Abstract
The transcriptional regulation of the human epidermal growth factor receptor-2 (HER2) contributes to an enhanced HER2 expression in HER2-positive breast cancers with HER2 gene amplification and HER2-low or HER2-negative breast cancers following radiotherapy or endocrine therapy, and this drives tumorigenesis and the resistance to therapy. Epigenetic mechanisms are critical for transcription regulation, however, such mechanisms in the transcription regulation of HER2 are limited to the involvement of tri-methylated histone 3 lysine 4 (H3K4me3) and acetylated histone 3 lysine 9 (H3K9ac) at the HER2 promoter region. Here, we report the identification of a novel enhancer in the HER2 3’ gene body, which we have termed HER2 gene body enhancer (HGE). The HGE starts from the 3’ end of intron 19 and extends into intron 22, possesses enhancer histone modification marks in specific cells and enhances the transcriptional activity of the HER2 promoters. We also found that TFAP2C, a known regulator of HER2, binds to HGE and is required for its enhancer function and that DNA methylation in the HGE region inhibits the histone modifications characterizing enhancer and is inversely correlated with HER2 expression in breast cancer samples. The identification of this novel enhancer sheds a light on the roles of epigenetic mechanisms in HER2 transcription, in both HER2-positive breast cancer samples and individuals with HER2-low or HER2-negative breast cancers undergoing radiotherapy or endocrine therapy.
Similar content being viewed by others
Introduction
Human epidermal growth factor receptor-2 (HER2)/Erb-B2 receptor tyrosine kinase 2 is a member of the erbB-like oncogene family, its overexpression occurs in approximately 20–30% of breast cancers1 and is strongly associated with poor prognosis.2 HER2 has roles in the development of HER2-positive breast cancers3, 4 and resistance to therapy in HER2-low or HER2-negative breast cancers, in which HER2 is transcriptionally upregulated by radiotherapy or by endocrine therapy.5, 6, 7 HER2 gene amplification is a major mechanism for HER2 overexpression, however, higher transcription rate of HER2 per gene copy was also observed in HER2-amplified breast cancer cells,8, 9, 10 that is, HER2 mRNA levels are 4- to 8-fold and 64- to 128-fold higher in HER2-overexpressing and HER2-amplified breast cancer cells, respectively, than would be expected from HER2 gene copy numbers.8 A run-on assay showed SKBR3 cells displayed about two fold HER2 transcription rate higher than in BT474 cells.11 Transcription factors such as TFAP2,12, 13 Sp1,14 PBP,15 YY1,16 ETS,17 YB-118 and EGR219 have been shown to positively regulate HER2, whereas MYB,20 FOXP3,21 GATA4,22 PEA3,23 MBP-1,24 NOTCH and RBP-Jk25 have negative effects. Most of these studies focused on the regulation of the originally characterized HER2 promoter (promoter 2),14, 26, 27 which has the dominant role in the overexpression of HER2 in breast cancers28 despite of the identification of an alternative promoter (promoter 1)29 (Figure 1a). Moreover, intron 1 enhancer when bound by PAX2 mediates transcriptional repression of HER2 by activated ER.5 Although these transcriptional mechanisms can explain HER2 regulation in part, the molecular basis of the increase in HER2 transcription in certain cancers remains unexplained.
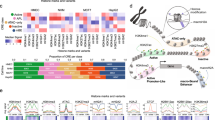
Epigenetic demarcation of the HGE. (a) Top: schematic illustration of the locations of the HER2 promoters (1 and 2), two C-terminal transcripts (CTF611 and CTF687), and the HGE region, as well as the exon composition of HER2, based on UCSC gene annotation (GRCh37/hg19). Bottom: enrichment of H3K4me1, H3K4me, H3K27ac and DNase I hypersensitivity signals, retrieved using the ENCODE Regulation Super-track Settings. Colors representing different cell types are shown in the legend. Chromatin immunoprecipitation (ChIP)-PCR amplicons are shown. NC: sequences to which nonspecific controls were generated. (b) Profile of TFAP2C binding to the HER2 gene in the indicated cell lines, as retrieved from ChIP-seq data (GSE36351). The maximum reads on the y axis represent normalized coverage (reads per million mapped). (c, d) TFAP2C binding and H3K4me1 and H3K27ac modification, in SKBR3 (c) and HCC1954 (d) cells, as assessed by ChIP. Enrichment was interpreted as a percentage of input. Fold change over input normalized to NC is shown. P1, HER2 promoter 1; P2, HER2 promoter 2; EHN1, intron 1 enhancer. Mean±s.d. was determined for three independent experiments, and the Student's t-test was used to calculate the significance. *P<0.05, **P<0.001.
Chromatin modifications can greatly influence transcriptional regulation and contribute to cancer development.30 H3K4me3 and H3K9ac, two histone marks typically associated with gene activation, were reported to be critical for inducing HER2 transcription through promoter 2.10 WDR5, a key component of the H3K4me3 methyltransferase complex, is essentially involved in this process.10 However, these mechanisms are common to general transcriptional activation. Thus, additional mechanisms may exist and specifically contribute to HER2 overexpression. We discovered a novel enhancer HER2 gene body enhancer (HGE) in the 3’ gene body of HER2.
The HGE activates promoters 1 and 2 in trans., and hence the TFAP2C-mediated transcriptional induction of HER2 expression. This novel regulatory mechanism of HER2 transcription contributes to the understanding of increased expression of HER2.
Results and discussion
Identification of a novel enhancer in HER2 locus
Enhancer interacts with promoter to recruit RNA polymerase II and regulate transcription.31 Chromatin signatures such as DNase I hypersensitivity sites32 and histone modifications, that are, H3K4me1 and H3K27ac can be used to predict putative enhancers.33, 34, 35 We took advantage of data from the Encyclopedia of DNA Elements (ENCODE) Consortium36 to search for novel regulatory element(s) that may contribute to the increase of HER2 transcription. With the ENCODE Regulation Super-track Settings, DNase I hypersensitivity sites, H3K4me1 and H3K27ac were found in the previously identified intron 1 enhancer5 (using the annotation of NM_001005862); 5-kb upstream of promoter 2; and in a previously undiscovered region within the 3’ gene body (Figure 1a). The 3’ gene body region starts from intron 19 and ends within intron 22 (based on NM_001289936) (Figure 1a) and, in addition to the above-described features, it contains binding sites for many transcription factors (including POLII, TEAD4, c-MYC and PHF8) identified in K562 cells (Supplementary Figure 1). We named this region as the HGE.
TFAP2C is known to positively regulate HER2 expression as it is required for the HER2 expression in BT474 cells;37 binds to and regulates HER2 promoter 2;12, 38, 39 mediates the repression of HER2 by estrogen;38 its expression is positively correlated with HER2 expression in primary breast cancers.37 We analyzed the TFAP2C occupancy data from four cell lines (GSE36351)40 and revealed that TFAP2C is enriched at both promoter 2 and intron 1 enhancer in all cell lines, with stronger enrichments in HER2-amplified SKBR3 and BT474 lines than in HER2-low MCF7 cells (Figure 1b). Importantly, TFAP2C also binds to the HGE in both SKBR3 and BT474 cells, with the stronger occupancy in SKBR3 cells than in BT474 cells (Figure 1b), which supports HGE as a candidate enhancer. Chromatin immunoprecipitation of TFAP2C, H3K4me1 and H3K27ac were performed in SKBR3 cells and TFAP2C occupancies were confirmed at promoters 1 and 2, the intron 1 enhancer and HGE (Figure 1c). H3K27ac is enriched at all four regions, whereas H3K4me1 enrichment was only obvious at HGE (Figure 1c). Chromatin immunoprecipitation experiments in another HER2-amplified breast cancer cell line, HCC1954 (Figure 1d), revealed that TFAP2C was significantly enriched at all four regions, however, its enrichments at both enhancers were dramatically lower than at promoter 2. H3K27ac was significantly enriched at both promoters and intron 1 enhancer, whereas, H3K4me1 was more enriched at promoter 2 compared with a slight increase at promoter 1 and HGE (Figure 1d). These data support the enhancer feature of the HGE and its cell type dependency. Importantly, a chromosome 17-wide binding data of ERRα and PGC-1β in SKBR3 cells showed binding of ERRα to HGE in addition to intron 1 enhancer and both promoters,41 indicating that the HGE can recruit additional transcription factors.
The HGE enhances the transcriptional activity of HER2 promoters
We tested the ability of the HGE to regulate the transcriptional activities of the HER2 promoters by luciferase reporter assay in 293T, SKBR3 and BT474 cells (Figure 2a). Both promoters (placed upstream of the Luciferase gene), but not the HGE (placed downstream of Luciferase gene), had basal activities in all cases (Figure 2a). Notably, in all three cell lines, the transcriptional activity associated with promoter 2 was stronger than that associated with promoter 1 (Figure 2a), consistent with a previous study.28 Importantly, when HGE was placed at the enhancer position in pGL3-basic vector, that is, 5’ to 3’ downstream of the Luciferase gene, it enhanced the transcriptional activities of both HER2 promoters in all three cell lines (Figure 2a). As H3K4me3 presents at HGE in K562 cells and HER2 C-terminal fragment (CTF) 687 uses exon 21 to initiate its transcription42 (Figure 1a), we engineered HGE upstream of the Luciferase gene. Only a minor transcriptional activity was detected in 293T cells (Figure 2b), suggesting that the transcriptional initiation capacity of HGE is much weaker compared with that of HER2 promoters. Interestingly, HGE, when placed immediately upstream of the Luciferase gene significantly interrupted the transcriptional activities of both promoters (Figure 2b). Meanwhile, the HGE maintained its enhancer function for both HER2 promoters when it was inversely inserted into the enhancer position, that is, 3’ to 5’ downstream of the Luciferase gene (Figure 2c). Moreover, when compared with Intron 1 enhancer, the HGE possesses similar enhancer function for promoter 1 but slightly weaker for promoter 2 (Figure 2c). These data support that the HGE has enhancer function comparable with the intron 1 enhancer.
The HGE enhances transcriptional activity of HER2 promoters. (a) Relative luciferase activities of reporter constructs illustrated at left, normalized to expression of Renilla luciferase from co-transfected pRL-TK-Rluc plasmid. (a) Relative luciferase activity elicited by HER2 promoter 1 (P1), HER2 promoter 2 (P2) and Luciferase (Luc), in 293T, SKBR3 and BT474 cells. # indicates the significance between each group versus pGL3-basic vector. (b) Relative luciferase activities of the pGL3-HGE and pGL3-HGE-promoters in 293T cells. Mean±s.d. was obtained from three independent experiments. The Student's t-test was used to calculate the significance between underlined groups. *P<0.05, **P<0.001. # indicates the significance between each group versus pGL3-basic vector. #P<0.05, ##P<0.001. (c) Comparison of the enhancement activities of HGE, reverse HGE and intron 1 enhancer by luciferase assay. R-HGE, reverse HGE; EHN1, intron 1 enhancer. Student’s t-test was used to calculate the significance between constructs with R-HGE or EHN1 versus HGE with each promoter separately. No significance was observed between R-HGE and HGE, whereas EHN1 significantly enhance transcriptional activities of the basic vector and HER2 promoter 2 compare with HGE and R-HGE.
TFAP2C regulates the enhancer function of HGE
To determine the minimal enhancer element of HGE, we generated a series of deletions including the deletion of exon 20 and intron 20 (T20), the deletion of exon 21 and intron 21 (T21) and the deletion of exon 22 and intron 22 (T22) (Figure 3a, left panel). The transcriptional activities of these constructs were analyzed using the luciferase assay in 293T and SKBR3 cells. The T21 deletion abolished the enhancement of transcriptional activity from promoter 2 in both cell lines, whereas the T20 deletion had no significant effect in either case, and the T22 deletion increased the transcriptional activity of promoter 2 (Figure 3a). These data suggested that exon 21 and intron 21 contain sequences involved in transcriptional activation, and exon 22 and intron 22 contain sequences involved in transcriptional repression. We next searched for the TFAP2C consensus sequence (GCCTGAGGG)43 and identified three closest potential TFAP2C-binding sites (GCCCCAGAG, GCCCTAGGG, GCCCAGGGC) (Figure 3b) located within intron 21. An electrophoretic mobility shift assay using in vitro translated TFAP2C and three oligonucleotide probes corresponding to the three potential TFPA2C-binding sites showed that TFAP2C binds to all the three probes (Figure 3b). The specificity of TFAP2C binding was further confirmed by competition of non-labeled oligos and the supershifts when cultured with a specific TFAP2C antibody (Figure 3b). TFAP2C silencing attenuated luciferase activity of the HGE enhancer in both 293T and SKBR3 cells (Figure 3c) and downregulated HER2 protein levels in SKBR3 cells. (Figure 3d). These data support our hypothesis that TFAP2C has an important role in regulating the enhancer function of the HGE.
TFAP2C positively regulates enhancer activity of the HGE. (a) Relative luciferase activities of reporter constructs bearing deletions in HGE region shown at left (T20: deletion of exon 20 and intron 20 in the pGL3-P2-HGE vector) in 293T and SKBR3 cells. Relative luciferase activities of the indicated constructs normalized to Renilla luciferase expression in cells co-transfected with pRL-TK-Rluc. (b) Schematic illustration of positions of three potential TFAP2C-binding sites corresponding to three probes for electrophoretic mobility shift assay (EMSA). Unlabeled 25-bp oligonucleotides or nonspecific competitor were added as indicated. + and − indicate the presence or absence of reaction components. Shifts, supershifts and free probes are indicated with arrows. (c) Fold change of relative luciferase activity of pGL3-P2-HGE normalized to that of pGL3-P2 vectors in 293T or SKBR3 cells transiently transfected with control (Ctrl) or TFAP2C siRNAs. (d) Expression of the indicated proteins as assessed by western blotting. Mean±s.d. in a and c were obtained from three independent experiments. Student's t-test was used to calculate the significance. *P<0.05, **P<0.001. (e, f). Six gRNAs were designed to introduce mutations in intron 21 as indicated in b. Virus infected (e) and transient transfection for 48 h (f) of CRISPR-Cas9-GFP vector with corresponding gRNAs in SKBR3 cells were assessed by western blotting. (g) RT-PCR used to determine a correlation between HER2 expression and expression of TFAP2C in panel of primary breast cancer samples (n=12). The correlation (r2) and significance (P-value) are shown.
We next carried out genomic editing using CRISPR-Cas9 system to determine the role of the TFAP2C-binding sites at the HGE in the regulation of HER2 expression. Six single-guide RNAs (gRNA) were designed to mutate or truncate TFAP2C-binding sites (Figure 3b). SKBR3 stable cell lines with the single or combined gRNA(s) were attempted to be established, however, only one stable cell line with gRNA6 was achieved. It is likely that the cell viability of SKBR3 cells depends on HER2 as knockdown HER2 in SKBR3 cells results in growth arrest and apoptosis.44 HER2 protein in this cell line was dramatically downregulated while cleaved PARP was induced (Figure 3e). The genotyping of SKBR3-gRNA6 showed heterogenous genomic compositions, that is, normal HGE region and extended mutations from the gRNA6-targeting site (Supplementary Figure 2). This data suggest that CRISPR-Cas9 system with these gRNAs introduced additional mutations in HER2 gene and may interfere with HER2 mRNA splicing, resulting in decreased HER2 expression and apoptosis of SKBR3 cells. In fact, a recent study show that genomic editing of HER2 gene using CRISPR-Cas9 system produced short truncated HER2 caused by alternative splicing of HER2 gene and inhibits cell proliferation in both SKBR3 and BT474 cells,45 consistent with an recent observation of off-target mutations introduced by CRISPR-Cas9 system.46
Considering the HER2-dependent cell viability of SKBR3 cells, the six gRNAs were transiently transfected to SKBR3 cells for 48 h and downregulation of HER2, phospho-AKT levels at various extents was found without inducing obvious apoptosis (Figure 3f). These data suggest that the gRNAs targeting intron 21 containing three TFAP2C-binding sites and the junction of exon 21 and intron 21 interfere with HER2 expression. We also examined the mRNA levels of TFAP2C and HER2 from 12 unidentifiable HER2-positive breast cancer samples and found positive correlation (r2=0.6073) between them (Figure 3g).
DNA methylation within the HGE inhibits the enhancer histone modifications and is inversely correlated with HER2 gene expression in breast cancer samples
The cell-type-dependent enrichments of TFAP2C, H3K4me1 and H3K27ac in the HGE region (Figures 1c and d) prompted us to investigate the underlying mechanism. DNA methylation is known to prevent TFAP2C from accessing the target promoter,47 and DNA methylation and certain histone modifications such as H3K4me3 are mutually exclusive.48 Thus, we hypothesized that DNA methylation within the HGE affects TFAP2C binding and enrichment of the enhancer histone modifications. Analysis of both Methyl 450 K bead array data (ENCODE/HAIB) and Reduced Representation Bisulfite Seq data (ENCODE/HudsonAlpha)36 showed that DNA hypomethylation in the HGE region is coincident with enrichments of transcription factors and enhancer histone modifications in K562 cells (Supplementary Figure 1). We performed a bisulfite sequencing assay to determine the DNA methylation status of 28 CpG sites within the HGE. The HGE is extensively DNA-methylated in MCF7, BT474, HCC1954, MDA-MB-231, MCF10A and ZR-75-1 cells. It is less methylated in K562 and hypomethylated in SKBR3 cells (Figure 4a). These data support our hypothesis that DNA methylation status is critical for the enrichments of TFAP2C, H3K4me1 and H3K27ac in the HGE region. The minor enrichments of TFAP2C at HGE in both BT474 and HCC1954 cells (Figures 1b and d) likely reflect the existence of minor cell populations possessing hypomethylated HGE or the cells toward complete establishment of DNA methylation during cell cycle. We next performed in vitro methylation of the pGL3-promoter constructs with and without HGE, using the CpG methyltransferase M.SssI49, 50 and found a strong decrease of enhanced luciferase activity of the methylated pGL3-promoters vector with HGE compared with that without HGE (Supplementary Figure 3).
DNA methylation determines the enhancer function of HGE and is inversely correlated with HER2 gene expression in breast cancers. (a) DNA methylation of the 28 CpG dinucleotides in the HGE region in multiple cell lines, assessed using the bisulfite sequencing. Each circle represents a CpG dinucleotide: open circle, unmethylated site; closed circle, methylated site; half-closed circle, semi-methylated site; circle with an x, mutated site. Exons are marked, and TFAP2C consensus nucleotides are indicated by arrows. (b) Indicated proteins from SKBR3 cells either transfected with CRISPR-dCas9-DNMT3A-EGFP carrying gRNA 1 or 6 and undergoing stable selection (left) or transiently transfected with the same plasmids for 48 h (right) were assessed by western blotting. (c) DNA methylation status of the HGE region in SKBR3 during the stable selection as assessed by bisulfide sequencing. (d) Correlation (Pearson's r) of all 47 DNA methylation sites around HER2 gene including HGE regions (red color) with HER2 expression in 839 breast invasive carcinoma samples was analyzed using MethHC.53 (e) The inverse correlation of average and each individual CpG sites located in HGE with HER2 expression in HER2-positive breast cancer samples grouped in hypomethylation (light) and hypermethylation (dark) is shown. Pearson r2 values of the correlations are displayed in each subgroup of samples. (f) Correlation between TFAP2C, ESRRA, PPARGC1B and HER2 mRNA level in HER2-positive breast cancer samples sub grouped by DNA methylation status of HGE region were plotted. Light and dark dots are designated to hypomethylation and hypermethylation in the HGE region, respectively.
Furthermore, we performed CRISPR-dCas9-guided specific DNA methylation51 at the HGE region in SKBR3 cells. The six gRNAs (Figure 3b) were inserted into pdCas9-DNMT3A-EGFP vector and again, stable cell lines using SKBR3 cells were failed to establish. However, we were able to collect cells from gRNA 1 and 6 during the attempt and found that the cells undergo apoptosis (Figure 4b, left panel). Importantly, the HGE region became partially methylated in SKBR3 cells that transfected with pooled gRNAs, validating the target-specific DNA methylation by the dCas9-DNMT3A system and indicating that the apoptosis might be caused at least partially by loss of HER2 (Figure 4c). The dCas9-DNMT3A-gRNA 1 and 6 transient transfected into SKBR3 cells for 48 h markedly downregulated HER2 expression, phospho-AKT, but not that of TFAP2C and no apparent apoptosis was observed (Figure 4b, right panel). These data suggest that the DNA methylation at the HGE region is important for repression of HER2 expression in SKBR3 cells.
DNA methylation is in general strongly correlated with HER2 expression (r2=0.5055868) in breast cancer.52 Using the methylation database MethHC,53 we analyzed the correlation of the DNA methylation status of 47 CpG probes and HER2 expression in 839 breast invasive carcinoma samples cataloged in The Cancer Genome Atlas. The general inverse correlation of DNA methylation in HER2 gene body and HER2 mRNA is stronger (r=−0.48) compared with that of the promoters (r=−0.22 and −0.19 for promoters 1 and 2, respectively) (Supplementary Figure 4). The correlation of the DNA methylation status of all 47 CpG probes with HER2 mRNA expression shows strong inverse correlation (r values ranging from −0.291 to −0.408) between the DNA methylation of the four HGE CpG sites and HER2 mRNA expression (Figure 4d). Further analysis in HER2-positive breast cancers revealed that the DNA hypomethylation of all four HGE CpG sites is inversely correlated with HER2 mRNA (Figure 4e). In the samples with hypomethylated HGE region, TFAP2C mRNA is positively associated with HER2 mRNA (Figure 4f). As discussed earlier that ERRα also binds to HGE region and regulates HER2 expression in SKBR3 cells,41 we also found a positive, but weaker association between the mRNA of the coding gene ESRRA and HER2 expression. In contrast, the expression of PPARGC1B does not show any association. These data suggest that the hypomethylation of the HGE region in breast cancers contributes to HER2 expression by gaining the accessibility of transcription factors such as TFAP2C and ERRα.
In sum, this study unveiled a novel regulatory mechanism by a 3’ gene body enhancer contributing to the transcriptional regulation of HER2. Further studies are sought to determine the role of this enhancer in the transcriptional upregulation of HER2 in HER2-low or HER2-negative breast cancers that undergo radiotherapy or endocrine therapy.
References
Slamon DJ, Clark GM, Wong SG, Levin WJ, Ullrich A, McGuire WL . Human breast cancer: correlation of relapse and survival with amplification of the HER-2/neu oncogene. Science 1987; 235: 177–182.
Dittrich A, Gautrey H, Browell D, Tyson-Capper A . The HER2 signaling network in breast cancer—like a spider in its web. J Mammary Gland Biol Neoplasia 2014; 19: 253–270.
Roskoski R Jr . The ErbB/HER family of protein-tyrosine kinases and cancer. Pharmacol Res 2014; 79: 34–74.
Baselga J, Swain SM . Novel anticancer targets: revisiting ERBB2 and discovering ERBB3. Nat Rev Cancer 2009; 9: 463–475.
Hurtado A, Holmes KA, Geistlinger TR, Hutcheson IR, Nicholson RI, Brown M et al. Regulation of ERBB2 by oestrogen receptor-PAX2 determines response to tamoxifen. Nature 2008; 456: 663–666.
Duru N, Fan M, Candas D, Menaa C, Liu HC, Nantajit D et al. HER2-associated radioresistance of breast cancer stem cells isolated from HER2-negative breast cancer cells. Clin Cancer Res 2012; 18: 6634–6647.
Cao N, Li S, Wang Z, Ahmed KM, Degnan ME, Fan M et al. NF-kappaB-mediated HER2 overexpression in radiation-adaptive resistance. Radiat Res 2009; 171: 9–21.
Kraus MH, Popescu NC, Amsbaugh SC, King CR . Overexpression of the EGF receptor-related proto-oncogene erbB-2 in human mammary tumor cell lines by different molecular mechanisms. EMBO J 1987; 6: 605–610.
Bofin AM, Ytterhus B, Martin C, O'Leary JJ, Hagmar BM . Detection and quantitation of HER-2 gene amplification and protein expression in breast carcinoma. Am J Clin Pathol 2004; 122: 110–119.
Mungamuri SK, Murk W, Grumolato L, Bernstein E, Aaronson SA . Chromatin modifications sequentially enhance ErbB2 expression in ErbB2-positive breast cancers. Cell Rep 2013; 5: 302–313.
Pasleau F, Grooteclaes M, Gol-Winkler R . Expression of the c-erbB2 gene in the BT474 human mammary tumor cell line: measurement of c-erbB2 mRNA half-life. Oncogene 1993; 8: 849–854.
Vernimmen D, Begon D, Salvador C, Gofflot S, Grooteclaes M, Winkler R . Identification of HTF (HER2 transcription factor) as an AP-2 (activator protein-2) transcription factor and contribution of the HTF binding site to ERBB2 gene overexpression. Biochem J 2003; 370: 323–329.
Delacroix L, Begon D, Chatel G, Jackers P, Winkler R . Distal ERBB2 promoter fragment displays specific transcriptional and nuclear binding activities in ERBB2 overexpressing breast cancer cells. DNA Cell Biol 2005; 24: 582–594.
Chen Y, Gill GN . Positive and negative regulatory elements in the human erbB-2 gene promoter. Oncogene 1994; 9: 2269–2276.
Chen Y, Gill GN . A heterodimeric nuclear protein complex binds two palindromic sequences in the proximal enhancer of the human erbB-2 gene. J Biol Chem 1996; 271: 5183–5188.
Begon DY, Delacroix L, Vernimmen D, Jackers P, Winkler R . Yin Yang 1 cooperates with activator protein 2 to stimulate ERBB2 gene expression in mammary cancer cells. J Biol Chem 2005; 280: 24428–24434.
Scott GK, Chang CH, Erny KM, Xu F, Fredericks WJ, Rauscher FJ 3rd et al. Ets regulation of the erbB2 promoter. Oncogene 2000; 19: 6490–6502.
Wu J, Lee C, Yokom D, Jiang H, Cheang MC, Yorida E et al. Disruption of the Y-box binding protein-1 results in suppression of the epidermal growth factor receptor and HER-2. Cancer Res 2006; 66: 4872–4879.
Dillon RL, Brown ST, Ling C, Shioda T, Muller WJ . An EGR2/CITED1 transcription factor complex and the 14-3-3sigma tumor suppressor are involved in regulating ErbB2 expression in a transgenic-mouse model of human breast cancer. Mol Cell Biol 2007; 27: 8648–8657.
Mizuguchi G, Kanei-Ishii C, Takahashi T, Yasukawa T, Nagase T, Horikoshi M et al. c-Myb repression of c-erbB-2 transcription by direct binding to the c-erbB-2 promoter. J Biol Chem 1995; 270: 9384–9389.
Zuo T, Wang L, Morrison C, Chang X, Zhang H, Li W et al. FOXP3 is an X-linked breast cancer suppressor gene and an important repressor of the HER-2/ErbB2 oncogene. Cell 2007; 129: 1275–1286.
Hua G, Zhu B, Rosa F, Deblon N, Adelaide J, Kahn-Perles B et al. A negative feedback regulatory loop associates the tyrosine kinase receptor ERBB2 and the transcription factor GATA4 in breast cancer cells. Mol Cancer Res 2009; 7: 402–414.
Xing X, Wang SC, Xia W, Zou Y, Shao R, Kwong KY et al. The ets protein PEA3 suppresses HER-2/neu overexpression and inhibits tumorigenesis. Nat Med 2000; 6: 189–195.
Contino F, Mazzarella C, Ferro A, Lo Presti M, Roz E, Lupo C et al. Negative transcriptional control of ERBB2 gene by MBP-1 and HDAC1: diagnostic implications in breast cancer. BMC Cancer 2013; 13: 81.
Chen Y, Fischer WH, Gill GN . Regulation of the ERBB-2 promoter by RBPJkappa and NOTCH. J Biol Chem 1997; 272: 14110–14114.
Tal M, King CR, Kraus MH, Ullrich A, Schlessinger J, Givol D . Human HER2 (neu) promoter: evidence for multiple mechanisms for transcriptional initiation. Mol Cell Biol 1987; 7: 2597–2601.
Grooteclaes M, Pasleau F, Dijkmans H, Berzi P, Albert A, Winkler-Gol R . The 6-kilobase c-erbB2 promoter contains positive and negative regulatory elements functional in human mammary cell lines. Cancer Res 1994; 54: 4193–4199.
Benz CC, Fedele V, Xu F, Ylstra B, Ginzinger D, Yu M et al. Altered promoter usage characterizes monoallelic transcription arising with ERBB2 amplification in human breast cancers. Genes Chromosomes Cancer 2006; 45: 983–994.
Nezu M, Sasaki H, Kuwahara Y, Ochiya T, Yamada Y, Sakamoto H et al. Identification of a novel promoter and exons of the c-ERBB-2 gene. Biochem Biophys Res Commun 1999; 258: 499–505.
Greer EL, Shi Y . Histone methylation: a dynamic mark in health, disease and inheritance. Nat Rev Genet 2012; 13: 343–357.
Malik S, Roeder RG . The metazoan mediator co-activator complex as an integrative hub for transcriptional regulation. Nat Rev Genet 2010; 11: 761–772.
Crawford GE, Holt IE, Whittle J, Webb BD, Tai D, Davis S et al. Genome-wide mapping of DNase hypersensitive sites using massively parallel signature sequencing (MPSS). Genome Res 2006; 16: 123–131.
Ong CT, Corces VG . Enhancer function: new insights into the regulation of tissue-specific gene expression. Nat Rev Genet 2011; 12: 283–293.
Rada-Iglesias A, Bajpai R, Swigut T, Brugmann SA, Flynn RA, Wysocka J . A unique chromatin signature uncovers early developmental enhancers in humans. Nature 2011; 470: 279–283.
Creyghton MP, Cheng AW, Welstead GG, Kooistra T, Carey BW, Steine EJ et al. Histone H3K27ac separates active from poised enhancers and predicts developmental state. Proc Natl Acad Sci USA 2010; 107: 21931–21936.
Consortium EP. An integrated encyclopedia of DNA elements in the human genome. Nature 2012; 489: 57–74.
Allouche A, Nolens G, Tancredi A, Delacroix L, Mardaga J, Fridman V et al. The combined immunodetection of AP-2alpha and YY1 transcription factors is associated with ERBB2 gene overexpression in primary breast tumors. Breast Cancer Res 2008; 10: R9.
Perissi V, Menini N, Cottone E, Capello D, Sacco M, Montaldo F et al. AP-2 transcription factors in the regulation of ERBB2 gene transcription by oestrogen. Oncogene 2000; 19: 280–288.
Bosher JM, Totty NF, Hsuan JJ, Williams T, Hurst HC . A family of AP-2 proteins regulates c-erbB-2 expression in mammary carcinoma. Oncogene 1996; 13: 1701–1707.
Kulak MV, Cyr AR, Woodfield GW, Bogachek M, Spanheimer PM, Li T et al. Transcriptional regulation of the GPX1 gene by TFAP2C and aberrant CpG methylation in human breast cancer. Oncogene 2013; 32: 4043–4051.
Deblois G, Chahrour G, Perry MC, Sylvain-Drolet G, Muller WJ, Giguere V . Transcriptional control of the ERBB2 amplicon by ERRalpha and PGC-1beta promotes mammary gland tumorigenesis. Cancer Res 2010; 70: 10277–10287.
Anido J, Scaltriti M, Bech Serra JJ, Santiago Josefat B, Todo FR, Baselga J et al. Biosynthesis of tumorigenic HER2 C-terminal fragments by alternative initiation of translation. EMBO J 2006; 25: 3234–3244.
Woodfield GW, Chen Y, Bair TB, Domann FE, Weigel RJ . Identification of primary gene targets of TFAP2C in hormone responsive breast carcinoma cells. Genes Chromosomes Cancer 2010; 49: 948–962.
Choudhury A, Charo J, Parapuram SK, Hunt RC, Hunt DM, Seliger B et al. Small interfering RNA (siRNA) inhibits the expression of the Her2/neu gene, upregulates HLA class I and induces apoptosis of Her2/neu positive tumor cell lines. Int J Cancer 2004; 108: 71–77.
Wang H, Sun W . CRISPR-mediated targeting of HER2 inhibits cell proliferation through a dominant negative mutation. Cancer Lett 2017; 385: 137–143.
Schaefer KA, Wu WH, Colgan DF, Tsang SH, Bassuk AG, Mahajan VB . Unexpected mutations after CRISPR-Cas9 editing in vivo. Nat Methods 2017; 14: 547–548.
Woodfield GW, Hitchler MJ, Chen Y, Domann FE, Weigel RJ . Interaction of TFAP2C with the estrogen receptor-alpha promoter is controlled by chromatin structure. Clin Cancer Res 2009; 15: 3672–3679.
Rose NR, Klose RJ . Understanding the relationship between DNA methylation and histone lysine methylation. Biochim Biophys Acta 2014; 1839: 1362–1372.
Renbaum P, Abrahamove D, Fainsod A, Wilson GG, Rottem S, Razin A . Cloning, characterization, and expression in Escherichia coli of the gene coding for the CpG DNA methylase from Spiroplasma sp. strain MQ1(M.SssI). Nucleic Acids Res 1990; 18: 1145–1152.
Tata PR, Tata NR, Kuhl M, Sirbu IO . Identification of a novel epigenetic regulatory region within the pluripotency associated microRNA cluster, EEmiRC. Nucleic Acids Res 2011; 39: 3574–3581.
Vojta A, Dobrinic P, Tadic V, Bockor L, Korac P, Julg B et al. Repurposing the CRISPR-Cas9 system for targeted DNA methylation. Nucleic Acids Res 2016; 44: 5615–5628.
Moarii M, Boeva V, Vert JP, Reyal F . Changes in correlation between promoter methylation and gene expression in cancer. BMC Genomics 2015; 16: 873.
Huang WY, Hsu SD, Huang HY, Sun YM, Chou CH, Weng SL et al. MethHC: a database of DNA methylation and gene expression in human cancer. Nucleic Acids Res 2015; 43: D856–D861.
Acknowledgements
We thank Dr Brad Amendt and his lab for helpful discussions, and the ENCODE Consortium and the ENCODE production laboratories for generating the relevant data sets. The results of the bioinformatical analysis are based, in whole or part, upon data generated by The Cancer Genome Atlas Research Network (http://cancergenome.nih.gov/). The K562 cell line was a kind gift from Dr Fenghuang Zhan. We also thank Drs Christine Blaumueller and Marie Gaine for editorial consultation. This work was supported by to HHQ start-up funds from the Department of Anatomy and Cell Biology, the Carver College of Medicine, University of Iowa; Carver Trust Young Investigator Award (01-224 to HHQ) from the Roy J Carver Charitable Trust; a Breast Cancer Research Award (to HHQ) by the Holden Comprehensive Cancer Center at University of Iowa; The NIH grant (P30 CA086862) to the Genomics and Flow Cytometry core facilities at the University of Iowa. NIH grants R01CA183702 (PI: RJW) and by a generous gift from the Kristen Olewine Milke Breast Cancer Research Fund (PI: RJW). NB was supported by NIH MD/PhD fellowship (F30 CA206255); WZ was supported by NIH grants CA200673, and CA203834, the V Scholar award, a Breast Cancer Research Award and an Oberley Award (National Cancer Institute Award P30 CA086862) from Holden Comprehensive Cancer Center at the University of Iowa.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Additional information
Supplementary Information accompanies this paper on the Oncogene website
Supplementary information
Rights and permissions
This work is licensed under a Creative Commons Attribution-NonCommercial-ShareAlike 4.0 International License. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in the credit line; if the material is not included under the Creative Commons license, users will need to obtain permission from the license holder to reproduce the material. To view a copy of this license, visit http://creativecommons.org/licenses/by-nc-sa/4.0/
About this article
Cite this article
Liu, Q., Kulak, M., Borcherding, N. et al. A novel HER2 gene body enhancer contributes to HER2 expression. Oncogene 37, 687–694 (2018). https://doi.org/10.1038/onc.2017.382
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/onc.2017.382
This article is cited by
-
DDM1-mediated gene body DNA methylation is associated with inducible activation of defense-related genes in Arabidopsis
Genome Biology (2023)
-
Solid-phase capture and profiling of open chromatin by spatial ATAC
Nature Biotechnology (2023)
-
Functional analysis of a putative HER2-associated expressed enhancer, Her2-Enhancer1, in breast cancer cells
Scientific Reports (2023)
-
FGFR blockade inhibits targeted therapy-tolerant persister in basal FGFR1- and FGF2-high cancers with driver oncogenes
npj Precision Oncology (2023)
-
TFAP2C regulates carbonic anhydrase XII in human breast cancer
Oncogene (2020)