Abstract
Interferon-γ (IFN-γ) and interleukin-4 (IL-4) are key effector cytokines for the differentiation of T helper type 1 and 2 (Th1 and Th2) cells. Both cytokines induce fate-decisive transcription factors such as GATA3 and TBX21 that antagonize the polarized development of opposite phenotypes by direct regulation of each other’s expression along with many other target genes. Although it is well established that mesenchymal cells directly respond to Th1 and Th2 cytokines, the nature of antagonistic differentiation programs in airway epithelial cells is only partially understood. In this study, primary normal human bronchial epithelial cells (NHBEs) were exposed to IL-4, IFN-γ, or both and genome-wide transcriptome analysis was performed. The study uncovers an antagonistic regulation pattern of IL-4 and IFN-γ in NHBEs, translating the Th1/Th2 antagonism directly in epithelial gene regulation. IL-4- and IFN-γ-induced transcription factor hubs form clusters, present in antagonistically and polarized gene regulation networks. Furthermore, the IL-4-dependent induction of IL-24 observed in rhinitis patients was downregulated by IFN-γ, and therefore IL-24 represents a potential biomarker of allergic inflammation and a Th2 polarized condition of the epithelium.
Similar content being viewed by others
INTRODUCTION
The airway epithelium plays an important role as physical barrier to the environment, counteracts invasive microbes, and deals with harmless components such as pollen or other particles. Defense mechanisms range from apoptosis to limit intracellular infections down to passive protective reactions such as membrane reinforcement or mucus production.1 By physical interaction of T cells with the epithelium, not only defense mechanisms but also regeneration or even immunosuppressive mechanisms are induced.2 These T cells originate from a common antigen-naive precursor cell that differentiates either into T helper type 1 or 2 (Th1 or Th2) cells or other T-cell subtypes in a process of antagonistic regulation that allows the phenotypic polarization. Specifically Th1-derived interferon-γ (IFN-γ) inhibits Th2 cell differentiation, whereas Th2-derived interleukin-4 (IL-4) inhibits the fate in favor of Th1 differentiation. These key cytokines are also recognized by airway epithelial cells, but whether airway epithelial cells respond to IL-4 and IFN-γ by antagonistic gene regulation is only partially understood. The resulting polarization in transcriptional regulation of epithelial cells has been described for fate decision in pancreas cell development.3 It is known that Th2-exposed epithelial cells can contribute to asthma by enhanced mucus production,4 epithelial hyperplasia,5 and enhanced matrix production.6 Other well-known pro-allergic type 2 epithelial cytokines, such as IL-33, IL-18BP, or IL-1RL1, act on innate cells and can enhance IL-4 production.7, 8, 9
Th1 cells were previously described to play a role in multiple inflammatory airway disease such as cystic fibrosis,10 sarcoidosis,11 pulmonary tuberculosis,12 bronchiolitis obliterans,13 and hypersensitivity pneumonitis.14 We and others have previously demonstrated that IFN-γ has substantial effect on airway epithelial cells such as the induction of CXCL9, CXCL10, CXCL11, MCP1, STAT1, ICAM, IL8, MHCI, and MHCII.2, 15 Because of the chemokine induction and expression of lymphocyte relevant genes such as ICAM and MHC, it is hypothesized that Th1-primed epithelium can trigger inflammation by recruitment and support antigen-specific defense mechanisms at the epithelial surface. However, it is not clear whether Th1/Th2 programs exist in airway epithelial cells, and hence the relevance of changed ratios of lung-infiltrating Th1 and Th2 cells is not known.2 In fact, it could be speculated that the infiltration of Th1 cells in an otherwise Th2-affected epithelium is beneficial. In addition, epithelial Th2 responses could be easily diagnostically assessed in disease conditions with unclear case history such as wheezing in newborns. This unmet clinical need can be noninvasively addressed by nasal lining fluids that contain abundant amount of epithelial cytokines with biomarker potential for diagnosis of allergic inflammation such as allergic rhinitis and allergic asthma.
We previously showed that explants from allergic inflammation (skin, upper and lower airways) contain “plastic” T-cell phenotypes secreting multiple cytokines in parallel, including IL-4 and IFN-γ,2 raising the question of the functional consequence on the side of airway epithelial inflammation.
Essential for the IL-4-mediated polarization is an intact IL-4 receptor (IL-4R) downstream signaling. Airway epithelial cells do not only recognize IL-4, but also express the receptor for its structural homolog IL13. Both cytokines share the IL-4Rα and the common γ chain (γc) or the IL-13Rα chain that activates the Janus kinase/signal transducer and activator of transcription (JAK/STAT) pathways. As a consequence, IL-4 is able to bind to two receptor (type I and type II) heterodimers consisting of either γc/IL-13Rα (type I) or IL-4Rα/IL-13Rα (type II), whereas binding of IL-13 is restricted to type II.16 The receptor complex that mediates IFN-γ signaling consists of two species-matched chains: IFN-γR1 and IFN-γR2. IFN-γR1 is the major binding subunit for IFN-γ, whereas IFN-γR2 increases the affinity of IFN-γR1 and is obligatory for transducing the IFN-γ signal.17 Like the IL-4R complex, the IFN-γ receptors are expressed by airway epithelial cells and induce proinflammatory chemokines18 and MHCII expression.2
The key hypothesis of this study was that IL-4 antagonizes IFN-γ-mediated gene regulation in airway epithelial cells. Therefore, we studied whether antagonistic gene regulation prevails over synergistic gene induction along with its associated transcriptional network.
This study shows for the first time that airway epithelial cells are subjected to a transcriptional program similar to the Th1/Th2 antagonism known in lymphocytes. Notably, airway epithelial cells express several IL-4- or IFN-γ-induced genes that are antagonized by IFN-γ and IL-4 respectively.
RESULTS
To understand the pathological consequences of T-cell plasticity on the human airway tissue, we coexposed normal human bronchial epithelial cells (NHBEs) with IL-4 and IFN-γ at the same time and discovered that IL-4 and IFN-γ antagonize airway epithelial gene regulation.
IL-4 and IFN-γ antagonistic and synergistic gene regulation
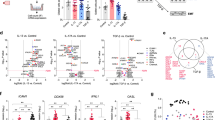
Airway epithelial cells were exposed to the cytokines alone or in combination for visualization of IL-4- and IFN-γ-induced antagonism. Although the response to IFN-γ is well established in epithelial cells, the response to IL-4 is more complex and was monitored before the whole genome transcriptome assessment. Immunocytofluorescence stainings were used as quality control to demonstrate the homogenous distribution of the IL-4Rα expression on the cell surface and cytoplasm of the majority of the cells (Supplementary Figure S1 online). In order to extract Th1/Th2-driven antagonistic gene regulation, NHBE cultures were exposed to IL-4 or IFN-γ and coexposure was additionally analyzed. The cytokine-induced signaling resulted in 1,501 entities induced by IL-4 and 2,412 entities by IFN-γ (Supplementary Figure S2, gene list in Supplementary Table S1). The Venn diagram analysis revealed the genes that are induced by IL-4 and downregulated by IFN-γ and that still show changes in the presence of both cytokines (Figure 1a, center cut set). We found that the majority of genes were downregulated following cytokine treatments (Figure 1a,c). A cut set of 48 transcripts was regulated in the presence of both IL-4 and IFN-γ (Figure 1b; gene list in Supplementary Table S2). These genes were hierarchically grouped into three clusters (clusters 1–3; Figure 1b) of which cluster 1 represents the strongest upregulated genes by IL-4 (Figure 1b). The genes highlighted in Figure 1b are the result of a hypothesis-free ranking of literature citation numbers for each gene. Thus, the most cited genes (PubMed) are ranking highest, whereas less cited are ranking lower and noncited the lowest. Consequently, less studied genes show lower ranking. Using this visualization, it is possible to get an unbiased view of the nature of the cluster on the current state of knowledge. For cluster 1, these included CCL26, ANO1 and RUNX2 as most literature-cited genes. The cluster 2 contained particularly IL-4-enhanced and IFN-γ-neutralized genes and included the transcription factor DLX5 (splice variant 1). Cluster 3 included IL-4-induced, IFN-γ-neutralized, or IFN-γ-antagonized genes containing DLX5 (splice variant 2) and MEX3B.
Interleukin-4 (IL-4) and interferon-γ (IFN-γ) antagonistic gene regulation. Normal human bronchial epithelial cells (NHBEs) were cultured in the presence of IL-4 or IFN-γ or IL-4 and IFN-γ for 6 h to study RNA whole transcriptome by array technology. (a) Comparison of 1,501 IL-4-upregulated entities, 3,782 IFN-γ-downregulated entities, and 7,264 IL-4/IFN-γ-regulated entities in a Venn diagram. A cut set of 48 antagonistic regulated entities was identified. The analysis is based on cultures of six genetically independent NHBEs of healthy donors. (b) A hierarchic clustering analysis of genes upregulated in comparison with untreated cells (medium) was performed, resulting in three different clusters. Labeled are six most cited genes in each cluster. (c) Comparison of 4,213 IL-4-downregulated entities, 2,412 IFN-γ-upregulated entities, and 7,264 IL-4/IFN-γ-regulated entities in a Venn diagram. A cut set of 70 antagonistic regulated entities was identified. (d) A hierarchic clustering analysis of genes upregulated by IFN-γ in comparison with untreated cells (medium) was performed, resulting in two different clusters. Labeled are the most cited genes in each cluster, except cluster 3 that only contained five genes. The color code indicates the abundance of transcripts ranging from low (blue) to high (red). Asterisks indicate two isoforms that were present in the analysis.
The cut set of 70 genes is covering the genes (Figure 1d; gene list in Supplementary Table S3) that were IL-4 downregulated and IFN-γ upregulated as well as regulated in the presence of IL-4 and IFN-γ at the same time. Again, these genes were hierarchically grouped into two clusters (Figure 1d, clusters 4 and 5). For cluster 4, this included IDO1 and LY96. The cluster 5 carried IFN-γ-induced and IL-4 downregulated gene expression and covered TNFAIP2 and IL22RA1.
In addition to antagonistic gene regulation, the synergistic induction of genes was investigated as well. For this purpose the cut set of genes was analyzed that are only induced in the presence of IL-4 and IFN-γ at the same time but not in the presence of only one of them. The cut set comprised 530 genes (Supplementary Figure 3a) in the case of IL-4- and IFN-γ-induced genes and 1,378 genes in the case of IL-4 and IFN-γ downregulated genes (Supplementary Figure 3c). However, a closer look reveals that both groups carry predominantly if not entirely those genes that escape the statistical thresholds of IL-4 or IFN-γ regulation, but that make it over these thresholds when an additive effect by the presence of both cytokines is generated. Among these entities are no genes with established immunoregulatory or epithelial function except SLC10A2 (sodium and bile channel) or RGS4 (G-protein regulator). Furthermore, a large number of not annotated genes were found (gene list in Supplementary Tables S4 and S5).
IL-4- and IFN-γ-induced secreted gene products
In order to extract specifically IL-4-inducible genes, all IFN-γ-regulated entities as well as all IL-4/IFN-γ-regulated entities were excluded from further analysis (Figure 2a). When IL-4-induced but not IFN-γ-regulated genes (540 entities) were filtered for specific secreted gene products, 22 entities were identified by Gene Ontology (GO) terms (Figure 2a,b; gene list in Supplementary Table S6). This group contained genes such as Wnt5a, CCL6, or IL24 (Figure 2b). The group IL-4-induced and IL-4/IFN-γ-regulated genes (368 entities, Figure 2a) contained among others TNFAIP6, CLDN5, and POSTN (gene list in Supplementary Table S7). Conversely, to extract IFN-γ-specific genes induced in airway epithelial cells, all IL-4-regulated entities as well as all IL-4/IFN-γ-regulated entities were excluded from analysis (Figure 2c). The remaining 462 IFN-γ-induced entities were filtered for secreted gene products as described above and 19 entities were identified respectively by GO terms, including CHI3L1 (YKL-40) and CCL17 (Figure 2c, d; gene list in Supplementary Table S8). The group of IFN-γ-induced and IL-4/IFN-γ-regulated genes (1,379 entities, Figure 2c) contained among others CXCL9, CXCL10, and CXCL11 (gene list in Supplementary Table S9). Several genes found to be significantly regulated by the array technology were validated real-time quantitative reverse transcription-PCR (Figure 3).
Biomarker of T helper type 1/2 (Th1/Th2) imprinted airway epithelial cells. Normal human bronchial epithelial cells (NHBEs) were cultured in the presence of interleukin-4 (IL-4) or interferon-γ (IFN-γ) or IL-4 and IFN-γ for 6 h to study RNA whole transcriptome by array technology. The analysis is based on cultures of six genetically independent NHBEs of healthy donors. (a) Comparison of 1,501 IL-4-upregulated entities, 6,219 IFN-γ-regulated entities, and 7,264 IL-4/IFN-γ-regulated entities in a Venn diagram. A cut set of 540 IL-4 only-induced entities was identified within the groups of IL-4-upregulated entities. The identified genes were listed in a ranking according to current literature using the “Genomatix” software and Gene Ontology-Terms (GO-Terms) 0007267, 0005125, 0008009, and 0005615 for identification of biomarkers resulting in a list of 22 entities. (b) Hierarchical ranking of identified IL-4-induced secreted biomarkers. Asterisks indicate two isoforms that were present in the analysis. (c) Comparison of 5,721 IL-4-regulated entities, 2,412 IFN-γ-upregulated entities, and 7,264 IL-4/IFN-γ-regulated entities in a Venn diagram. A cut set of 462 IFN-γ only induced entities was identified within the groups of IFN-γ-upregulated entities. A filter for the functions of the identified genes was listed by GO terms for identification of biomarkers resulting in a list of 22 entities. (d) Hierarchical ranking of identified IFN-γ-induced secreted biomarkers. Asterisks indicate two isoforms that were present in the analysis. The color code indicates the abundance of transcripts ranging from low (blue) to high (red).
Validation of in vitro gene expression changes. Normal human bronchial epithelial cells (NHBEs) were cultured in the presence of interleukin-4 (IL-4) or interferon-γ (IFN-γ) for 6 h to study RNA expression by real-time quantitative reverse transcription–PCR (qRT-PCR) confirming array results for ANO1, IL22RA1, IL24, and CCL26, expressed as log2 gene expression changes (ΔΔCt). The analysis is based on cultures of six genetically independent NHBEs of healthy donors. Data have been log2 transformed to get a normal distribution, and the Shapiro test for normal distribution was performed. The genes are normally distributed on a 1% significance threshold.
Transcriptional mechanisms underlying IL4 and IFN-γ antagonism in epithelial gene expression
Transcriptional antagonism and specifically the polarization of a transcriptional program are accompanied by direct counterregulation of one transcription factor against the other as it is observed for GATA3 and TBX21 or GATA3 and FOXP3.19 As a result, a cellular phenotype may successively develop into either one or the other phenotype while sparing “in-between” phenotypes. To identify transcriptional regulators in this context, IL-4- or IFN-γ-induced genes were filtered for transcriptional regulators in conjunction with either IL-4- or IFN-γ-induced genes. Respectively, 80 and 102 entities were identified by GO terms and Genomatix curated annotations (Figure 4; gene list in Supplementary Tables S10 and S11). In the group of IL-4-induced transcriptional regulators, a group of 15 entities was identified containing NFE2, GATA3, RARB, FoxQ1, and AhR (Figure 4). We therefore analyzed whether GATA3 and TBX21 could potentially account for the antagonistic gene expression. It is interesting to note that TBX21 and GATA3 are also expressed in airway epithelial cells, and IL-4Rα, STAT-6, and GATA-3 are present in a majority of cells (Supplementary Figure S1 a–c). We performed a bioinformatics analysis of all antagonized gene products and conducted a pathway analysis based on transcription factor binding site involving literature evidence as well as predicted binding sites in respective promoters (Figure 5). The resulting network visualizes that GATA3 could be interlinked with 38 genes, and TBX21 could only be connected with 10 gene promoters. However, other IL-4- or IFN-γ-induced transcription factors are frequently connected such as Meis1, Hey2 that link with 23 or 38 IL-4-induced/IFN-γ-antagonized genes, respectively, and TEAD4 connecting with 36 IFN-γ-induced/IL-4-antagonized genes (Supplementary Table S12).
Antagonistic transcriptional regulation. Normal human bronchial epithelial cells (NHBEs) were cultured in the presence of interleukin-4 (IL-4) or interferon-γ (IFN-γ) or IL-4 and IFN-γ for 6 h to study RNA whole transcriptome by array technology. The analysis is based on cultures of six genetically independent NHBEs of healthy donors. (a) Comparison of 1,501 IL-4-upregulated entities, 3,782 IFN-γ-downregulated entities, and 7,264 IL-4/IFN-γ-regulated entities in a Venn diagram. A cut set of 768 entities was identified within the groups of IL-4-upregulated and IL-4/IFN-γ-regulated entities. The identified genes were listed in a ranking according to current literature using the "Genomatix" software and Gene Ontology-Terms (GO-Terms) 0003676 and 0044212 for transcription factors resulting in a list of 80 entities. (b) Hierarchical ranking of identified IL-4-induced transcription factors, containing IL-4-master transcription factor GATA3. (c) Comparison of 4,213 IL-4-downregulated entities, 2,412 IFN-γ-upregulated entities, and 7,264 IL-4/IFN-γ-regulated entities in a Venn diagram. A cut set of 1,779 entities was identified within the groups of IL-4-upregulated and IL-4/IFN-γ-regulated entities. (d) Hierarchical ranking of identified IFN-γ-induced transcription factors, containing IFN-γ-master transcription factor TBX21. The color code indicates the abundance of transcripts ranging from low (blue) to high (red).
Schematic illustration of the transcriptional network. Transcriptional network of interleukin-4 (IL-4; left)- and interferon-γ (IFN-γ)-induced transcription factors (right) and genes of the IL-4/IFN-γ antagonism of epithelial cells. The network connects genes that are known to be transcriptionally regulated or that contain transcription factor binding sites (Genomatix database) of the respective factor. Green lines originate from IL-4- and red lines from IFN-γ-induced transcription factors. Black lines show intertranscription factor connections. The figure shows numerous intertranscription factor lines that may be critical for polarized gene regulation. GATA3 appears to be connected to many type 2, and also several type 1 genes. On the side of IFN-γ-induced transcription factors it appears that TEAD4, MEF2C, and ZEB1 have hub character, whereas TBX21 is connected with only a few genes.
Th1/Th2-specific gene expression changes in vivo
In order to show the physiologic relevance of identified genes under well-known Th2 conditions during allergic inflammation in nasal mucosa, allergic subjects sensitized for grass pollen and healthy controls were recruited. Nasal airway epithelial cells were obtained through nasal scrapings from the inferior turbinates. We refrained to obtain bronchial cells for ethical reasons and instead used nasal epithelial cells (scrapings) on the basis of previous studies showing comparable regulation patterns of epithelial cells derived from upper vs. lower airways.20 The scrapings were subjected to RNA isolation and real-time quantitative reverse transcription–PCR was performed for the prominent targets, in particular those that are secreted, and those known to be involved in transcriptional regulation in type 2 differentiation and disease: IL7, IL24, IL18BP, IL22RA1, IL33, ANO1, IL1RL1 (sST2; IL33R), AhR, GATA3, TBX21, and RUNX2 (Figure 6). Significant increased gene expression changes in allergic subjects in comparison with healthy control subjects were shown for IL7 (log2 expression change: 1.98, 1.57 to 5.33; P<0.05), IL24 (log2 expression change: 3.09, −1.12 to 4.02; P<0.05), ANO-1 (log2 expression change: 1.56, −0.18 to 3.79; P<0.01), IL18BP (log2 expression change: 2.41, 1.84 to 3.23; P<0.05), IL1RL1 (sST2, IL33R; log2 expression change: 2.31, −0.42 to 4.78; P<0.05), IL22RA1 (log2 expression change: 1.02, −0.27 to 4.20; P<0.05), AhR (log2 expression change: 1.08, 0.54 to 2.01; P<0.05), GATA3 (log2 expression change: 1.64, 0.67 to 3.18; P<0.05), and RUNX2 (log2 expression change: 1.35, 0.99 to 2.45; P<0.05). A trend for a decreased expression of TBX21 was discovered in allergic subjects (log2 expression change: −1.33, −3.47 to 0.92) compared with healthy control subjects. No significant changes were shown for the expression of IL33 in allergic subjects (log2 expression change: 1.57, 0.24 to 4.12) in comparison with healthy controls.
Gene expression levels for selected transcripts. Nasal scrapings of asthmatic patients (n=8) as well as healthy controls (n=12) were subjected to RNA isolation and real-time quantitative reverse transcription–PCR (qRT-PCR) was performed. Gene expression changes (ΔΔCt) have been log2-transformed to get a normal distribution. Significant differences could be shown for IL7, IL24, ANO1, IL18BP, IL1RL1 (sST2; IL33R), IL22RA1, AhR, GATA3, and RUNX2.
Biomarker of Th2-primed epithelial cells
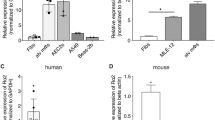
Among the identified IL-4-induced, secreted gene products, IL-24 has to be highlighted as potential Th2 biomarker, as this protein belongs to the IL-10 superfamily. Currently, it is not described to be regulated by other epithelial activation processes such as IL8 that is not only induced by IL-4 but also by IL-17. Western blot analysis was performed for IL-24 in supernatants of in vitro culture of NHBEs after stimulation with IL-4, IFN-γ, and IL-4/IFN-γ for 6 and 24 h (Figure 7a). IL-24 was detected at 6 and 24 h in IL-4-stimulated samples but not in medium control or IFN-γ-subjected samples. As epithelial cells are able to secrete cytokines into the airway lumen, we hypothesized that IL-24 could also be detectable in nasal secretions. Therefore, secreted protein levels of nasal secretions of allergic asthma patients collected in and out of season were measured by electrochemiluminescence detection for IL-24 and compared with healthy controls. A significantly upregulated secretion of IL-24 was detected for patients in season (121.70 pg ml−1, 61.91–242.80; P<0.01) in comparison with healthy controls (49.33 pg ml−1, 31.98–71.13), whereas only a minor, nonsignificant increase of IL-24 was observed in the out-of-season group (93.85 pg ml−1, 28.15–145.40) compared with healthy controls (Figure 7b).
Interleukin-24 (IL-24) protein secretion in vitro and in vivo. (a) A signal for IL-24 secretion in vitro was detected by western blot analysis after 6 and 24 h of stimulation with IL-4 but not for interferon-γ (IFN-γ) or IL-4/IFN-γ cultivation of normal human bronchial epithelial cells (NHBEs). There was no detectable signal in medium controls. (b) Significantly upregulated secretion of IL-24 of patients in season (n=13) in comparison with healthy controls (n=14) was detected. An increased secretion of IL-24 could be detected for both patient groups compared with healthy controls.
DISCUSSION
The objective of this study was to investigate whether IL-4 and IFN-γ are subject to antagonistic gene regulation in airway epithelial cells. The study reveals a large number of IL-4-induced and IFN-γ-inhibited genes. In fact, IL-4-inducible genes of cluster 1 such as CCL26 (Eotaxin-3), ANO1, or RUNX2 are dampened in the presence of IFN-γ. The example of CCL26 is not only reproducing previous results that demonstrated that CCL-26 is an IL-4-inducible, proinflammatory chemokine in allergic rhinitis,21 but also that epithelial chemokines can act as biomarkers for bullous pemphigoid and asthma with eosinophilic infiltration into the airways.21, 22 ANO1 represents another example that has a major impact on basal and stimulated chloride conductance in airway epithelial cells23 and serves as an indicator for epithelial proliferation and migration in the repair processes.24 As this study shows that IFN-γ inhibits CCL-26-mediated chemotaxis and chloride conductance, it can be proposed that IFN-γ may ameliorate eosinophilic infiltration and tracheal cartilage deposition as well as repair processes. This study reveals 46 additional IL-4-inducible and IFN-γ-inhibited genes that are yet not known to be under control of this IL-4–IFN-γ antagonism. Most of these genes have not been implicated in Th2-driven diseases, except Calpain-14, a calcium-sensitive protease that together with TSLP is genetically associated with eosinophilic esophagitis.25
Conversely, genes of clusters 4 and 5 such as IDO1 or IL22RA1 were previously reported to be IFN-γ inducible and are downregulated by IL-4. The IL-4-mediated suppression of IDO1, a potential mediator of tolerance, was previously observed in house dust mite-stimulated dendritic cell cultures.26 Similarly, IL22RA1 has been shown to be related to potentially tolerogenic downregulation of MHCI and MHCII in airway epithelial cells, and thus IL-4-mediated downregulation can also contribute to break tolerance.2, 27 Taken together, it appears that important genes are subject to antagonistic regulation as it is known from T-cell differentiation and further substantiate the concept of IL-4 as tolerance breaking mediator that was previously demonstrated on the level of T cells.19 Future studies are necessary to show that IL-4 counteracts tolerance also at the level of epithelial surfaces.
In contrast, the search for genes synergistically induced by IL-4 and IFN-γ, but not by isolated cytokines, did not reveal strongly induced clusters. However, it is clear that there are numerous genes that are insensitive to antagonistic regulation. These nonantagonized genes are particularly interesting, as they may provide robust indicators for Th2-mediated immune responses, even if other, IFN-γ-inducing inflammatory processes are disguising the pathology. Narrowing the view on genes that are secreted and detectable in secretions, 22 IL-4-induced genes were identified, including CSF3, CXCL6, IL24, and WNT5a. In addition, there is a small group of genes that is IL-4 induced and survives the presence of IFN-γ in the co-culture situation, such as TNFAIP6, CLDN5, and POSTN, and all of these genes are known to be IL-4 regulated.28, 29, 30 Conversely, IFN-γ induced 19 genes with no influence of IL-4 such as CHI3L1 (Chitinase-3-Like Protein 1 or YKL-40) that is known to be induced by IFN-γ nasal epithelial cells.31 Of note, there is also a considerable number of genes that are induced by IFN-γ alone and regulated by the combination of IL-4 and IFN-γ. In this group, IFN-γ dominantly induces well-known IFN-γ-responsive genes such as CXCL9, CXCL10, and CXCL11, whereas IL-4 has a neglectable effect.32
The physiological relevance of these antagonized and nonantagonized genes in human disease conditions was assessed in nasal scrapings of in-season allergic rhinitis patients and healthy subjects. On the basis of previous studies showing comparable regulation patterns of epithelial cells derived from upper vs. lower airways,20 we anticipate that nasal scrapings containing mainly nasal epithelial cells provide an ethically appropriate surrogate biomarker source for validation of the targets identified in bronchial epithelial cells. IL24 and ANO1, the latter a biomarker known from cystic fibrosis, are showing distinct expression in allergic patients during the allergy season but not in healthy individuals. In addition, the secreted protein levels of IL-24 showed a significant increase for patients in season compared with healthy controls, whereas this difference is lost out of the allergen season. Current studies are initiated to validate the IL-10 family member IL-24 in large cohorts and may provide assistance to physicians to use immunomodulatory drugs in the treatment of rhinitis in anticipation of a Th2 response, for example, in wheezing newborns or in conditions where the allergic impact is not certain.
The antagonistic nature of the IL-4 and IFN-γ has been shown to depend on the crossregulation of TBX21 and GATA3 in lymphocyte differentiation. Both genes were previously detected in airway epithelial cells33 and confirmed to be homogenously expressed in NHBEs in this study along with other essential signaling components IL4Rα and STAT6 (Supplementary Information and data not shown, respectively). Based on this finding we considered that a transcription network is underlying epithelial response profile of GATA3 and TBX21. Strikingly, all IL-4-inducible, GATA3-co-clustered genes were downregulated by IFN-γ. RUNX2 was previously described to be downregulated by IFN-γ in mesenchymal stem cells, whereas its inducibility by IL-4 is unknown so far.34 As RUNX2 enhances TGF-β-induced SMAD signaling, it appears possible that IL-4 induction of RUNX2 promotes airway remodeling.35 IL-4 signaling was also shown to synergize with TGF-β signaling that is enhancing the IL-4-inducible FOXQ1 in alternatively (IL-4-) activated macrophages, also defined as “hidden type 2 inflammation”.36
The IL-4-mediated induction of the Aryl hydrocarbon receptor (AhR) is confirming previous studies in B cells37 and builds the bridge to the key function of AhR in inducing IL22 (ref. 38, 39) that in turn promotes epithelial repair mechanisms.40, 41 Furthermore, AhR is inhibiting GATA3,42 and this could not only create a negative feedback loop, but also allow educt inhibition by IFN-γ-induced IDO metabolites such as kynurenine.38, 39, 43
Besides the connection of GATA3 to the AhR, GATA3 interconnects with many other IL-4- and IFN-γ-regulated genes, highlighting GATA3 as hub gene, as it is known from the polarization network of T cells. TBX21, which is an equally important hub gene in T cells, is less interconnected in airway epithelial cells. In addition, the network analysis highlights that IL-4 also regulates hub genes Meis1 and Hey2 that are involved in transcriptional regulation of many genes, being a cofactor for HOX genes44 and HEY2 as a Notch-downstream target,42 that in turn is known to regulate airway epithelial differentiation.43, 45 Future studies will have to confirm the role of these hub genes in the polarization of epithelial cells in Th2-driven diseases and airway remodeling.
Taken together, this study suggests that type 1-primed and type 2-primed airway epithelial cells exist and are pre-commissioned to respond along proallergic pathology. Future studies will need to define whether this behavior is epigenetically imprinted as it is the case for T cells and whether this knowledge can be used to exit proallergic disease conditions.
METHODS
Patients. Healthy subjects (n=12) and allergic patients in season (n=8) aged between 20 and 65 years in good health (forced expiratory volume in one second 1% >70%) with a history of clinically significant hay fever during the grass-pollen season since more than 2 years were included. Nasal scrapings were performed in the Allergy Section, Department of Otolaryngology, TUM School of Medicine (Munich, Germany). In addition, nasal secretions from healthy controls (n=14) and allergic and asthmatic patients in and out of season (n=13) were collected as previously described.46 The full list of inclusion and exclusion criteria is available in the Supplementary Information. Each participant provided written informed consent. The study was approved by the local ethics committee.
Cell culture. Primary NHBEs (Lonza, Walkersville, MD) of six genetically independent donors were grown as monolayers in 100% humidity and 5% CO2 at 37 °C in serum-free defined growth media (BEGM, Lonza). NHBEs (passage 3) were used at ∼80% confluence in 6-well plates. To avoid gene expression changes or influences on the IL-4 or IFN-γ signaling induced by growth factors in the BEGM medium, cells were rested in basal medium (BEBM) for 12 h, then stimulated with recombinant human IL-4 at 50 ng ml−1 (R&D Systems, Minneapolis, MN) and human IFN-γ at 10 ng ml−1 (R&D Systems) in BEBM medium for 6 h at the indicated concentrations or medium alone or the same-time presence of both cytokines, IL-4 at 50 ng ml−1 and IFN-γ at 10 ng ml−1. For RNA analysis, harvested cells were lysed in RLT buffer (Qiagen, Hilden, Germany) containing 1% β-mercaptoethanol (Roth, Karlsruhe, Germany) directly in the cell culture well. For western blot analysis, cells were cultured in the same conditions, but for 6 h and an additional 24 h time point.
RNA isolation and whole genome microarray. Total RNA was extracted using RNeasy Mini Kit (Qiagen) with on-column DNase digestion (Qiagen) for avoiding DNA contaminations. RNA quantification and quality assessments were performed by ultraviolet–visible spectrophotometry (Nanodrop Technologies, Wilmington, DE) and the RNA 6000 Nano Chip Kit with the Agilent 2100 Bioanalyzer (Agilent Technologies, Waldbronn, Germany). The RNA quality of all samples reached a RNA integrity number (developed by Agilent Technologies) of 9.1. Total RNA was amplified and Cy3 labeled by using the one-color Low Input Quick Amp Labeling Kit (Agilent Technologies) according to the manufacturer’s protocol. Hybridization to SurePrint G3 Human Gene Expression 8x60K Microarrays (Agilent Technologies) was performed with the Gene Expression Hybridization Kit (Agilent Technologies).
Data analysis strategy. Data analysis was performed using the Genespring Software GX 12.5 (Agilent Technologies) under minimal data reduction constraints (1.2-fold change and P≤0.05 cutoff). Upon data import a standard baseline transformation to the median of all values was performed, including log transformation and computation of fold changes (log2(A/B)=log2(A)−log2(B)). Subsequently, a principle component analysis was conducted and revealed a homogenous component distribution. Compromised array signals (array spot is nonuniform if pixel noise of feature exceeds threshold or above saturation threshold) were excluded from further analysis. These threshold settings were verified looking at genes known to be regulated by the stimuli, e.g., HLADRs for IFN-γ or CCL-26 for IL-4.37 Genes regulated more than 1.2-fold were further analyzed by using the paired Student’s t-test and filtered for P-value (P<0.05) that represents a rather permissive threshold. The significantly regulated genes were summarized in entity lists (see Supplementary Tables). These entity lists were analyzed for overlaps using Venn diagrams. Manhattan cityblock on entities (Ward’s linkage) was used to cluster changes in gene expression. The functions of the identified genes were listed according to current literature using the Genomatix software (Munich, Germany) and GO terms 0003676 and 0044212 for transcription factors and GO terms 0007267, 0005125, 0008009, and 0005615 for identification of biomarkers. The gene expression network was generated using the Genomatix Pathway System (GePS) that integrates information extracted from public and proprietary databases to display canonical pathways or to create and extend networks based on literature data. Data displayed are focusing on binding sites of transcription factor families that were more than twofold regulated by IL-4 or IFN-γ extracted from the transcriptome data sets.
Reverse transcription and quantitative real-time PCR. Isolated total RNA was subjected to reverse transcription using a high-capacity cDNA kit (Applied Biosystems, Foster City, CA) following the manufacturer’s instructions. Real-time PCR profiles were visualized using the commercially available FastStart Universal SYBR Green Mastermix (Roche, Basel, Suisse) and quantified by the ViiA 7 Real-Time PCR System (Applied Biosystems). The specific primers used in the real-time PCR are listed in Supplementary Table S13. The amount of ANO-1, IL-22RA1, IL-24, and CCL-26 mRNA expression was normalized with endogenous control 18S, GAPDH, and β-actin (housekeeping gene index, ΔCt values) and the relative quantification and calculation of range of confidence was performed using the comparative threshold cycle (2−ΔΔCt) method (relative gene expression) as previously described.2 All amplifications were carried out at least in duplicate.
Protein measurement by multi-array technology. Protein levels of IL-24 in nasal secretions were detected in duplicates by MesoScaleDiscovery’s (MSD) Multi-Array technology (MSD Mesoscale, Rockville, MD), a plate-based technology that enables the detection of biomarkers utilizing an electrochemiluminescence detection.
Western blot analysis. A Bradford assay was used to quantify amounts of protein concentrations of NHBE culture supernatants to ensure that equal amounts were loaded per lane of the sodium dodecyl sulfate–polyacrylamide gel electrophoresis (for details see Supplementary Information).
Immunofluorescent staining of NHBEs. For immunofluorescent staining of NHBEs of three genetically independent donors, 15 × 104 cells were cultured on a sterilized, removable 12-well chamber (ibidi GmbH, Martinsried, Germany). At confluency of 50–60%, cells were fixed for 10 min with 4% paraformaldehyde at room temperature, followed by fixation with methanol, prechilled at −20 °C and washed three times with phosphate-buffered saline. The primary antibody was diluted as 1:250 (IL-4Rα, R&D, Abdingdon, UK) and added overnight at 4 °C in a humid chamber. When incubation was finished, cells were washed again three times with phosphate-buffered saline and incubated with diluted secondary antibody (goat anti-mouse AlexaFluor 488, 1:300 (Life Technologies, Waltham, MA) with added DAPI D1306 (Life Technologies) for 90 min at room temperature in a humid chamber. After incubation, cells were washed three times and slides were covered with coverslips (Schott glass, Mainz, Germany) using VectaShield Mounting Medium (Vector Laboratories, Burlingame, CA). Slides were analyzed with LEICA confocal microscope (LEICA, Wetzlar, Germany).
Statistical analysis. Expression data of in vitro and in vivo gene expression have been log2 transformed. One-sample Wilcoxon rank-sum tests were used to determine significant changes in expression of in vitro samples of the real-time PCR analysis (R Statistical Programming Language, Vienna, Austria). For in vivo data, two-sample Wilcoxon-rank sum tests were used to test for significant differences between control and patient samples. Samples of each group are summarized taking the median. The fold change has been computed to get absolute differences. Mann–Whitney tests were used to determine significant changes of protein levels in nasal secretions. Statistically significant differences were defined as P values *P<0.05, **P<0.01, and ***P<0.001.
References
Holgate, S.T. Epithelium dysfunction in asthma. J. Allergy Clin. Immunol. 120, 1233–1244 (2007).
Pennino, D. et al. IL-22 suppresses IFN-gamma-mediated lung inflammation in asthmatic patients. J. Allergy Clin. Immunol. 131, 562–570 (2013).
Schaffer, A.E., Freude, K.K., Nelson, S.B. & Sander, M. Nkx6 transcription factors and Ptf1a function as antagonistic lineage determinants in multipotent pancreatic progenitors. Dev. Cell. 18, 1022–1029 (2010).
Cohn, L., Homer, R.J., Marinov, A., Rankin, J. & Bottomly, K. Induction of airway mucus production By T helper 2 (Th2) cells: a critical role for interleukin 4 in cell recruitment but not mucus production. J. Exp. Med. 186, 1737–1747 (1997).
Hurst, S.D. et al. New IL-17 family members promote Th1 or Th2 responses in the lung: in vivo function of the novel cytokine IL-25. J. Immunol. 169, 443–453 (2002).
Liu, X. et al. Th2 cytokine regulation of type I collagen gel contraction mediated by human lung mesenchymal cells. Am. J. Physiol. Lung Cell. Mol. Physiol. 282, L1049–L1056 (2002).
Akhabir, L. et al. Lung expression quantitative trait loci data set identifies important functional polymorphisms in the asthma-associated IL1RL1 region. J. Allergy Clin. Immunol. 134, 729–731 (2014).
Kamekura, R. et al. The role of IL-33 and its receptor ST2 in human nasal epithelium with allergic rhinitis. Clin. Exp. Allergy 42, 218–228 (2012).
Saglani, S. et al. IL-33 promotes airway remodeling in pediatric patients with severe steroid-resistant asthma. J. Allergy Clin. Immunol. 132, 676–685 (2013).
Eickmeier, O. et al. Sputum biomarker profiles in cystic fibrosis (CF) and chronic obstructive pulmonary disease (COPD) and association between pulmonary function. Cytokine 50, 152–157 (2010).
Mollers, M. et al. Intracellular cytokine repertoire in different T cell subsets from patients with sarcoidosis. Thorax 56, 487–493 (2001).
Lopez-Maderuelo, D. et al. Interferon-gamma and interleukin-10 gene polymorphisms in pulmonary tuberculosis. Am. J. Respir. Crit. Care Med. 167, 970–975 (2003).
Rosewich, M. et al. Airway inflammation in children and adolescents with bronchiolitis obliterans. Cytokine 73, 156–162 (2015).
Yamasaki, H., Ando, M., Brazer, W., Center, D.M. & Cruikshank, W.W. Polarized type 1 cytokine profile in bronchoalveolar lavage T cells of patients with hypersensitivity pneumonitis. J. Immunol. 163, 3516–3523 (1999).
Pawliczak, R. et al. Influence of IFN-gamma on gene expression in normal human bronchial epithelial cells: modulation of IFN-gamma effects by dexamethasone. Physiol. Genomics 23, 28–45 (2005).
LaPorte, S.L. et al. Molecular and structural basis of cytokine receptor pleiotropy in the interleukin-4/13 system. Cell 132, 259–272 (2008).
Tau, G. & Rothman, P. Biologic functions of the IFN-gamma receptors. Allergy 54, 1233–1251 (1999).
Konno, S. et al. Interferon-gamma enhances rhinovirus-induced RANTES secretion by airway epithelial cells. Am. J. Respir. Cell Mol. Biol. 26, 594–601 (2002).
Mantel, P.Y. et al. GATA3-driven Th2 responses inhibit TGF-beta1-induced FOXP3 expression and the formation of regulatory T cells. PLoS Biol. 5, e329 (2007).
Braunstahl, G.J. et al. Nasal allergen provocation induces adhesion molecule expression and tissue eosinophilia in upper and lower airways. J. Allergy Clin. Immunol. 107, 469–476 (2001).
Baumann, R. et al. Comparison of the nasal release of IL-4, IL-10, IL-17, CCL13/MCP-4, and CCL26/eotaxin-3 in allergic rhinitis during season and after allergen challenge. Am. J. Rhinol. Allergy 27, 266–272 (2013).
Gunther, C., Wozel, G., Meurer, M. & Pfeiffer, C. Up-regulation of CCL11 and CCL26 is associated with activated eosinophils in bullous pemphigoid. Clin. Exp. Immunol. 166, 145–153 (2011).
Jang, Y. & Oh, U. Anoctamin 1 in secretory epithelia. Cell Calcium 55, 355–361 (2014).
Ruffin, M. et al. Anoctamin 1 dysregulation alters bronchial epithelial repair in cystic fibrosis. Biochim. Biophys. Acta 1832, 2340–2351 (2013).
Rothenberg, M.E. Molecular, genetic, and cellular bases for treating eosinophilic esophagitis. Gastroenterology 148, 1143–1157 (2015).
Maneechotesuwan, K. et al. Der p 1 suppresses indoleamine 2, 3-dioxygenase in dendritic cells from house dust mite-sensitive patients with asthma. J. Allergy Clin. Immunol. 123, 239–248 (2009).
Schmidt-Weber, C.B. Th1 7 and treg cells innovate the TH1/TH2 concept and allergy research. Chem. Immunol. Allergy 94, 1–7 (2008).
Ueta, M., Mizushima, K., Yokoi, N., Naito, Y. & Kinoshita, S. Expression of the interleukin-4 receptor alpha in human conjunctival epithelial cells. Br. J. Ophthalmol. 94, 1239–1243 (2010).
Dalmasso, A.P. et al. Interleukin-4 induces up-regulation of endothelial cell claudin-5 through activation of FoxO1: role in protection from complement-mediated injury. J. Biol. Chem. 289, 838–847 (2014).
Takayama, G. et al. Periostin: a novel component of subepithelial fibrosis of bronchial asthma downstream of IL-4 and IL-13 signals. J. Allergy Clin. Immunol. 118, 98–104 (2006).
Park, S.J. et al. Increased expression of YKL-40 in mild and moderate/severe persistent allergic rhinitis and its possible contribution to remodeling of nasal mucosa. Am. J. Rhinol. Allergy 27, 372–380 (2013).
Sauty, A. et al. The T cell-specific CXC chemokines IP-10, Mig, and I-TAC are expressed by activated human bronchial epithelial cells. J. Immunol. 162, 3549–3558 (1999).
Chou, J., Provot, S. & Werb, Z. GATA3 in development and cancer differentiation: cells GATA have it!. J. Cell. Physiol. 222, 42–49 (2010).
Duque, G. et al. Autocrine regulation of interferon gamma in mesenchymal stem cells plays a role in early osteoblastogenesis. Stem Cells 27, 550–558 (2009).
Zhang, X. et al. Runx2-Smad signaling impacts the progression of tumor-induced bone disease. Int. J. Cancer 136, 1321–1332 (2015).
Gratchev, A. et al. Novel monocyte biomarkers of atherogenic conditions. Curr. Pharm. Des. 19, 5859–5864 (2013).
Tanaka, G. et al. Induction and activation of the aryl hydrocarbon receptor by IL-4 in B cells. Int. Immunol. 17, 797–805 (2005).
Brembilla, N.C. et al. In vivo dioxin favors interleukin-22 production by human CD4+ T cells in an aryl hydrocarbon receptor (AhR)-dependent manner. PLoS One 6, e18741 (2011).
Zelante, T. et al. Tryptophan catabolites from microbiota engage aryl hydrocarbon receptor and balance mucosal reactivity via interleukin-22. Immunity 39, 372–385 (2013).
Eyerich, S., Eyerich, K., Cavani, A. & Schmidt-Weber, C. IL-17 and IL-22: siblings, not twins. Trends Immunol. 31, 354–361 (2010).
Eyerich, K. & Eyerich, S. Th22 cells in allergic disease. Allergo J. Int. 24, 1–7 (2015).
Negishi, T. et al. Effects of aryl hydrocarbon receptor signaling on the modulation of TH1/TH2 balance. J. Immunol. 175, 7348–7356 (2005).
Opitz, C.A. et al. An endogenous tumour-promoting ligand of the human aryl hydrocarbon receptor. Nature 478, 197–203 (2011).
Bomgardner, D., Hinton, B.T. & Turner, T.T. 5' hox genes and meis 1, a hox-DNA binding cofactor, are expressed in the adult mouse epididymis. Biol. Reprod. 68, 644–650 (2003).
Niessen, K. & Karsan, A. Notch signaling in cardiac development. Circ. Res. 102, 1169–1181 (2008).
Dumitru, A.F. et al. Petasol butenoate complex (Ze 339) relieves allergic rhinitis-induced nasal obstruction more effectively than desloratadine. J. Allergy Clin. Immunol. 127, 1515–1521 (2011).
Acknowledgements
This study was funded by the German Center for Lung Research (DZL).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
CSW received support for research projects from PLS Design, LETI, Zeller AG and Allergopharma and accepted honoraria for consultancy and seminars from LETI, Allergopharma and Leo Pharma.
Additional information
SUPPLEMENTARY MATERIAL is linked to the online version of the paper
Rights and permissions
About this article
Cite this article
Zissler, U., Chaker, A., Effner, R. et al. Interleukin-4 and interferon-γ orchestrate an epithelial polarization in the airways. Mucosal Immunol 9, 917–926 (2016). https://doi.org/10.1038/mi.2015.110
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/mi.2015.110
This article is cited by
-
Interferons as negative regulators of ILC2s in allergic lung inflammation and respiratory viral infections
Journal of Molecular Medicine (2023)
-
TAFA4-IL-10 axis potentiate immunotherapy for airway allergy by induction of specific regulatory T cells
npj Vaccines (2022)