Abstract
In this paper we present a results of first comprehensive study of the complete mitogenomes in the Buryats with regard to their belonging to the main regional (eastern and western Buryats); tribal (Khori, Ekhirid, Bulagad, and Khongodor), and ethno-territorial (Aginsk, Alar, Balagansk, Barguzin, Ida, Khorinsk, Kuda, Selenga, Verkholensk, Olkhon, Tunka, and Shenehen Buryats) groups. The analysis of molecular variation performed using regional, tribal, and ethno-territorial divisions of the Buryats showed lack of genetic differentiation at all levels. Nonetheless, the complete mitogenome analysis revealed a very high level of genetic diversity in the Buryats which is the highest among Siberian populations and comparable to that in populations of eastern and western Asia. The AMOVA and MDS analyses results imply to a strong genetic similarity between the Buryats and eastern Asian populations of Chinese and Japanese, suggesting their origin on the basis of common maternal ancestry components. Several new Buryat-specific branches of haplogroup G (G2a2a, G2a1i, G2a5a) display signals of dispersals dating to 2.6–6.6 kya with a possible origin in eastern Asia, thus testifying Bronze Age and Neolithic arrival of ancestral eastern Asian component to the South Siberia region.
Similar content being viewed by others
Introduction
The Buryats, numbering about 461,000, are the second largest indigenous group in Siberia, concentrated mostly in their homeland, the Buryat Republic, located in the south-central part of Siberia along the eastern shore of Lake Baikal. They also live in Zabaikal and Irkutsk Regions of Russia, Mongolia (in the northern part of Hentei Aimak) and China (Hulunbuir Aimak of Inner Mongolia Autonomous Region). The Buryats living west of Lake Baikal are often referred to as western (Irkutsk or Cis-Baikal) Buryats and living to the east—as eastern (Transbaikal) Buryats. The modern Buryats are represented by four major tribes (Khori, Ekhirid, Bulagad, and Khongodor) and a variety of other tribes, coming from Mongolia and Dzungaria [1]. The Buryats are often subdivided by territorial principle into Aginsk, Alar, Balagansk, Barguzin, Ida, Khorinsk, Kuda, Selenga, Verkholensk, Olkhon, Tunka, and Shenehen Buryats. The Buryats are typical representatives of the Central Asian anthropological type of Mongoloids, though the Buryats of northern regions of Buryat Republic demonstrate a notable admixture of Baikal anthropological type [2, 3]. The Buryat language belongs to the northern subgroup of the Mongolian group of the Altai language family and divides into a number of dialects.
According to archeological and ethnological data, the formation of the Buryats took place in the Cis-Baikal region—in addition to indigenous Buryat-Mongolian (Bulagad, Khori, Ekhirit, Khongodor) tribes, the Buryats assimilated also some other population groups, including Mongolic Barghuts, Oirots, Songols, Sartuls, Khatagins and others, as well as Tungusic and Turkic ones, inhabited the territories of modern Buryat–Mongolia [4, 5]. It is considered that some proto-Buryat tribes were already formed in the Neolithic and the Bronze Age (2500–1300 years BC). At that time, the tribes of pastoralists-farmers coexisted with the tribes of hunters. In the late Bronze Age, the territories of Central Asia, including the Baikal region, were occupied by the proto-Turkic and proto-Mongolic tribes of so-called “slabs”.
Starting with the third century BC the population of the Cis-Baikal region and Transbaikalia became part of the Central Asian Empires, i.e., the Xiongnu, Xianbei, Rouran, and others [4]. From the seventh to ninth centuries AD the Cis-Baikal and Transbaikal regions were inhabited by Kurykans and Bayegu (Bayyrku) tribes, which were part of tribal confederation under the Uyghur nobility, who created Uyghur Khaganate. It is believed that from ninth to tenth centuries AD the western tribes ancestral to the Buryats were members of Uch-Kurykan tribal Union, whereas the eastern tribes were part of powerful Khori-Tumat Union. Then, in the12th–13th centuries AD under the pressure of the Mongolian tribes most of Kurykans have moved towards to the middle reaches of the Lena River, whereas some of them, together with Bayegu, were assimilated by newly arrived people.
Since the end of the 17th century the tribal groups of Bulagads, Ekhirits, and Khongodors are known in the Cis-Baikal region and Khori in the Transbaikal region. During this period, Transbaikal was part of northern Mongolia, and Cis-Baikal region was dependent on western Mongolia and numerous nomadic groups of Mongols roamed from Mongolia to Transbaikal region and back. Notably, that administrative-territorial disunity and the lack of economic, cultural and religious identity were the main obstacle to the Buryat tribe consolidation, which was completed only by the end of the 19th century.
In light of complex population history of the Buryats and important role the Baikal region has played in the peopling of Siberia, the analysis of genetic variation in the Buryats has been conducted in several studies using different sets of allozyme, mitochondrial DNA (mtDNA), Y-chromosome and genome-wide markers [6,7,8,9,10,11,12,13,14,15,16,17,18,19,20,21].
The mtDNA variability in the Buryats (among other Siberian populations) has been studied mainly to investigate the human colonization process of northern Asia and dispersals to the Americas [7, 9, 15] or particularly to understand the northward expansion of the Yakuts [10, 12]. All of these studies mainly utilized the phylogeographic method, using pooled samples to reveal a maximum of mtDNA diversity. The only local Baikal region population originated from the Barguzin Valley has been studied in detail to elucidate the micro-evolutionary history of the Buryats [16]. However, all of those studies are hindered by using the insufficient set of mtDNA markers (control-region sequence data combined with RFLP analysis of coding regions) that significantly restricted the correct definition of phylogeographic patterns and performing of accurate molecular dating.
Recently, the significant importance of complete mtDNA sequencing has been emphasized, as far as the comprehensive genealogical resolution of complete mitogenomes along with adequate sampling can provide a detailed reconstruction of genetic history both for populations in general and specific lineages in particular [15, 22,23,24,25,26,27,28,29,30]. Although data on the variability of complete mitogenomes from several Siberian populations has been published recently [27], the Buryat mtDNAs were characterized only for some haplogroups [15, 23, 24, 28]. Thus, no extensive population studies were performed comparing complete mtDNAs from different tribal, ethno-territorial or regional groups of the Buryats so far.
In this study, we present for the first time an extensive data set of complete mitogenomes from 172 Buryat individuals (including 92 new sequences), covering the main regional (eastern and western Buryats), tribal (Khori, Ekhirid, Bulagad, and Khongodor), ethno-territorial (Aginsk, Alar, Balagansk, Barguzin, Ida, Khorinsk, Kuda, Selenga, Verkholensk, Olkhon, Tunka, and Shenehen Buryats) groups and administrative territories (Buryat Republic, Zabaikal and Irkutsk Regions of Russia; Hulunbuir Aimak of Inner Mongolia Autonomous Region of China), inhabited by modern Buryats. The aim of our study is to characterize the genetic structure, differentiation and diversity of the Buryats as well as to provide insights into South Siberia genetic history.
Materials and methods
Sample collection and mtDNA sequencing
Blood samples were collected from 92 unrelated Buryat individuals, coming from different villages of Irkutsk Region, the Buryat Republic, and Zabaikal Region of Russia, as well as from Hulunbuir Aimak of Inner Mongolia Autonomous Region of China, thus encompassing all territories inhabited by modern Buryats, and from five Koryaks residing in Severo-Evensk district of Magadan Region of Russia (Supplementary Fig. S1). Only a few samples were chosen from each locality, limiting the probability of sampling relatives. All of these individuals were paternally and maternally unrelated and originated from the area considered for this study. All subjects provided written informed consent for the sample collection and subsequent analysis. Detailed information on the geographical origin, tribe, and ethno-territorial affiliation of the samples can be found in Supplementary Table S1.
Genomic DNA was extracted using a standard phenol/chloroform procedure. The complete mtDNA sequencing was performed, as previously described [31] using an ABI 3500xL Genetic Analyzer. Complete mtDNA sequences were analyzed using SeqScape 2.7 (Applied Biosystems) and compared with the revised Cambridge reference sequence [32].
The study was approved by the Ethics Committee of the Institute of Biological Problems of the North, Russian Academy of Sciences, Magadan, Russia (statement no. 003/013 from 24 January, 2013).
Statistical analysis and molecular dating
The mtDNA phylogenies were reconstructed based on the data obtained in this study and published earlier [15, 23, 24, 27, 28, 33, 34], as well as on all available data at PhyloTree, Build 17 [35]. The most-parsimonious trees of the complete mtDNA sequences were reconstructed using the Network 4.5.1.0 [36] and mtPhyl software (http://eltsov.org). The haplogroup assignment for each sample was established using mtPhyl program in accordance with the nomenclature that suggested by van Oven and Kayser [35] and Derenko et al. [28]. The coalescence dates were estimated using rho statistics and their error ranges [37]. The mutation rates based on mtDNA complete genome data (one mutation every 3624 years [38] and one mutation every 2202 years [39]) and coding region substitutions (one mutation every 4610 years) [40] were used. Nucleotide positions (nps) with point indels and/or transversions located between nps 16180–16193, 303–315, 522–524, 573–576, as well as unstable position 16519 were excluded from the analysis. Concerns about the accuracy of coalescence dates estimations using rho statistics were raised earlier [41], but recent studies based on complete mtDNA variability data show high similarity between rho statistics and maximum likelihood results [29, 42, 43].
The Bayesian skyline plots (BSPs) [44] were obtained for the Buryat complete mtDNAs using BEAST 1.7.5 with a strict molecular clock and the HKY+I+G model of nucleotide substitutions [45, 46]. This model was selected as the best-fit model of nucleotide substitution by the Bayesian Information Criterion scores using MEGA 5.05 [47]. The estimated value of the shape parameter of the gamma distribution was 0.05. The strict molecular clock was chosen due to preliminary analysis performed with an uncorrelated lognormal relaxed clock indicated that this model cannot be rejected because the frequency histogram of the coefficient of variation is abutting against zero [48]. Each MCMC sample was based on a run of 60,000,000 generations sampled every 6000 steps, with the first 6,000,000 generations regarded as burn-in. Three independent runs were carried out with the mutation rates from Soares et al. [38] and Posth et al. [39] (1.665 × 10−8 and 2.74 × 10−8, respectively). The BSPs were generated for the entire Buryat population, for eastern and western Buryats separately, as well as for some subclades of mtDNA tree, and the plots were visualized with Tracer v1.4.
The basic parameters of genetic diversity were calculated using DnaSP 5.10.01 [49]. The analysis of molecular variance (AMOVA) was performed by means of Arlequin 3.5.1.2 software [50]. The statistical significance of Fst values was estimated by 10,000 permutations. For multi-dimensional scaling (MDS) 51] the STATISTICA 10 (StatSoft Inc., Tulsa, OK, USA) was used. Published population data on complete mitogenome variation in Chinese and Japanese [52], Evens, Koryaks, Yakuts, Udegey, Nivkhs, Evenks, and Yukaghirs [27] were selected for comparisons. An exact test for population differentiation has been used for estimation of between-population differences based on mtDNA haplogroup frequencies [53].
The GenBank accession numbers for the 97 novel complete mtDNA sequences (92 Buryat mtDNAs and 5 haplogroup G Koryak mtDNAs) reported in this paper are MF043424-MF043520. The assignment of samples to their GenBank accession numbers is shown in Supplementary Table S1.
Results
Phylogeny of the Buryat complete mitogenome tree and haplogroup distribution
To elucidate the origin, genetic structure and diversity of the Buryats we have analyzed 172 complete mtDNA genomes, 92 of which are new. The haplogroup assignment for each sample based on the nomenclature of Phylotree.org (Build 17) [35] and Derenko et al. [28] data is shown in Supplementary Table S1; the phylogenetic relationships of mtDNA lineages found in the Buryats are depicted in detail in Supplementary Fig. S2.
A total of 107 different subhaplogroups or paragroups (unclassified lineages within a clade) were found, falling into 53 main haplogroups belonging to macrohaplogroups M, N, and R. Fifteen subhaplogroups were described here for the first time (highlighted red in Supplementary Table S1 and Supplementary Fig. S2), whereas for two previously known subhaplogroups (D4c2a and Z1a1b) we redefined diagnostic markers that allow the better resolution of the haplogroup structure. Haplogroup frequencies for eastern, western Buryats and the entire Buryat mtDNA data are given in Table 1. As can be seen, two regional groups of the Buryats are characterized by the same most typical eastern Eurasian mtDNA haplogroups (A, B, CZ, D, F, G, M7, and N9) with the similar frequency distribution of the majority of their subhaplogroups. Both regional groups showed the low frequencies of haplogroups A (3.7% and 5.6%, respectively), B (2.4% and 3.3%, respectively), F (2.4% and 4.4%, respectively), M7 (3.7% and 1.1%, respectively), and Y (1.2% and 2.2%, respectively) and pronounced frequencies of haplogroups C (15.9% and 18.9%, respectively), D (39% and 28.9%, respectively), and G (8.5% and 16.7%, respectively). Haplogroups N9 and Z were detected only in western Buryats at low frequencies (1.1% and 2.2%, respectively). Both eastern and western Buryats demonstrated a diverse collection of the western Eurasian mtDNA lineages that belong to haplogroups H, HV, U, and X, accounting together for 23.2% and 15.6% of mtDNAs, respectively (Table 1).
Genetic differentiation
Between-population comparisons for mtDNA haplogroup and subhaplogroup distributions show a lack of statistically significant differences between two regional groups of Buryats (an exact test for population differentiation, p=0.48) thus testifying to a considerable genetic homogeneity of the Buryats. Fst values estimated based on frequencies of either the twelve main (A, B, CZ, D, F, G, M7, N9, H, HV, U, and X2e) or all 59 mtDNA haplogroups (Table 1) also showed the absence of differentiation between eastern and western Buryats (Fst = 0.0026, p=0.28 and Fst = −0.00073, p=0.56, respectively).
Population pairwise Fst values calculated from complete mtDNA sequences among different tribes and ethno-territorial groups of the Buryats are summarized in Supplementary Table S2. The regional, tribal, and ethno-territorial affiliations of Buryat samples analyzed are as shown in Supplementary Table S1. The analysis of molecular variation performed using two regional (eastern and western), three (Khori, Ekhirid/Bulagad, and Khongodor) or four tribal (Khori, Ekhirid, Bulagad, and Khongodor) and eleven ethno-territorial (Shenen, Aginsk, Khorinsk, Barguzin, Ida, Olkhon, Kuda, Selenga, Balagansk, Tunka, and Alar) groups of the Buryats showed a lack of genetic differentiation at all levels as far as all pairs of the compared groups exhibited insignificant genetic distances.
Genetic diversity, between-population relationships and demographic analysis
Summary statistics depicting the parameters of genetic diversity in the Buryats compared with other Siberian and Asian ethnic groups are presented in Table 2. In general, the genetic diversity in the Buryats is very high, with 141 haplotypes found among 172 individuals. A total of 639 variable sites were found in the Buryats, and the number of mean pairwise differences was 33.21. As can be seen, the Buryats (along with the Yakuts) exhibit the similar values of haplotype and nucleotide diversity which are the highest among Siberian populations studied. Both populations demonstrate an excess of low-frequency variants that (together with significantly negative values for Tajima’s D) points to a recent population expansion. For other Siberian populations (Evenks, Evens, Koryaks, Nivkhs, Udegeys, and Yukaghirs) neutrality test gave non-significant values, thus rejecting the population growth hypothesis and implying to the marked effects of drift and/or the small population sample sizes. In general, the level of genetic variation in the Buryats exceeds that in the majority of Siberian populations investigated but is comparable to that in eastern and western Asian populations.
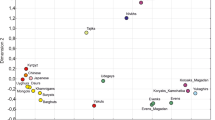
To visualize the relationships between the Buryats and other Siberian and eastern Asian populations, MDS plot was constructed based on the pairwise Fst values (Table 3). The pairwise Fst comparisons show that eastern and western Buryats were indistinguishable from each other, but both of them differ significantly from other Siberian and eastern Asian populations. For all other pairs of populations the pairwise Fst values were statistically significant, with the exceptions of the Yukaghirs, who were closely associated with the Evens and Evenks.
The results of our MDS analysis show that eastern and western Buryats cluster close to each other and to Chinese and Japanese from eastern Asia, whereas other Siberian populations are positioned rather distantly from them and scattered on the plot (Fig. 1). Thus, the Evenks and Evens are clustered together with the Yukaghirs, and all of them are clearly separated from Koryaks and Yakuts, who occupies an intermediate position on the plot. Meanwhile, the Udegeys are located in close proximity to the Koryaks/Yakuts grouping, whereas the Nivkhs displays the outlying position on the plot. Generally, the results of our MDS analysis do not cluster the populations investigated according to their linguistic affinity and/or geographic proximity. Thus, despite the close genetic similarity of the Tungusic-speaking Evenks and Evens, both of them located quite distantly from the Udegeys, who also speak Tungusic language. On the other hand, the Koryaks and Evens from northeastern Asia are clearly separated from each other on the plot, even though association between them has been revealed earlier based on control region mtDNA variability data [24] Similarly, the Udegeys and Nivkhs from the Lower Amur region of Siberia are located quite far from each other, although close genetic affinity between them and other ethnic groups from the Lower Amur region, Sakhalin and Hokkaido Islands has been postulated previously [25].
The BSP obtained for the Buryats using the mutation rate of Soares et al. [38] indicates a continuous increase of population size with a population expansion around 18–20 kya followed by a small but noticeable both in the total sample of Buryats and in western Buryats decrease around ~5 kya (Fig. 2). As could be expected the use of faster rate proposed by Posth et al. [39] leads to younger time estimates of population expansion and decline which could have occurred around 11–12 kya and around 3 kya, respectively (Supplementary Fig. S3). In general, these patterns coincide with those of the Evenks and Evens demonstrating a prominent decline in population size around 1–2 kya, which is not followed by an increase as well [27]. In contrast, other Siberian populations show some other different patterns. So, Yakuts demonstrate a sharp increase in population size around 1 kya followed the decline 3–5 kya, whereas Koryaks, Nivkh, Yukaghirs, and Udegeys are characterized by relatively flat curves with a relatively smooth decline close to the present [27].
BSP indicating the median of the hypothetical effective population size through time generated based on complete mtDNA genome data for the Buryats and the mutation rate of 1.665 × 10−8 substitutions per nucleotide per year [38]. Maximum time (x axis) corresponds to the median posterior estimate of the genealogy root-height
New insights from complete mtDNA genomes
The phylogeny of haplogroup G
Our analyses at the maximal resolution considerably contributed to refine the phylogeny of haplogroup G, the third most common haplogroups in the Buryats. Despite of the relatively low frequencies over most of its range, haplogroup G shows an extremely wide geographic distribution being found in all northern Asian regional groups, eastern and central Asia, as well as in some populations of western, southern and mainland southeastern Asia, the Volga-Ural region, the Caucasus and Europe. Haplogroup G is detected at 8.1% overall in northern Asia, though it reaches 18.6% in northeastern Asian regional group where its highest frequencies being detected in Itelmens (69.6%), Koryaks (41.6%), Chuvantsi (28.1%), and Chukchi (22.6%). This haplogroup is found at relatively high frequencies in the Negidals of Siberian Far East (27.3%) and Todjins of the East Sayan region of south Siberia (18.8%). Haplogroup G is characteristic of all Mongolic-speaking populations, being detected with similar frequencies of 8–12% in Buryats, Mongolians, Kalmyks, Khamnigans and Barghuts (Supplementary Table S3). The frequency of this haplogroup retains as high as 7–11% in the majority of eastern Asian populations and declines to 1–8% in central Asia, and to 0.4–2% in western and southern Asia. Noteworthy, haplogroup G presents as rarely as 1–5.7% in the Volga-Ural region and the Caucasus, and it occurs with a very low frequency (0.2–0.4%) in some populations of Europe (Supplementary Table S3).
In this paper we present the reconstructed phylogeny of haplogroup G based on 235 complete mitogenomes, twenty four of which are new, with the age estimates for the main nodes. The most parsimonious tree confirmed subclades G1-G4 and added considerable internal variation to G1b and G2a2′3′4′5′6′7 with one and five additional branches, respectively (Supplementary Fig. S4). As shown, haplogroup G has a likely pre-LGM/LGM time depth with an overall coalescence time estimated as 21–37 kya. Two other major subclades, G1, and G3 have a relatively similar age estimates of 14–23 kya, whereas G2, the most represented of G clades, has an older coalescence time calculated as 19–32 kya. The forth main branch of the tree, G4 has a youngest coalescence time estimates of 7–15 kya (Fig. 3; Supplementary Fig. S4).
Complete mtDNA phylogenetic tree of haplogroup G. This schematic tree is based on phylogenetic tree presented in Supplementary Fig. S4. Time estimates (in kya) shown for mtDNA subclusters are based on complete mtDNA genome [38, 39] and the coding region [40] clock. The size of each circle is proportional to the number of individuals sharing the corresponding haplotype, with the smallest size corresponding to one individual
While all haplogroup G subclades are detected in eastern Asia, so that eastern Asian mitogenomes occur throughout the tree, some of them are found to be characteristic for northern Asian populations, i.e., G2a1i, G2a2b, G2a2c, G2a5a, G2a6, G2a7, and G1b. For example, Khamnigan, Barghut, and two Buryats samples (10_Khm, 114_Bt, 53_IM, 542_Br) bearing variants 1808, 8766 and 12825 as well as a series of private mutations likely belong to a novel branch of haplogroup G2a1–G2a1i. Ten of the new Buryat sequences, as well as seven previously published sequences (five Yakuts from central Siberia, one Pole and one individual of unknown origin) clustered into common G2a2-branch with at least two novel Buryat-specific subclades – G2a2b and G2a2c (Supplementary Fig. S4). Inside haplogroup G2a5 one more subclade, G2a5a, specific for the Buryats has been found. It is characterized by back mutation at np 16227 which seems to be diagnostic for this subclade. Two other Buryat samples (609_Br and 544_Br) formed a novel branch with a defining motif 151-237-1189-6899insC-9932-10804-12372-16129-16274 and back mutation at np 16227 that we called “G2a6”. One new Buryat mitogenome and one previously published Chinese sequence shared 3351-4721-7604 motif and indicated additional branch on the same phylogenetic level – G2a7. It should be noted that the estimates of coalescence time for the Buryat-specific G-lineages do not exceed 7 kya, ranging from 0.4 to 1.5 kya for G2a2c, from 3.5 to 6.6 kya for G2a1i, from 3.6 to 6.2 kya for G2a2a, and from 2.6 to 4.3 kya for G2a5 (using different mutation rates; Supplementary Fig. S4).
The addition of five complete mitogenomes from the Koryaks has allowed us to update the phylogeny of northeastern Asian-specific subgroup G1b (Fig. 3; Supplementary Fig. S4). The complete mtDNA based phylogeny testifies that G1b can be additionally divided into G1b1, found predominantly in the Koryaks and Evens; G1b2, largely restricted to the Chukchi and Koryaks; G1b3 and G1b4 detected in the Evens/Chukchi and Yukaghirs/Evens, respectively. As can be seen, a few mtDNA genomes of the Nivkhs do not form separate branch(es) within G1b but on the contrary scattered through this subclade. By the way, the relatively large amount of internal G1b diversity can testify that this haplogroup arose directly in northeastern Asia or more specifically in the Sea of Okhotsk region as it has been supposed earlier [54] or in the Far East region of northern Asia. But to prove the latter more complete mtDNA genome data from populations of Ulchi, Nivkhs, Negidals, Oroks, and Nanai should be generated. Meanwhile, the phylogeny presented in Supplementary Fig. S4 gives an information about the evolutionary age of the G1b node estimated as 7–12 kya (with different mutation rates), thus attesting to the Holocene origin of this subcluster. The BSP constructed from the overall G1b mitogenome data indicates that this haplogroup remained constant throughout the past 5–8 kya (Supplementary Fig. S5), thus pointing to the lack of recent demographic growth events (dated at 1–2 kya) which are shown to be characteristic for other northeastern Asian-specific mtDNA lineages such as A2a, A2b, and D2a [29].
Discussion
In this study we present the results of first comprehensive study of the complete mitogenomes in the Buryats with regard to their belonging to the main regional (eastern and western Buryats), tribal (Khori, Ekhirid, Bulagad, and Khongodor), ethno-territorial (Aginsk, Alar, Balagansk, Barguzin, Ida, Khorinsk, Kuda, Selenga, Verkholensk, Olkhon, Tunka, and Shenehen Buryats) groups and administrative territories (Buryat Republic, Zabaikal and Irkutsk Regions of Russia; Hulunbuir Aimak of Inner Mongolia Autonomous Region of China). Overall, the gene pool of Buryats consists of diverse set of 107 mitochondrial lineages which fall into 41 eastern Eurasian and 18 western Eurasian subhaplogroups. Eastern Eurasian component, composed of principal haplogroups A, B, CZ, D, F, G, M7, and N9, accounts for 80.8% of all samples, whereas contribution from western Eurasian haplogroups (H, HV, U, and X) is estimated as 19.2%. The western Eurasian influence is slightly more pronounced in eastern Buryats (23.2%) than in western Buryats (15.6%). Despite some regional differences in haplogroup and subhaplogroup frequencies exist, all of them are statistically insignificant pointing to the lack of genetic differences between eastern and western Buryats.
The subsequent analysis of molecular variation performed using regional, tribal and ethno-territorial divisions of the Buryats showed lack of genetic differentiation at all levels as well. This seems to be the principal characteristic of the Buryats regarding complete mtDNA variability data. Such situation was not observed when studying Y-chromosome diversity in the Buryats. On the contrary, the significant level of differentiation was registered with the partition of the Buryats studied into three regional groups (western, eastern and southern) as well as into eight territorial groups in accordance with the geographical place of origin [19]. This can be explained by a lower male vs. female effective population sizes as it was done by comparisons of mtDNA and Y-chromosome variability in regional populations [55] or by variability of the autosomes and X chromosome [56]. However, other studies explain this discrepancy by a higher female migration rate (via patrilocality, the trend for a wife to move into her husband’s natal household) [57]. Particularly, specific cultural behaviors (especially patrilocality and patrilinearity) are assumed to have a significant impact on the sex-specific genetic structure of populations of Central and Northern Asia [58].
In general, the complete mitogenome analysis revealed a very high level of genetic diversity in the Buryats which is the highest among Siberian populations and equivalent to that in populations of eastern and western Asia. The AMOVA and MDS analyses results did not connect any regional or linguistic group of populations in the Siberia implying to a strong genetic similarity of the Buryats and the eastern Asian populations of Chinese and Japanese, thus suggesting their origin on the basis of common maternal ancestry components. The pronounced impact of eastern Asian component into gene pool of the Buryats is also evident from the genome-wide SNP and whole genome sequence data analyses [18, 21, 59]. Thus, the genome-wide ADMIXTURE data points to a high fraction of eastern Asian ancestry component in the Buryats (47%), which may have appeared in their gene pool through at least two admixture events: an older admixture event of around 2.7 kya coincided with the Slab Grave Culture emergence in Mongolia which is associated with the ancestors of the Xiongnu [21] and a more recent admixture event of around 800–600 ya, consistent with the expansion of Mongols led by Genghis Khan [18, 21]. However, our mtDNA results suggest that the process is likely to have been more complex. Several branches of haplogroups G (G2a2a, G2a1i, G2a5a) and D (D4b1a2a1b, D4c2a, D4c2b-6914, D4j8) [23], which were found to be the Buryat-specific, display signals of dispersals dating to 2.6–6.6 kya and 2.2–7.5 kya, respectively, with a possible origin in eastern Asia, thus testifying earlier (Bronze Age and Neolithic) arrival of ancestral eastern Asian component to South Siberia region.
Notably, the results of our MDS analysis testify a close genetic affinity of the Buryats to the eastern Asian populations of Chinese and Japanese than to those from Siberia. This is consistent with the results of our earlier study showing a striking resemblance of all Mongolic-speaking populations studied (Mongolians, Buryats, Khamnigans, Kalmyks, and Barghuts) and implying to their origin on a common maternal ancestral basis [24]. In contrast, genome-wide associated study does not group the Mongolic-speaking Buryats with Mongolians, who, in turns, fall close to Han Chinese and Japanese, but shows closer affinities of the Buryats to the Turkic-speaking Tuvinians and Altaians [21]. Moreover, it has been shown recently that Turkic- and Mongolic-speaking populations from southern Siberia and Mongolia have an extraordinary large number of a long chromosomal tracts which are identical by descent with Turkic peoples from all over western Eurasia. The data obtained suggest that the only Tuvinians, Buryats and Mongols from southern Siberia and Mongolia are autochthonous to their current residence and thus these territories may be considered as a source of recent large-scale nomadic migrations [60]. The another opposite tendency has recently been found in Y-STR diversity study of different ethno-linguistic groups, including the Mongolic-speaking populations from northeastern China, Mongolia, Siberia, and eastern Europe. The results revealed that the Mongolic-speaking populations are clustered into two pronounced groups—the Buryats, Khamnigans, and Barghuts are grouped together (though located quite distantly from all other Siberian populations), but not with the Mongols and Kalmyks, who are closely related to each other and to the Altaian Kazakhs and Chinese Manchu [20]. On the contrary, the analysis of Y-chromosome haplogroup frequencies points to the high genetic differences between the Buryats and other south Siberian populations which are much higher than differences between the Buryats and Mongolians [19].
Summing up, it remains difficult to make inferences regarding the identification of specific mtDNA lineages in the Buryat mitochondrial gene pool and correlation them with geography without adding the representative complete mitogenome data set from Mongolia and neighboring regions. Moreover, to provide a comprehensive mtDNA portrait of northern Asia in a whole, including an assessment of genetic contribution from other regions (mainly eastern Asia), and to evaluate the impact of migrations during the Late Glacial and subsequent periods in shaping diversity and genetic structure in northern Asia, the complete mitogenome data on ancient specimens (still absent for northern and eastern Asia) will be extremely helpful.
References
Bowles GT. The People of Asia. London: Weidenfeld & Nicolson; 1977.
Zolotareva IM. Ethnic anthropology of Buryats. Ethnographic Collection. 1960;1:11–30.
Alexseev VP, Gokhman II. Anthropology of Soviet Asia. Moscow: Nauka; 1984. (in Russian)
Levin MG, Potapov LP. The Peoples of Siberia. Chicago: University of Chicago Press; 1964. (in Russian)
Nimaev DD. The Problems of Buryat Ethnogenesys. Novosibirsk: Nauka; 1988. (in Russian)
Novoradovsky AG, Spitsyn VA, Duggirala R, Crawford MH. Population genetics and structure of Buryats from the Lake Baikal Region of Siberia. Hum Biol. 1993;65:689–710.
Derenko MV, Malyarchuk BA, Dambueva IK, Shaikhaev GO, Dorzhu CM, Nimaev DD, et al. Mitochondrial DNA variation in two South Siberian Aboriginal populations: implications for the genetic history of North Asia. Hum Biol. 2000;72:945–73.
Karafet TM, Osipova LP, Gubina MA, Posukh OL, Zegura SL, Hammer MF. High levels of Y-chromosome differentiation among native Siberian populations and the genetic signature of a boreal hunter-gatherer way of life. Hum Biol. 2002;74:761–89.
Derenko MV, Grzybowski T, Malyarchuk BA, Dambueva IK, Denisova GA, Czarny J, et al. Diversity of mitochondrial DNA lineages in South Siberia. Ann Hum Genet. 2003;67:391–411.
Pakendorf B, Wiebe V, Tarskaia LA, Spitsyn VA, Soodyall H, Rodewald A, et al. Mitochondrial DNA evidence for admixed origins of central Siberian populations. Am J Phys Anthropol. 2003;120:211–24.
Derenko M, Malyarchuk B, Denisova GA, Wozniak M, Dambueva I, Dorzhu C, et al. Contrasting patterns of Y-chromosome variation in South Siberian populations from Baikal and Altai-Sayan regions. Hum Genet. 2006;118:591–604.
Pakendorf B, Novgorodov IN, Osakovskij VL, Danilova AP, Protod’jakonov AP, Stoneking M. Investigating the effects of prehistoric migrations in Siberia: genetic variation and the origins of Yakut. Hum Genet. 2006;120:334–53.
Woźniak M, Derenko M, Malyarchuk B, Dambueva I, Grzybowski T, Miścicka-Sliwka D. Allelic and haplotypic frequencies at 11 Y-STR loci in Buryats from South-East Siberia. Forensic Sci Int. 2006;164:271–5.
Derenko M, Malyarchuk B, Denisova G, Wozniak M, Grzybowski T, Dambueva I, et al. Y-chromosome haplogroup N dispersals from south Siberia to Europe. J Hum Genet. 2007;52:763–70.
Derenko M, Malyarchuk B, Grzybowski T, Denisova G, Dambueva I, Perkova M, et al. Phylogeographic analysis of mitochondrial DNA in northern Asian populations. Am J Hum Genet. 2007;81:1025–41.
Gibert M, Theves C, Ricaut FX, Dambueva I, Bazarov B, Moral P, et al. mtDNA variation in the Buryat population of the Barguzin Valley: New insights into the micro-evolutionary history of the Baikal area. Ann Hum Biol. 2010;37:501–23.
Malyarchuk B, Derenko M, Denisova G, Wozniak M, Grzybowski T, Dambueva I, et al. Phylogeography of the Y-chromosome haplogroup C in northern Eurasia. Ann Hum Genet. 2010;74:539–46.
Cardona A, Pagani L, Antao T, Lawson DJ, Eichstaedt CA, Yngvadottir B, et al. Genome-wide analysis of cold adaptation in indigenous Siberian populations. PLoS ONE. 2014;9:e98076. https://doi.org/10.1371/journal.pone.0098076
Kharkov VN, Khamina KV, Medvedeva OF, Simonova KV, Eremina ER, Stepanov VA. Gene pool of Buryats: Clinal variability and territorial subdivision based on data of Y-chromosome markers. Russ J Genet. 2014;50:180–90.
Malyarchuk BA, Derenko M, Denisova G, Woźniak M, Rogalla U, Dambueva I, et al. Y chromosome haplotype diversity in Mongolic-speaking populations and gene conversion at the duplicated STR DYS385a,b in haplogroup C3-M407. J Hum Genet. 2016;61:491–6.
Pugach I, Matveev R, Spitsyn V, Makarov S, Novgorodov I, Osakovsky V, et al. The complex admixture history and recent southern origins of Siberian populations. Mol Biol Evol. 2016;33:1777–95.
Volodko NV, Starikovskaya EB, Mazunin IO, Eltsov NP, Naidenko PV, Wallace DC, et al. Mitochondrial genome diversity in arctic Siberians, with particular reference to the evolutionary history of Beringia and Pleistocenic peopling of the Americas. Am J Hum Genet. 2008;82:1084–100.
Derenko M, Malyarchuk B, Grzybowski T, Denisova G, Rogalla U, Perkova M, et al. Origin and post-glacial dispersal of mitochondrial DNA haplogroups C and D in northern Asia. PLoS ONE. 2010;5:e15214. https://doi.org/10.1371/journal.pone.0015214
Derenko M, Malyarchuk B, Denisova G, Perkova M, Rogalla U, Grzybowski T, et al. Complete mitochondrial DNA analysis of eastern Eurasian haplogroups rarely found in populations of northern Asia and eastern Europe. PLoS ONE. 2012;7:e32179. https://doi.org/10.1371/journal.pone.0032179
Sukernik RI, Volodko NV, Mazunin IO, Eltsov NP, Dryomov SV, Starikovskaya EB. Mitochondrial genome diversity in the Tubalar, Even, and Ulchi: contribution to prehistory of native Siberians and their affinities to Native Americans. Am J Phys Anthropol. 2012;148:123–38.
Derenko M, Malyarchuk B, Bahmanimehr A, Denisova G, Perkova M, Farjadian S, et al. Complete mitochondrial DNA diversity in Iranians. PLoS ONE. 2013;8:e80673. https://doi.org/10.1371/journal.pone.0080673
Duggan AT, Whitten M, Wiebe V, Crawford M, Butthof A, Spitsyn V, et al. Investigating the prehistory of Tungusic peoples of Siberia and the Amur-Ussuri region with complete mtDNA genome sequences and Y-chromosomal markers. PLoS ONE. 2013;8:e83570. https://doi.org/10.1371/journal.pone.0083570
Derenko M, Malyarchuk B, Denisova G, Perkova M, Litvinov A, Grzybowski T, et al. Western Eurasian ancestry in modern Siberians based on mitogenomic data. BMC Evol Biol. 2014;14:217. https://doi.org/10.1186/s12862-014-0217-9
Dryomov SV, Nazhmidenova AM, Shalaurova SA, Morozov IV, Tabarev AV, Starikovskaya EB, et al. Mitochondrial genome diversity at the bering strait area highlights prehistoric human migrations from Siberia to northern North America. Eur J Hum Genet. 2015;23:1399–404.
Kivisild T, Maternal ancestry and population history from whole mitochondrial genomes. Investig Genet. 2015;6:3. https://doi.org/10.1186/s13323-015-0022-2
Torroni A, Rengo C, Guida V, Cruciani F, Sellitto D, Coppa A, et al. Do the four clades of the mtDNA haplogroup L2 evolve at different rates? Am J Hum Genet. 2001;69:1348–56.
Andrews RM, Kubacka I, Chinnery PF, Lightowlers RN, Turnbull DM, Howell N. Reanalysis and revision of the Cambridge reference sequence for human mitochondrial DNA. Nat Genet. 1999;23:147.
Malyarchuk B, Derenko M, Grzybowski T, Perkova M, Rogalla U, Vanecek T, et al. The peopling of Europe from the mitochondrial haplogroup U5 perspective. PLoS ONE. 2010;5:e10285. https://doi.org/10.1371/journal.pone.0010285
Pankratov V, Litvinov S, Kassian A, Shulhin D, Tchebotarev L, Yunusbayev B, et al. East Eurasian ancestry in the middle of Europe: genetic footprints of Steppe nomads in the genomes of Belarusian Lipka Tatars. Sci. Rep. 2016;6:30197. https://doi.org/10.1038/srep30197
van Oven M, Kayser M. Updated comprehensive phylogenetic tree of global human mitochondrial DNA variation. Hum Mutat. 2009;30:386–94.
Bandelt HJ, Forster P, Röhl A. Median-joining networks for inferring intraspecific phylogenies. Mol Biol Evol. 1999;16:37–48.
Saillard J, Forster P, Lynnerup N, Bandelt HJ, Nørby S. mtDNA variation among Greenland Eskimos: the edge of the Beringian expansion. Am J Hum Genet. 2000;67:718–26.
Soares P, Ermini L, Thomson N, Mormina M, Rito T, Röhl A, et al. Correcting for purifying selection: an improved human mitochondrial molecular clock. Am J Hum Genet. 2009;84:740–59.
Posth C, Renaud G, Mittnik A, Drucker DG, Rougier H, Cupillard C, et al. Pleistocene mitochondrial genomes suggest a single major dispersal of non-Africans and a Late Glacial population turnover in Europe. Curr Biol. 2016;26:827–33.
Perego UA, Achilli A, Angerhofer N, Accetturo M, Pala M, Olivieri A, et al. Distinctive Paleo-Indian migration routes from Beringia marked by two rare mtDNA haplogroups. Curr Biol. 2009;19:1–8.
Cox MP. Accuracy of molecular dating with the Rho statistic: deviations from coalescent expectations under a range of demographic models. Hum Biol. 2008;80:335–57.
Olivieri A, Pala M, Gandini F, Hooshiar Kashani B, Perego UA, Woodward SR, et al. Mitogenomes from two uncommon haplogroups mark late glacial/postglacial expansions from the Near East and neolithic dispersals within Europe. PLoS ONE. 2013;8:e70492. https://doi.org/10.1371/journal.pone.0070492
Gomes SM, Bodner M, Souto L, Zimmermann B, Huber G, Strobl C, et al. Human settlement history between Sunda and Sahul: a focus on East Timor (Timor-Leste) and the Pleistocenic mtDNA diversity. BMC Genomics. 2015;16:70. https://doi.org/10.1186/s12864-014-1201-x
Drummond AJ, Rambaut A, Shapiro B, Pybus OG. Bayesian coalescent inference of past population dynamics from molecular sequences. Mol Biol Evol. 2005;22:1185–92.
Drummond AJ, Rambaut A, BEAST: Bayesian evolutionary analysis by sampling trees. BMC Evol Biol. 2007;7:214. https://doi.org/10.1186/1471-2148-7-214
Soares P, Alshamali F, Pereira JB, Fernandes V, Silva NM, Afonso C, et al. The expansion of mtDNA haplogroup L3 within and out of Africa. Mol Biol Evol. 2012;29:915–27.
Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S. MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol. 2011;28:2731–9.
Drummond, AJ, Ho, SYW, Rawlence, N & Rambaut, A. A rough guide to BEAST 1.4. (2007). http://beast.bio.ed.ac.uk/.
Librado P, Rozas J. DnaSPv5: a software for comprehensive analysis of DNA polymorphism data. Bioinformatics. 2009;25:1451–2.
Schneider S, Roessli D, Excoffier L. Arlequin ver 2.000: A Software for Population Genetics Data Analysis. Switzerland: Genetics and Biometry Laboratory, University of Geneva; 2000.
Kruskal JB. Multidimensional scaling by optimizing goodness of fit to a nonmetric hypothesis. Psychometrika. 1964;29:1–27.
Zheng H-X, Yan S, Qin Z-D, Wang Y, Tan J-Z, Li H, et al. Major population expansion of East Asians began before Neolithic time: evidence of mtDNA genomes. PLoS ONE. 2011;6:e25835. https://doi.org/10.1371/journal.pone.0025835
Raymond M, Rousset F. An exact test for population differentiation. Evolution. 1995;49:1280–3.
Schurr TG, Sukernik RI, Starikovskaya YB, Wallace DC. Mitochondrial DNA variation in Koryaks and Itel’men: population replacement in the Okhotsk Sea-Bering Sea region during the Neolithic. Am J Phys Anthropol. 1999;108:1–39.
Lippold S, Xu H, Ko A, Li M, Renaud G, Butthof A, et al. Human paternal and maternal demographic histories: insights from high-resolution Y chromosome and mtDNA sequences. Investig Genet.2014;5:13. https://doi.org/10.1186/2041-2223-5-13
Hammer MF, Mendez FL, Cox MP, Woerner AE, Wall JD. Sex-biased evolutionary forces shape genomic patterns of human diversity. PLoS Genet. 2008;4:e1000202. https://doi.org/10.1371/journal.pgen.1000202
Seielstad MT, Minch E, Cavalli-Sforza LL. Genetic evidence for a higher female migration rate in humans. Nat Genet. 1998;20:278–80.
Marchi N, Hegay T, Mennecier P, Georges M, Laurent R, Whitten M, et al. Sex-specific genetic diversity is shaped by cultural factors in Inner Asian human populations. Am J Phys Anthropol. 2017;162:627–40.
Pagani L, Lawson DJ, Jagoda E, Mörseburg A, Eriksson A, Mitt M, et al. Genomic analyses inform on migration events during the peopling of Eurasia. Nature. 2016;538:238–42.
Yunusbayev B, Metspalu M, Metspalu E, Valeev A, Litvinov S, Valiev R, et al. The genetic legacy of the expansion of Turkic-speaking nomads across Eurasia. PLoS Genet. 2015;11:e1005068. https://doi.org/10.1371/journal.pgen.1005068
Acknowledgements
We thank all the donors for providing samples for this project. This study was supported by Program “Basic research for the development of the Russian Arctic” of the Presidium of Russian Academy of Sciences and by grant of the Ministry of Education and Science of the Russian Federation (N 14.W03.31.0016).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of Interest
The authors declare no competing interests.
Rights and permissions
About this article
Cite this article
Derenko, M., Denisova, G., Malyarchuk, B. et al. Mitogenomic diversity and differentiation of the Buryats. J Hum Genet 63, 71–81 (2018). https://doi.org/10.1038/s10038-017-0370-2
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s10038-017-0370-2
This article is cited by
-
Genetic diversity, phylogeography, and maternal origin of yak (Bos grunniens)
BMC Genomics (2024)
-
Mitogenomics of the Koryaks and Evens of the northern coast of the Sea of Okhotsk
Journal of Human Genetics (2023)
-
Mitogenomics of modern Mongolic-speaking populations
Molecular Genetics and Genomics (2022)
-
Ancient DNA from Tubo Kingdom-related tombs in northeastern Tibetan Plateau revealed their genetic affinity to both Tibeto-Burman and Altaic populations
Molecular Genetics and Genomics (2022)
-
Mitogenomic diversity and phylogeny analysis of yak (Bos grunniens)
BMC Genomics (2021)