeled with SpectrumGreen as a control probe for 9p. Samples with two fusion signals (red and green colocalization) were classified as normal. The presence of more than three fusion signals was deemed amplification, and more than three SpectrumGreen signals resulted in a polyploid classification [15].
DNA extraction, polymerase chain reaction (PCR), and sanger sequencing
DNA was isolated from three 5-µm-thick sections of formalin-fixed paraffin-embedded (FFPE) tumor tissue with a minimum of 70% neoplastic cells using the QIAamp FFPE DNA Tissue Kit (Qiagen, Germantown, MD, USA) according to the manufacturer’s instructions. Selected exons and mutation hotspots of TP53 (exons 5–10) were investigated for mutations by PCR and sequencing. The primer sequences and PCR conditions are listed in Supplementary Table 2. The PCR products were routinely purified and sequenced in both directions using the BigDye Terminator version 3.1 Cycle Sequencing Kit (Applied Biosystems, Forster City, CA, USA). At least two independent PCR and sequencing experiments were used to confirm mutations, which were shown to not be single-nucleotide polymorphisms through the NCBI dbSNP database.
Whole-exome sequencing (WES) and large-panel sequencing
We selected six clinical cases of SLBCL that had sufficiently high DNA quality for WES. A QIAamp DNA FFPE Tissue Kit (Qiagen Inc., Hilden, Germany) was used to extract genomic DNA according to the manufacturer’s recommendations. Enrichment of 334,378 targeted exonic regions of 20,965 genes was performed using an Agilent SureSelect Human All Exon Kit (Santa Clara, CA, USA). The WES libraries were then amplified and quality checked, and Illumina HiSeq platform sequencing (Novogene Bioinformatics Technology Co., Ltd., Beijing, China) was carried out. One case was assessed using large-panel sequencing of 446 genes. A Prep DNA/RNA FFPE Kit (Qiagen Inc., Hilden, Germany) was used to extract genomic DNA and total RNA. A total of 100 ng of DNA was sheared to provide average fragments of 200 bp. Hybridization-captured, adapter ligation-based libraries were synthesized using a HemasalusD/DR (Shihe, Nanjing, China), which was designed to enrich panel genes that are important for the pathogenesis of hematologic tumors. These gene libraries were then amplified and quality checked, and HiSeq2500 platform sequencing (Illumina) was performed. The average sequencing depth of the target regions was >100×.
Genomic analysis
After generating raw data through base calling, paired-end reads were trimmed to remove stretches of low-quality bases (<Q10) and adapters in the sequences. Then, Burrows–Wheeler Aligner (http://biobwa.sourceforge.net/) and Samblaster (https://github.com/GregoryFaust/samblaster) were used to align the paired-end reads on the human reference genome GRCh37 (hg19). The Genome Analysis Toolkit (https://software.broadinstitute.org/gatk/) and SAMtools (http://www.htslib.org/) were used to perform variant calling. To maximize sensitivity in heterogeneous SLBCL specimens, validation was performed to detect basic replacement and short insertion and deletions (indels) with a threshold depth of coverage >5× and a variant allele frequency ≥10%. Furthermore, point mutations and small insertions in genes were annotated for their functional effect on their respective proteins via ANNOVAR (version 2013 Aug 23) (http://www.openbioinformatics.org/annovar/). Selected nonsynonymous and nonsense single-nucleotide variations (SNVs), frameshift/nonframeshift indels, and splice region mutations were detected. Variants present in the Exome Aggregation Consortium (http://exac.broadinstitute.org/) or 1000 Genomes (https://www.1000genomes.org/) databases with a frequency >1%, which indicated potential germline variants, were excluded from the analysis. All indels and SNVs predicted to be deleterious by at least one of four prediction tools (SIFT, PolyPhen-2_HDIV, Mutation Taster, CADD) were included in further analyses. The screened genes were compared with the COSMIC database.
Statistical analysis
Fisher’s exact and Mann–Whitney U tests were employed to compare patient characteristics across different subgroups. The χ2 test was used for correlation analysis of categorical variables. Additionally, OS was defined as the interval between the date of initial diagnosis and the time of death due to any cause or the date of last follow-up. Survival analysis was performed using Kaplan–Meier curves and compared using the log-rank test. Comparative test differences were considered significant if the 2-tailed P value was <0.05. SPSS (version 23.0; IBM Inc., Armonk, NY) was used for all statistical analyses.
Results
Patients
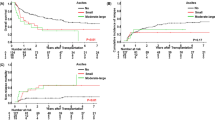
Table 1 summarizes the major clinical characteristics, treatment, and follow-up information for all 17 patients. The study group included 17 patients with SLBCL, including 7 men and 10 women, with a median age of 57 years (range, 43 to 80 years). All patients presented with de novo disease. Complete information, including clinical examination and radiologic studies, was recorded for 15 patients. Nine (60%) patients had B symptoms, and 11 (73.3%) showed elevated serum lactate dehydrogenase (LDH) levels. Eleven (73.3%) patients had Ann Arbor stage III or IV disease, and 10 (66.6%) patients had an IPI score ≥3. Since one patient was newly diagnosed and not treated, chemotherapy treatment and response were known for 14 patients. After the initial combined chemotherapy treatment, only 5/14 (35.7%) patients demonstrated a complete response (CR); 6 (42.9%) patients had a partial response, and 3 (21.4%) had progressive disease. Because one patient was lost to follow-up, clinical outcomes were available for 13 patients treated with aggressive chemotherapy regimens. At the time of last clinical follow-up (range, 1–50 mo), the median follow-up time was 16 months, and the median OS was 18 months. The 1-year OS rate was 69.2%, and the 2-year OS rate was 34.6% (Fig. 1).
Morphologic and immunophenotypic features
All 17 tumors occurred in the lymph nodes and included cervical (n = 11), inguinal (n = 2), supraclavicular (n = 2), submandibular (n = 1), and inguinal (n = 1) tumors. All cases exhibited a sinusoidal growth pattern of tumor cell distribution to degrees varying from 30 to 100% (Fig. 2a), often associated with residual atrophic lymphoid tissue. The lymphatic sinuses were dilated, and the sinus walls were covered with a complete monolayer of elongated cells (Fig. 2b). Large pleomorphic Reed–Sternberg (RS)-like tumor cells with abundant cytoplasms grew focally in a nest-like pattern and displayed round, kidney-shaped or polygonal nuclei. The nuclear membranes were clear, and 1 to several small nucleoli or 1 to 2 prominent nuclei were observed. Fifteen of 17 (88.2%) cases exhibited numerous anaplastic multinucleated tumor cells (n = 8) or scattered anaplastic tumor cells (n = 7) (Table 2, Supplementary Fig. 1).
Case 5 showed evidence of a sinusoidal growth pattern (a H&E, ×200; b H&E, ×400). The tumor cells of case 5 were positive for CD30 (c), BCL2 (d), BCL6 (e), and MYC (f) (×400). The p53 staining showed a diffuse strong positive pattern in case 6 (g) (×400). The tumor cells and nontumor cells were PD-L1-positive in case 6 (h) (×400).
The IHC results are listed in Table 2. All cases were strongly positive for CD20 and negative for CD5 and CD15. All 17 cases were negative for EBV RNA as determined by ISH. According to the Hans algorithm, 16/17 (94.1%) cases demonstrated a non-GCB immunophenotype, whereas one (5.9%) case demonstrated a GCB immunophenotype. The Ki-67 proliferation rate was generally high, with a median of 80% and 16/17 (93.8%) ranging from 70 to 90%. CD30 expression was variable, with 16 of 17 (93.8%) cases being positive, ranging from strong positivity in most neoplastic cells (Fig. 2c) to heterogeneous expression. CD30+ cells ranged from 23 to 90%, and the median value was 68%. Thirteen of 15 (86.7%) and 13/14 (92.9%) cases were positive for BCL2 (Fig. 2d) and MYC (Fig. 2f) expression, respectively, and 12 of 14 (85.7%) cases were positive for BCL2 and MYC (double expression). p53 staining patterns were classified into three groups as follows: four cases had diffuse strong positive expression (Fig. 2g), six cases had variable expression, and six cases were completely negative. With a cutoff value of 50%, 10/16 (63.3%) cases were p53-positive. Eleven of 14 (78.6%) cases demonstrated PD-L1 positivity on tumor cells (7 cases were strongly positive and 4 cases weakly positive), 5 of 11 (45.5%) had a PD-L1 TPS of 1–49%, and 6 of 11 (54.5%) had a PD-L1 TPS of ≥50% (Fig. 2h).
Molecular genetic characteristics
The FISH and mutation status results are listed in Table 2 and Fig. 3a. Extra copies of MYC were found in 12 of 16 (75%) patients (Fig. 3b). Regarding the 12/16 (75%) cases that harbored BCL2 abnormalities, 1 case showed a break-apart signal, and 11 cases carried an extra copy (Fig. 3b). An extra copy of BCL6 was found in 12/16 (75%) patients, including 6 patients who exhibited both a break-apart signal and an extra copy (Fig. 3b). Seven cases harbored PD-L1/L2 abnormalities, four cases had amplification, and another three cases carried amplifications with polyploidy. PD-L1/L2 was amplified in clusters in cases 8 and 9. A positive correlation was observed between PD-L1/L2 abnormalities and protein expression (r = 0.592, P = 0.033 < 0.05). Furthermore, 10/15 (66.7%) patients exhibited concurrent abnormalities in MYC and BCL2 and/or BCL6, and MYC, BCL2 and BCL6 triple abnormalities were found in 9/15 (60%) cases.
The FISH analysis showed extra copy signals for BCL2, BCL6, MYC, and amplification signals for PD-L1/L2, respectively, PD-L1/L2 is amplified in clusters in case 8, and case 12 showed that both break apart and extra copy signals were found for BCL6 (a) (×400). The summaries of the FISH analysis for BCL6, BCL2, MYC, and PD-L1/L2 (b). The distribution of the TP53 mutations is shown. The lines represent the position of the mutations (c).
With respect to mutation status, TP53 exons 5–10 were successfully amplified in 13/17 cases. Eight TP53 gene missense mutations were detected in 13 patients, including 1 who carried 2 gene alterations (Fig. 3c, Supplementary Fig. 2). All missense mutations occurred in the DNA-binding domain containing two mutations in Loop-L2 (codons 164–194), two mutations in Loop-L3 (codons 237–250), and two mutations in the LSH helix motif (codons 272–287). p53 expression was associated (r = 0.854, P = 0.0002 < 0.001) with TP53 missense mutations.
WES was performed for six cases of SLBCL; a large-panel sequencing of 446 panel genes that are important for the pathogenesis of hematologic tumors was performed in case 7. Candidate oncogenes with potentially deleterious mutations were analyzed in the coding sequence and splicing regions. For WES, an average of 69.2 million reads was obtained for each sample, and the average coverage depth of the target area of each sample was 71–160×. A total of 2134 mutations in 360 genes were identified in the six SLBCL patients (Supplementary Table 3). Missense mutations were the most frequent at 2005/2314 (86.6%) (Fig. 4a). All samples harbored nonsynonymous somatic alterations, with a maximum of 438 alterations detected in case 2 (Fig. 4b). WES (six cases) and large-panel sequencing (one case) results revealed that some genes had high mutation frequencies, such as TP53 (4/7), MYD88 (3/7), KMT2D (3/7), CREBBP (3/7), and PIM1 (3/7) (Fig. 4c). Compared with those of common DLBCLs in COSMIC data, the mutation frequencies of these genes in SLBCLs were obviously higher (Fig. 4d). However, no mutations in MLL2, Bcl-2, B2M, EZH2, and GNA13 were found in SLBCL based on WES and large-panel sequencing.
WES in six SLBCL patients showed variant mutation patterns with a predominance of missense mutations (a). Every specimen exhibited different mutation patterns, with a maximum of 438 alterations detected in case 2 (b). WES and large-panel sequencing identified 16 recurrently and significantly mutated genes for seven SLBCL patients (c). Common mutations in sinusoidal large B-cell lymphomas and their comparison with diffuse large B-cell lymphomas in COSMIC data (d).
Prognostic factors
The results of the above clinicopathological and genetic factors related to OS are shown in Supplementary Table 4. Univariate analysis demonstrated that patients with an IPI score ≥4, BCL6 abnormalities, and concurrent abnormalities of MYC and BCL2 and/or BCL6 had a significantly poorer OS than those who were negative for these features (P < 0.05; Fig. 5). All factors predictive of OS by univariate analysis were applied in multivariate analysis. Because of the small sample size, no independent prognostic factors were associated with OS.
SLBCL versus DLBCL without sinusoidal features (common DLBCL)
We compared patients with SLBCL (n = 17) to those with DLBCL without sinusoidal features (n = 40) (Table 3) and found that the common DLBCL patient group showed similar distributions of age and sex. However, patients with SLBCL presented more often with B symptoms, high-stage disease, high IPI scores and elevated serum LDH levels than patients with common DLBCL (P < 0.05). Because five patients with common DLBCL were lost to follow-up, clinical outcomes were available for 35 patients. At the time of last clinical follow-up of DLBCL (range, 1–94 mo), the median follow-up time was 41 months. The OS rate of patients with SLBCL was significantly worse than that of patients with common DLBCL (Fig. 1, P < 0.001). Cases of SLBCL more often showed a non-GCB immunophenotype and more frequently expressed CD30, MYC, BCL2, and PD-L1 than those of common DLBCL (P < 0.05). Moreover, SLBCL more often had TP53 abnormalities and concurrent abnormalities in MYC and BCL2 and/or BCL6 (P < 0.05). Patients with SLBCL also showed a trend of more MYC/BCL2 double expression and triple abnormalities of MYC, BCL2, and BCL6 (P < 0.05) than those with common DLBCL.
Discussion
We report 17 cases of a highly unusual variant of DLBCL characterized by the existence of a mainly sinusoidal infiltrative pattern. Conspicuous RS-like binucleated and multinucleated tumor cells were also present in some cases. In our study, despite intensive chemotherapy, patients with SLBCL displayed poor clinical outcomes, with 34.6% of patients being alive at 2 years and a median OS as short as 18 months; these survival statistics were much worse than those of patients with common DLBCL. Similar to that found by Coupland [3], the outcome of DLBCL was better than that of SLBCL. Patients with SLBCL have numerous poor prognostic features, and most of the patients in this study had high IPI scores. There was also a high frequency of patients in advanced Ann Arbor stages. These clinical findings suggest that SLBCL is a more aggressive disease than common DLBCL.
The present series of 17 cases of SLBCL revealed a high frequency of TP53 mutation, presenting in more than 50% of all cases, as well as concurrent abnormalities of MYC, BCL2, and/or BCL6 in ~66.7% of patients and triple abnormalities of MYC, BCL2, and BCL6 in ~60% of patients. Compared with typical cases of DLBCL, SLBCL has a high incidence of two- or three-gene abnormalities, which may be related to its poor prognosis.
In these SLBCL cases, numerous anaplastic multinucleated tumor cells or scattered anaplastic tumor cells were found in 15/17 (88.2%) cases. In the study of Asano et al., A-DLBCL exhibited a sinusoidal pattern in 16/18 cases [4]. In addition, we [16] reported 35 cases of A-DLBCL that showed at least focal evidence of a sinusoidal growth pattern in 12 of the 35. SLBCL and A-DLBCL display many clinicopathological and molecular feature similarities, such as high expression of p53, MYC, BCL2, and PD-L1; a non-GCB immunophenotype; a high frequency of TP53 mutations; concurrent MYC and BCL2 and/or BCL6 abnormalities; PD-L1/L2 amplification and/or polyploidy; and a poor prognosis [16]. Although SLBCL and A-DLBCL show many overlapping features, some differences do exist. In our study, the rate of CD30 positivity was significantly higher in SLBCL (16/17 cases) than in A-DLBCL (18/35, 51.4%) cases) [16], and all SLBCL cases were negative for CD5.
CD30 expression is present in ~14–25% of patients with common DLBCL [17, 18]. In this study, CD30 (16/17) was more frequently expressed in SLBCL than in common DLBCL. Brentuximab vedotin, an anti-CD30 antibody–drug conjugate, has been shown to be effective in patients with relapsed/refractory DLBCL with variable CD30 expression [19]. Given the relatively high expression of CD30 in patients with SLBCL, brentuximab vedotin is a potential treatment choice.
The PD-1/PD-L1 pathway is an inhibitory immune checkpoint with the ability to suppress T-cell immune activity [20], which thus allows tumor cells to escape from T-cell-mediated tumor-specific immunity by suppressing the immune activity of tumor-specific CD8+ T cells and thereby promoting tumor development [21]. Various studies have shown an association between increased PD-L1 expression and poor prognosis in several cancers, such as lung cancer [22], ovarian cancer [23], melanoma [24], and DLBCL [25]. In the present study, 7/13 (53.8%) cases had PD-L1/L2 amplification, and 11/14 SLBCLs (78.6%) expressed PD-L1. Amplification of PD-L1 is likely the cause of high PD-L1 expression. Several immunotherapeutic drugs for PD-L1 (durvalumab, atezolizumab and avelumab) and PD-1 (nivolumab and pembrolizumab) have been developed for clinical treatment [26] and may have certain effects on PD-L1 + SLBCL and improve clinical outcomes.
Suzuki et al. [27, 28] reported neoplastic PD-L1 (nPD-L1) expression in 35% of intravascular large B-cell lymphomas (IVLBCLs) by PD-L1 as determined immunostaining (clone SP142). IVLBCL is a rare variant of extranodal DLBCL, characterized by the proliferation of large tumor cells within the lumina of small or intermediate-sized blood vessels and capillaries [1, 29, 30]. However, our SLBCL cases showed tumor cells that were mainly located in the lymph node sinus, and there was no obvious evidence of extranodal vascular infiltration.
nPD-L1 expression in SLBCL with anaplastic features also raises the issue of differential diagnoses with nodal gray-zone lymphoma (GZL), which is unclassifiable and has features falling between those of DLBCL and CHL [1]. CHL-like GZL and LBCL-like GZL were divided into two subgroups (namely, groups 0 and 1 for cHL-like cases and groups 2 and 3 for LBCL-like cases) by Traverse-Glehen et al. [31], and SLBCL differs from these four subtypes. GZL group 0 and 1 cases showed a classic Hodgkin-like morphology, that is different from obvious sinusoidal morphology of SLBCL. GZL group 2 and 3 cases showed a large B-cell-like morphology with RS-like cells on an inflammatory background. Tumor cells present diffuse and strong expression of CD15 [31], and despite that SLBCL showed a large B-cell-like morphology, it had no inflammatory background, showed an obvious sinusoidal growth pattern, and was negative for CD15 staining.
According to WES and large-panel sequencing, the mutation frequencies of TP53, MYD88, KMT2D, CREBBP, and PIM1 in SLBCL were higher than those observed in DLBCL based on COSMIC data. The TP53 pathway is essential for suppressing tumors and favorable prognoses for patients with lymphoma [32]. Currently, treatment methods attempt to restore TP53 function and maintain TP53-dependent and TP53-independent apoptosis [32]. Further research to clarify the underlying mechanisms may lead to effective therapeutic intervention. Oncogenic MYD88 mutations, most notably the Leu 265 Pro (L265P) mutation, have been identified as potential drivers of various B-cell non-Hodgkin lymphomas [33, 34]. Mutation of MYD88 is related to activation of the NFκB pathway [35], and inhibitors of this pathway, such as ibrutinib, might serve as advantageous therapeutic targets for conventional chemotherapeutic intervention in SLBCLs [36]. KMT2D and CREBBP act as tumor suppressor genes, and their early loss facilitates lymphomagenesis by remodeling the epigenetic landscape of cancer precursor cells [37, 38], which may be related to the pathogenesis of SLBCL. PIM1 possesses oncogenic functions and is overexpressed in various types of cancer [39, 40], and its inhibition provides a new option for therapy. Mutations in these genes underlying the pathogenesis of SLBCL may provide new treatment options. Because next-generation sequencing was carried out for only seven patients, more cases are needed to confirm the results.
In conclusion, SLBCL displays many genetic alterations and biological features that differ substantially from common DLBCL. Our results show that SLBCL exhibits a non-GCB immunophenotype; has high expression of CD30, PD-L1, MYC, and BCL2; and has high frequencies of TP53 mutations, concurrent MYC and BCL2 and/or BCL6 abnormalities, and PD-L1/L2 amplification and/or polyploidy. WES showed high mutation frequencies of TP53, MYD88, KMT2D, CREBBP, and PIM1 in SLBCL. As most patients with SLBCL follow an aggressive disease course and have a poor prognosis, the recognition of SLBCL is important. In the future, novel treatment for patients with SLBCL is an urgent issue that warrants investigation.
References
Cazzola M. Introduction to a review series: the 2016 revision of the WHO classification of tumors of hematopoietic and lymphoid tissues. Blood. 2016;127:2361–4.
Liu Y, Barta SK. Diffuse large B-cell lymphoma: 2019 update on diagnosis, risk stratification, and treatment. Am J Hematol. 2019;94:604–16.
Lai R, Medeiros LJ, Dabbagh L, Formenti KS, Coupland RW. Sinusoidal CD30-positive large B-cell lymphoma: a morphologic mimic of anaplastic large cell lymphoma. Mod Pathol. 2000;13:223–8.
Megahed NA, Kohno K, Sakakibara A, Eladl AE, Elsayed AA, Wu CC, et al. Anaplastic variant of diffuse large B-cell lymphoma: Reappraisal as a nodal disease with sinusoidal involvement. Pathol Int. 2019;69:697–705.
Bhatt NS, Kelly ME, Batdorf B, Gheorghe G. Sinusoidal CD30+ diffuse large B-cell lymphoma can masquerade as anaplastic large cell lymphoma in pediatric posttransplant lymphoproliferative disorders. Pediatr Blood Cancer. 2017;64:1–3.
Goteri G, Costagliola A, Tassetti A, Stramazzotti D, Morroni M, Rupoli S, et al. Diffuse large B-cell lymphoma with Homer-Wright rosettes, sinusoidal growth pattern, and CD30 expression: a possible overlap between microvillous lymphomas and sinusoidal CD30-positive large B-cell lymphomas. Pathol Res Pract. 2009;205:279–82.
Wang E, Foo WC, Huang Q. A sinusoidal large cell lymphoma with expression of CD30, CD15, and multiple B-cell antigens: a classical Hodgkin lymphoma with sinusoidal infiltrating pattern or a sinusoidal CD30 positive large B-cell lymphoma with CD15? Leuk Lymphoma. 2010;51:1148–51.
Ashrafi F, Klein C, Poorpooneh M, Sherkat R, Khoshnevisan R. A case report of sinusoidal diffuse large B-cell lymphoma in a STK4 deficient patient. Medicine. 2020;99:e18601.
Yau D, Aron M, Siddiqi IN. Diffuse large B-cell lymphoma with striking intrasinusoidal pseudoglandular growth pattern as a diagnostic dilemma mimicking metastatic poorly differentiated pancreatic adenocarcinoma in an intraabdominal lymph node. Int J Surg Pathol. 2019;27:181–4.
Li X, Lu H, Yang J, Shi D, Zhu X, Xu Y, et al. A clinicopathologic study of CD30-positive sinusoidal large B-cell lymphoma. Zhonghua Bing Li Xue Za Zhi. 2002;31:305–8.
Swerdlow SH, Campo E, Pileri SA, Harris NL, Stein H, Siebert R, et al. The 2016 revision of the World Health Organization classification of lymphoid neoplasms. Blood. 2016;127:2375–90.
Arber DA, Orazi A, Hasserjian R, Thiele J, Borowitz MJ, Le Beau MM, et al. The 2016 revision to the World Health Organization classification of myeloid neoplasms and acute leukemia. Blood. 2016;127:2391–405.
Herbst RS, Baas P, Perez-Gracia JL, Felip E, Kim DW, Han JY, et al. Use of archival versus newly collected tumor samples for assessing PD-L1 expression and overall survival: an updated analysis of KEYNOTE-010 trial. Ann Oncol. 2019;30:281–9.
Hans CP, Weisenburger DD, Greiner TC, Gascoyne RD, Delabie J, Ott G, et al. Confirmation of the molecular classification of diffuse large B-cell lymphoma by immunohistochemistry using a tissue microarray. Blood. 2004;103:275–82.
Georgiou K, Chen L, Berglund M, Ren W, de Miranda NF, Lisboa S, et al. Genetic basis of PD-L1 overexpression in diffuse large B-cell lymphomas. Blood. 2016;127:3026–34.
Li M, Liu Y, Wang Y, Chen G, Chen Q, Xiao H, et al. Anaplastic variant of diffuse large B-cell lymphoma displays intricate genetic alterations and distinct biological features. Am J Surg Pathol. 2017;41:1322–32.
Hu S, Xu-Monette ZY, Balasubramanyam A, Manyam GC, Visco C, Tzankov A, et al. CD30 expression defines a novel subgroup of diffuse large B-cell lymphoma with favorable prognosis and distinct gene expression signature: a report from the International DLBCL Rituximab-CHOP Consortium Program Study. Blood. 2013;121:2715–24.
Slack GW, Steidl C, Sehn LH, Gascoyne RD. CD30 expression in de novo diffuse large B-cell lymphoma: a population-based study from British Columbia. Br J Haematol. 2014;167:608–17.
Jacobsen ED, Sharman JP, Oki Y, Advani RH, Winter JN, Bello CM, et al. Brentuximab vedotin demonstrates objective responses in a phase 2 study of relapsed/refractory DLBCL with variable CD30 expression. Blood. 2015;125:1394–402.
Sun C, Mezzadra R, Schumacher TN. Regulation and function of the PD-L1 checkpoint. Immunity. 2018;48:434–52.
Daassi D, Mahoney KM, Freeman GJ. The importance of exosomal PDL1 in tumour immune evasion. Nat Rev Immunol. 2020;20:209–15.
Mu CY, Huang JA, Chen Y, Chen C, Zhang XG. High expression of PD-L1 in lung cancer may contribute to poor prognosis and tumor cells immune escape through suppressing tumor infiltrating dendritic cells maturation. Med Oncol. 2011;28:682–88.
Clark CA, Gupta HB, Sareddy G, Pandeswara S, Lao S, Yuan B, et al. Tumor-intrinsic PD-L1 signals regulate cell growth, pathogenesis, and autophagy in ovarian cancer and melanoma. Cancer Res. 2016;76:6964–74.
Hino R, Kabashima K, Kato Y, Yagi H, Nakamura M, Honjo T, et al. Tumor cell expression of programmed cell death-1 ligand 1 is a prognostic factor for malignant melanoma. Cancer. 2010;116:1757–66.
Rossille D, Gressier M, Damotte D, Maucort-Boulch D, Pangault C, Semana G, et al. High level of soluble programmed cell death ligand 1 in blood impacts overall survival in aggressive diffuse large B-Cell lymphoma: results from a French multicenter clinical trial. Leukemia. 2014;28:2367–75.
Zhang J, Medeiros LJ, Young KH. Cancer immunotherapy in diffuse large B-cell lymphoma. Front Oncol. 2018;8:351.
Suzuki Y, Kohno K, Matsue K, Sakakibara A, Ishikawa E, Shimada S, et al. PD-L1 (SP142) expression in neoplastic cells predicts a poor prognosis for patients with intravascular large B-cell lymphoma treated with rituximab-based multi-agent chemotherapy. Cancer Med. 2020;9:4768–76.
Sakakibara A, Inagaki Y, Imaoka E, Sakai Y, Ito M, Ishikawa E, et al. Divergence and heterogeneity of neoplastic PD-L1 expression: two autopsy case reports of intravascular large B-cell lymphoma. Pathol Int. 2019;69:148–54.
Ponzoni M, Campo E, Nakamura S. Intravascular large B-cell lymphoma: a chameleon with multiple faces and many masks. Blood. 2018;132:1561–7.
Shimada K, Kinoshita T, Naoe T, Nakamura S. Presentation and management of intravascular large B-cell lymphoma. Lancet Oncol. 2009;10:895–902.
Sarkozy C, Copie-Bergman C, Damotte D, Ben-Neriah S, Burroni B, Cornillon J, et al. Gray-zone lymphoma between cHL and large B-cell lymphoma: a histopathologic series from the LYSA. Am J Surg Pathol. 2019;43:341–51.
Levine AJ, Vosburgh E. P53 mutations in lymphomas: position matters. Blood. 2008;112:2997–8.
Ngo VN, Young RM, Schmitz R, Jhavar S, Xiao W, Lim KH, et al. Oncogenically active MYD88 mutations in human lymphoma. Nature. 2011;470:115–9.
Schmitz R, Wright GW, Huang DW, Johnson CA, Phelan JD, Wang JQ, et al. Genetics and pathogenesis of diffuse large B-cell lymphoma. N. Engl J Med. 2018;378:1396–407.
Lim KH, Yang Y, Staudt LM. Pathogenetic importance and therapeutic implications of NF-kappaB in lymphoid malignancies. Immunol Rev. 2012;246:359–78.
Goy A, Ramchandren R, Ghosh N, Munoz J, Morgan DS, Dang NH, et al. Ibrutinib plus lenalidomide and rituximab has promising activity in relapsed/refractory non-germinal center B-cell-like DLBCL. Blood. 2019;134:1024–36.
Zhang J, Dominguez-Sola D, Hussein S, Lee JE, Holmes AB, Bansal M, et al. Disruption of KMT2D perturbs germinal center B cell development and promotes lymphomagenesis. Nat Med. 2015;21:1190–98.
Zhang J, Vlasevska S, Wells VA, Nataraj S, Holmes AB, Duval R, et al. The CREBBP acetyltransferase is a haploinsufficient tumor suppressor in B-cell lymphoma. Cancer Discov. 2017;7:322–37.
Kuo HP, Ezell SA, Hsieh S, Schweighofer KJ, Cheung LW, Wu S, et al. The role of PIM1 in the ibrutinib-resistant ABC subtype of diffuse large B-cell lymphoma. Am J Cancer Res. 2016;6:2489–501.
Merkel AL, Meggers E, Ocker M. PIM1 kinase as a target for cancer therapy. Expert Opin Investig Drugs. 2012;21:425–36.
Acknowledgements
We thank Dr. L Jeffrey Medeiros of Department of Hematopathology, The University of Texas MD Anderson Cancer Center, for editing the English text of a draft of this manuscript.
Funding
This work was supported by the National Science Foundation of China (grant numbers 81570180).
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Ethics approval and consent to participate
The study was conducted in accordance with institutional ethical regulations and was approved by the Xi Jing Hospital IRB, and informed consent was obtained from each patient involved in our research.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
About this article
Cite this article
Xu, J., Li, P., Chai, J. et al. The clinicopathological and molecular features of sinusoidal large B-cell lymphoma. Mod Pathol 34, 922–933 (2021). https://doi.org/10.1038/s41379-020-00685-7
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41379-020-00685-7