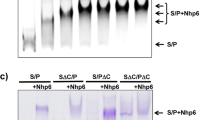
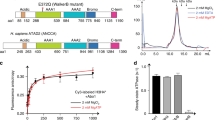
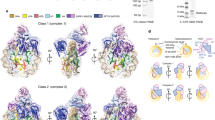
Abstract
DNA accessibility to regulatory proteins is substantially influenced by nucleosome structure and dynamics. The facilitates chromatin transcription (FACT) complex increases the accessibility of nucleosomal DNA, but the mechanism and extent of its nucleosome reorganization activity are unknown. Here we determined the effects of FACT from the yeast Saccharomyces cerevisiae on single nucleosomes by using single-particle Förster resonance energy transfer (spFRET) microscopy. FACT binding results in dramatic ATP-independent, symmetrical and reversible DNA uncoiling that affects at least 70% of the DNA within a nucleosome, occurs without apparent loss of histones and proceeds via an 'all-or-none' mechanism. A mutated version of FACT is defective in uncoiling, and a histone mutation that suppresses phenotypes caused by this FACT mutation in vivo restores the uncoiling activity in vitro. Thus, FACT-dependent nucleosome unfolding modulates the accessibility of nucleosomal DNA, and this activity is an important function of FACT in vivo.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$189.00 per year
only $15.75 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout






Similar content being viewed by others
Accession codes
References
Luger, K., Mäder, A.W., Richmond, R.K., Sargent, D.F. & Richmond, T.J. Crystal structure of the nucleosome core particle at 2.8 A resolution. Nature 389, 251–260 (1997).
Shaytan, A.K., Landsman, D. & Panchenko, A.R. Nucleosome adaptability conferred by sequence and structural variations in histone H2A-H2B dimers. Curr. Opin. Struct. Biol. 32, 48–57 (2015).
Kulaeva, O.I., Hsieh, F.-K., Chang, H.-W., Luse, D.S. & Studitsky, V.M. Mechanism of transcription through a nucleosome by RNA polymerase II. Biochim. Biophys. Acta 1829, 76–83 (2013).
Gurard-Levin, Z.A., Quivy, J.-P. & Almouzni, G. Histone chaperones: assisting histone traffic and nucleosome dynamics. Annu. Rev. Biochem. 83, 487–517 (2014).
Park, Y.-J. & Luger, K. Histone chaperones in nucleosome eviction and histone exchange. Curr. Opin. Struct. Biol. 18, 282–289 (2008).
Ransom, M., Dennehey, B.K. & Tyler, J.K. Chaperoning histones during DNA replication and repair. Cell 140, 183–195 (2010).
Belotserkovskaya, R., Saunders, A., Lis, J.T. & Reinberg, D. Transcription through chromatin: understanding a complex FACT. Biochim. Biophys. Acta 1677, 87–99 (2004).
Formosa, T. The role of FACT in making and breaking nucleosomes. Biochim. Biophys. Acta 1819, 247–255 (2012).
Hondele, M. & Ladurner, A.G. Catch me if you can: how the histone chaperone FACT capitalizes on nucleosome breathing. Nucleus 4, 443–449 (2013).
Winkler, D.D. & Luger, K. The histone chaperone FACT: structural insights and mechanisms for nucleosome reorganization. J. Biol. Chem. 286, 18369–18374 (2011).
Brewster, N.K., Johnston, G.C. & Singer, R.A. A bipartite yeast SSRP1 analog comprised of Pob3 and Nhp6 proteins modulates transcription. Mol. Cell. Biol. 21, 3491–3502 (2001).
Formosa, T. et al. Spt16-Pob3 and the HMG protein Nhp6 combine to form the nucleosome-binding factor SPN. EMBO J. 20, 3506–3517 (2001).
Reinberg, D. & Sims, R.J. III. de FACTo nucleosome dynamics. J. Biol. Chem. 281, 23297–23301 (2006).
Formosa, T. FACT and the reorganized nucleosome. Mol. Biosyst. 4, 1085–1093 (2008).
Mandemaker, I.K., Vermeulen, W. & Marteijn, J.A. Gearing up chromatin: a role for chromatin remodeling during the transcriptional restart upon DNA damage. Nucleus 5, 203–210 (2014).
Erkina, T.Y. & Erkine, A. ASF1 and the SWI/SNF complex interact functionally during nucleosome displacement, while FACT is required for nucleosome reassembly at yeast heat shock gene promoters during sustained stress. Cell Stress Chaperones 20, 355–369 (2015).
Takahata, S., Yu, Y. & Stillman, D.J. FACT and Asf1 regulate nucleosome dynamics and coactivator binding at the HO promoter. Mol. Cell 34, 405–415 (2009).
Cheung, V. et al. Chromatin- and transcription-related factors repress transcription from within coding regions throughout the Saccharomyces cerevisiae genome. PLoS Biol. 6, e277 (2008).
Jamai, A., Imoberdorf, R.M. & Strubin, M. Continuous histone H2B and transcription-dependent histone H3 exchange in yeast cells outside of replication. Mol. Cell 25, 345–355 (2007).
Voth, W.P. et al. A role for FACT in repopulation of nucleosomes at inducible genes. PLoS One 9, e84092 (2014).
Hsieh, F.-K. et al. Histone chaperone FACT action during transcription through chromatin by RNA polymerase II. Proc. Natl. Acad. Sci. USA 110, 7654–7659 (2013).
Belotserkovskaya, R. et al. FACT facilitates transcription-dependent nucleosome alteration. Science 301, 1090–1093 (2003).
Winkler, D.D., Muthurajan, U.M., Hieb, A.R. & Luger, K. Histone chaperone FACT coordinates nucleosome interaction through multiple synergistic binding events. J. Biol. Chem. 286, 41883–41892 (2011).
Stuwe, T. et al. The FACT Spt16 “peptidase” domain is a histone H3-H4 binding module. Proc. Natl. Acad. Sci. USA 105, 8884–8889 (2008).
Hondele, M. et al. Structural basis of histone H2A–H2B recognition by the essential chaperone FACT. Nature 499, 111–114 (2013).
Kemble, D.J., McCullough, L.L., Whitby, F.G., Formosa, T. & Hill, C.P. FACT disrupts nucleosome structure by binding H2A-H2B with conserved peptide motifs. Mol. Cell 60, 294–306 (2015).
Jamai, A., Puglisi, A. & Strubin, M. Histone chaperone spt16 promotes redeposition of the original h3-h4 histones evicted by elongating RNA polymerase. Mol. Cell 35, 377–383 (2009).
Xin, H. et al. yFACT induces global accessibility of nucleosomal DNA without H2A-H2B displacement. Mol. Cell 35, 365–376 (2009).
Gaykalova, D.A. et al. Structural analysis of nucleosomal barrier to transcription. Proc. Natl. Acad. Sci. USA 112, E5787–E5795 (2015).
Kulaeva, O.I. et al. Mechanism of chromatin remodeling and recovery during passage of RNA polymerase II. Nat. Struct. Mol. Biol. 16, 1272–1278 (2009).
Vasudevan, D., Chua, E.Y.D. & Davey, C.A. Crystal structures of nucleosome core particles containing the '601' strong positioning sequence. J. Mol. Biol. 403, 1–10 (2010).
Morozov, A.V. et al. Using DNA mechanics to predict in vitro nucleosome positions and formation energies. Nucleic Acids Res. 37, 4707–4722 (2009).
Kudryashova, K.S. et al. Preparation of mononucleosomal templates for analysis of transcription with RNA polymerase using spFRET. Methods Mol. Biol. 1288, 395–412 (2015).
Gurunathan, K. & Levitus, M. Single-molecule fluorescence studies of nucleosome dynamics. Curr. Pharm. Biotechnol. 10, 559–568 (2009).
Koopmans, W.J.A., Brehm, A., Logie, C., Schmidt, T. & van Noort, J. Single-pair FRET microscopy reveals mononucleosome dynamics. J. Fluoresc. 17, 785–795 (2007).
Kireeva, M.L. et al. Nucleosome remodeling induced by RNA polymerase II: loss of the H2A/H2B dimer during transcription. Mol. Cell 9, 541–552 (2002).
McCullough, L. et al. Insight into the mechanism of nucleosome reorganization from histone mutants that suppress defects in the FACT histone chaperone. Genetics 188, 835–846 (2011).
Böhm, V. et al. Nucleosome accessibility governed by the dimer/tetramer interface. Nucleic Acids Res. 39, 3093–3102 (2011).
Zlatanova, J., Bishop, T.C., Victor, J.-M., Jackson, V. & van Holde, K. The nucleosome family: dynamic and growing. Structure 17, 160–171 (2009).
Tsunaka, Y., Fujiwara, Y., Oyama, T., Hirose, S. & Morikawa, K. Integrated molecular mechanism directing nucleosome reorganization by human FACT. Genes Dev. 30, 673–686 (2016).
Ruone, S., Rhoades, A.R. & Formosa, T. Multiple Nhp6 molecules are required to recruit Spt16-Pob3 to form yFACT complexes and to reorganize nucleosomes. J. Biol. Chem. 278, 45288–45295 (2003).
Paull, T.T. & Johnson, R.C. DNA looping by Saccharomyces cerevisiae high mobility group proteins NHP6A/B: consequences for nucleoprotein complex assembly and chromatin condensation. J. Biol. Chem. 270, 8744–8754 (1995).
Biswas, D., Yu, Y., Prall, M., Formosa, T. & Stillman, D.J. The yeast FACT complex has a role in transcriptional initiation. Mol. Cell. Biol. 25, 5812–5822 (2005).
Wittmeyer, J., Joss, L. & Formosa, T. Spt16 and Pob3 of Saccharomyces cerevisiae form an essential, abundant heterodimer that is nuclear, chromatin-associated, and copurifies with DNA polymerase alpha. Biochemistry 38, 8961–8971 (1999).
Formosa, T. et al. Defects in SPT16 or POB3 (yFACT) in Saccharomyces cerevisiae cause dependence on the Hir/Hpc pathway: polymerase passage may degrade chromatin structure. Genetics 162, 1557–1571 (2002).
Rhoades, A.R., Ruone, S. & Formosa, T. Structural features of nucleosomes reorganized by yeast FACT and its HMG box component, Nhp6. Mol. Cell. Biol. 24, 3907–3917 (2004).
Bondarenko, V.A. et al. Nucleosomes can form a polar barrier to transcript elongation by RNA polymerase II. Mol. Cell 24, 469–479 (2006).
Kudryashova, K.S. et al. Development of fluorescently labeled mononucleosomes to study transcription mechanisms by method of microscopy of single complexes. Moscow Univ. Biol. Sci. Bull. 70, 189–193 (2015).
Buning, R. & van Noort, J. Single-pair FRET experiments on nucleosome conformational dynamics. Biochimie 92, 1729–1740 (2010).
Davey, C.A., Sargent, D.F., Luger, K., Maeder, A.W. & Richmond, T.J. Solvent mediated interactions in the structure of the nucleosome core particle at 1.9 a resolution. J. Mol. Biol. 319, 1097–1113 (2002).
Lu, X.-J. & Olson, W.K. 3DNA: a versatile, integrated software system for the analysis, rebuilding and visualization of three-dimensional nucleic-acid structures. Nat. Protoc. 3, 1213–1227 (2008).
Klose, D. et al. Simulation vs. reality: a comparison of in silico distance predictions with DEER and FRET measurements. PLoS One 7, e39492 (2012).
Acknowledgements
We thank D. Gaykalova for help with designing the fluorescent probes. This work was supported by NIH grants GM58650 to V.M.S. and R01GM064649 to T.F., and by the Program of the Presidium of the Russian Academy of Sciences 'Basic Research for the Development of Biomedical Technologies' (FIMT-2014-011). Part of this work was performed with the equipment of the Center for Collective Use 'Genom' of the Institute of Molecular Biology, Russian Academy of Sciences (http://www.eimb.ru/RUSSIAN_NEW/INSTITUTE/ccu_genome_c.php/) supported by the Ministry of Education and Science of the Russian Federation (agreement no. 14.621.21.0001, unique project identification number RFMEFI62114X0001). Development and applications of spFRET were supported by the Russian Science Foundation grant 14-24-00031. Facilities of the Supercomputing Center of Lomonosov Moscow State University were used for the modeling of FRET in nucleosomes. LSM710-Confocor3 microscope was granted by the M.V. Lomonosov Moscow State University Program of Development.
Author information
Authors and Affiliations
Contributions
M.E.V. constructed templates, designed and performed spFRET experiments, analyzed spFRET data, performed gel-shift and native gel experiments and wrote the manuscript; G.A.A. designed and performed computer modeling, developed a program for spFRET raw data analyses and contributed to writing the manuscript; K.S.K. designed and managed spFRET experiments; N.S.G. designed and performed fluorescence-marker gel-shift experiments, interpreted results and designed some experiments; A.K.S. designed computer modeling and wrote the manuscript; O.I.K. designed templates and analyzed the data; L.L.M. purified yFACT and some histones; T.F. wrote the manuscript and interpreted data; P.G.G. interpreted data; M.P.K. interpreted data; V.M.S. purified donor chromatin, designed experiments, interpreted results and wrote the manuscript; A.V.F. designed spFRET experiments, interpreted results and wrote the manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Integrated supplementary information
Supplementary Figure 1 Analysis of FRET efficiency in unfolded nucleosomes by molecular modeling.
On the left: Color-coded maps of predicted FRET efficiencies for molecular models of labeled nucleosomes with different degree of DNA uncoiling (see Online Methods). The red and blue areas on the color maps for all labels correspond to the structures where all three label pairs are in low or high FRET conformations, respectively. Shaded rectangles correspond to native structures, in which conformations are close to those expected from the crystal structure. On the right: Models of nucleosome conformations corresponding to positions numbered in cyan on the graphs.
Supplementary information
Supplementary Text and Figures
Supplementary Figure 1 (PDF 224 kb)
Supplementary Data Set 1
Uncropped gel images (PDF 385 kb)
Models of FACT-dependent uncoiling of nucleosomal DNA
Previously suggested models of nucleosome unfolding. Unfolding of intact nucleosomes could occur via: DNA uncoiling from an intact histone octamer, DNA uncoiling accompanied by octamer disassembly, without or with disruption of the H3:H3 dimer interface, opening of the (H3-H4) dimer-dimer interface without further DNA uncoiling. (MOV 8027 kb)
Rights and permissions
About this article
Cite this article
Valieva, M., Armeev, G., Kudryashova, K. et al. Large-scale ATP-independent nucleosome unfolding by a histone chaperone. Nat Struct Mol Biol 23, 1111–1116 (2016). https://doi.org/10.1038/nsmb.3321
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nsmb.3321
This article is cited by
-
Electron microscopy analysis of ATP-independent nucleosome unfolding by FACT
Communications Biology (2022)
-
The multifaceted histone chaperone RbAp46/48 in Plasmodium falciparum: structural insights, production, and characterization
Parasitology Research (2020)
-
The Arabidopsis histone chaperone FACT is required for stress-induced expression of anthocyanin biosynthetic genes
Plant Molecular Biology (2018)
-
Recent Perspectives on the Roles of Histone Chaperones in Transcription Regulation
Current Molecular Biology Reports (2017)