Abstract
Next-generation sequencing (NGS) technologies have enabled the application of broad-scale sequencing in microbial biodiversity and metagenome studies. Biodiversity is usually targeted by classifying 16S ribosomal RNA genes, while metagenomic approaches target metabolic genes. However, both approaches remain isolated, as long as the taxonomic and functional information cannot be interrelated. Techniques like self-organizing maps (SOMs) have been applied to cluster metagenomes into taxon-specific bins in order to link biodiversity with functions, but have not been applied to broad-scale NGS-based metagenomics yet. Here, we provide a novel implementation, demonstrate its potential and practicability, and provide a web-based service for public usage. Evaluation with published data sets mimicking varyingly complex habitats resulted into classification specificities and sensitivities of close to 100% to above 90% from phylum to genus level for assemblies exceeding 8 kb for low and medium complexity data. When applied to five real-world metagenomes of medium complexity from direct pyrosequencing of marine subsurface waters, classifications of assemblies above 2.5 kb were in good agreement with fluorescence in situ hybridizations, indicating that biodiversity was mostly retained within the metagenomes, and confirming high classification specificities. This was validated by two protein-based classifications (PBCs) methods. SOMs were able to retrieve the relevant taxa down to the genus level, while surpassing PBCs in resolution. In order to make the approach accessible to a broad audience, we implemented a feature-rich web-based SOM application named TaxSOM, which is freely available at http://www.megx.net/toolbox/taxsom. TaxSOM can classify reads or assemblies exceeding 2.5 kb with high accuracy and thus assists in linking biodiversity and functions in metagenome studies, which is a precondition to study microbial ecology in a holistic fashion.
Similar content being viewed by others
Introduction
The launch of next-generation sequencing (NGS) was nothing less than a paradigm shift in environmental molecular microbiology. The dramatic drop in sequencing costs that followed has resulted in an unprecedented rate of growth in microbial genome sequences. This development has spurred the establishment of sequencing initiatives aiming to explore the realm of microbial genomes in more targeted ways than before, for example, by focusing on specific habitats or taxa. For example, the ‘Marine Microbiology Initiative’ of the Gordon and Betty Moore foundation has contributed almost 200 draft genomes from marine habitats, and the ‘Genomic Encyclopedia for Bacteria and Archaea’ project of the Joint Genome Institute and the German Collection of Microorganisms and Cell Cultures (DSMZ) has begun to systematically fill the remaining gaps in the prokaryotic branches of the tree of life by aiming to sequence at least one representative from all clades (Wu et al., 2009). The introduction of NGS has also propelled metagenomic community-sequencing approaches, which led to dedicated initiatives as well. For marine habitats, the ‘International Census of Marine Microbes’ is focusing on extending microbial biodiversity knowledge by the large-scale sequencing of 16S ribosomal RNA V6 hyper-variable regions (Sogin et al., 2006; Huse et al., 2008), while integration of the wealth of metagenomic data from different sources is at focus of the ‘Community Cyberinfrastructure for Advanced Marine Microbial Ecology Research and Analysis’ (Camera) project (Seshadri et al., 2007). Similar data integration projects have been established in the field of medical microbiology, such as the NIH Human Microbiome Project (Peterson et al., 2009). Only a few years after the introduction of NGS, our picture of the microbial world is already becoming much clearer.
In spite of the advancements in DNA sequencing, currently available technologies still restrict low-cost full genome sequencing to cultivable strains. This requirement severely limits the application of NGS technologies to microbial biodiversity studies, because only a minor fraction (typically <1%) of the microbial species in a given habitat can be cultivated with current techniques (Amann et al., 1995; Huber et al., 2007). It is anticipated that progress in single-cell isolation techniques (Ochman, 2007) and single molecule sequencing (Gupta, 2008; Clarke et al., 2009; Eid et al., 2009) will soon overcome this limitation. For now, however, metagenomics the sequencing of DNA from an environmental sample without previous species separation or cultivation, is the method of choice for obtaining longer fragments from the genomes of the vast majority of as-yet uncultured microorganisms.
In the classical metagenome approach, genomic libraries are constructed by cloning fragmented environmental DNA into vectors that are subsequently amplified in ultra-competent host cells (Schloss and Handelsman, 2003). Once a metagenome library is constructed, it can be screened for inserts carrying specific genes or metabolic activities. These strategies have been termed sequence- and function-driven approaches (Schloss and Handelsman, 2003) and are used to select dedicated inserts from the library for full-length sequencing. These approaches, however, have the inherent disadvantage of limiting obtainable sequence information to the few genes adjacent to the respective target genes.
With the advent of NGS it has become feasible to omit the cloning step and sequence environmental DNA directly. In particular, if the target organism is in high abundance or even dominates a habitat, the sheer power of NGS allows for obtaining longer genomic fragments by direct sequencing and assembly of extracted environmental DNA. In contrast, direct DNA sequencing of habitats with high overall biodiversity or low-abundance target species mostly yields sequences harboring partial or single genes and relatively few longer assemblies with multiple genes. When a specific microorganism or function is desired, therefore, the classical metagenome approach is still much more favorable. If, however, community function in low to medium biodiverse habitats as a whole is at focus, then direct sequencing is a viable approach. Although brute-force direct sequencing of such microbial communities does not yield individual genomes, it often yields longer assemblies of the most abundant species and a wealth of sequences that can be taxonomically clustered into bins (taxobins) and subsequently mined for functions. This approach requires, of course, methods that allow these sequences to be taxonomically classified with reasonable accuracy.
In general, taxonomic classification of metagenomic DNA fragments can be achieved either on the level of the encoded genes or on the level of the DNA sequence themselves.
An introduction to gene-level taxonomic classification is beyond the scope of this article. In brief, they are either based on the post-processing of BLASTP (Altschul et al., 1990) searches as in Phylogena (Hanekamp et al., 2007) or MEGAN (Huson et al., 2007), or on the post-processing of Pfam searches (Sonnhammer et al., 1997) as in CARMA (Krause et al., 2008).
Taxonomic classification of DNA sequences on the level of base composition is still unintuitive to many biologists. However, not only the genes but also the DNA itself—including non-coding regions—is subjected to various evolutionary forces (Karlin et al., 1998), like species-specific codon preference, constraints because of DNA superstructure and G+C content maintenance, and biases that are introduced by the replication machinery. As a result, DNA carries a fingerprint-like species-specific signature in its base composition that is most pronounced in the patterns of statistical over- and underrepresentation of short oligonucleotides from tetra- to hexanucleotides (McHardy et al., 2007). As the factors that give rise to these fingerprints are inheritable, they also carry a detectable albeit weak phylogenetic signal (Pride et al., 2003). The first work on genomic DNA signatures dates back to well before the genomic area started with the sequencing of the first complete bacterial genome (Fleischmann et al., 1995) and was pioneered among others by Samuel Karlin et al. (Burge et al., 1992). At first, scientists have investigated this phenomenon with rather simplistic methods like dinucleotide or tetranucleotide relative abundances (Karlin and Ladunga, 1994; Karlin et al., 1994, 1998; Karlin and Burge, 1995; Karlin, 1998). Later, however, a whole variety of different methods have been applied to oligonucleotide signatures, such as Markov models (Rocha et al., 1998; Pride et al., 2003; Reva and Tümmler, 2004; Teeling et al., 2004), frequency chaos game representations (Deschavanne et al., 1999) and Bayesian classifiers (Sandberg et al., 2001). More recently, machine-learning algorithms have been applied to the task. These can be subdivided into supervised algorithms like support vector machines, and unsupervised algorithms like kernelized nearest-neighbor approaches and self-organizing maps (SOMs). Support vector machines have been used in PhyloPhythia (McHardy et al., 2007), a kernelized nearest-neighbor approach in TACOA (Diaz et al., 2009) and SOMs have been used in a variety of different variants, like batch-learning SOMs (BLSOMs) (Abe et al., 2003, 2005), growing SOMs (GSOMs) (Chan et al., 2008a, 2008b), hyperbolic SOMs (Martin et al., 2008) and emergent SOMs (Dick et al., 2009). One of the most recent DNA-based approaches to taxonomically classifying metagenomic DNA fragments is the usage of interpolated context models, as implemented in Phymm and PhymmBL (Brady and Salzberg, 2009).
Here, we explore the practical application of a novel implementation of GSOMs and BLSOMs for the taxonomic classification of metagenome data sets. We first demonstrate the performance of both SOM variants on the basis of previously published simulated metagenomes (Mavromatis et al., 2007) as well as data from complete microbial genomes. Then, we demonstrate how SOMs can be applied to real-world metagenomes for an overall taxonomic profiling as well as to follow community composition shifts over time. Our SOM implementation is termed TaxSOM and has been made available as a free and feature-rich web-service at http://www.megx.net/toolbox/taxsom.
Materials and methods
Implementation
TaxSOM has been implemented in the C++ programming language using the ocount2 (http://www.promedici.de/ocount2), Lapack++ (http://lapackpp.sourceforge.net), MySQL++ (http://tangentsoft.net/mysql++/) and Boost (http://www.boost.org) C++ libraries. Ocount2 has been used for oligonucleotide counting and Markov model-based z-transformations. Lapack++ has been used for Eigenvector transformation and other matrix operations for principal components analysis, MySQL++ for handling MySQL queries and Boost for parsing program options and serialization of computed SOMs. Boost Python libraries were used to provide an easy way for wrapping TaxSOMs C++ functions and make them accessible in the Python programming language. TaxSOM's web-interface was implemented in PHP (Hypertext Preprocessor) in conjunction with some Python scripts for data processing and with scalable vector graphics for SOM visualizations.
SOM specificity and sensitivities
Specificity (true positives/(true positives+false positives)) and sensitivity (true positives/(true positives+false negatives)) were used as classification accuracy measures. A classification was considered as a true positive, when a query sequence was classified on a SOM node representing only sequences of the query's taxonomic affiliation. It was considered as false positive, when a query sequences was classified on a node representing only sequences from different taxonomic affiliation. Classification of sequences that ought to be classified but were matching ambiguous nodes representing multiple taxa were treated as false negatives. In addition, the F-measure value, which is the harmonic mean of specificity and sensitivity, was used (see Supplementary Tables 1, 2, and 3).
Simulated metagenome data sets for evaluation
In order to evaluate the accuracy of TaxSOM's GSOM and BLSOM implementations for taxonomic DNA sequence classification, we used three previously published simulated data sets (simLC, simMC and simHC) of varying complexities (Mavromatis et al., 2007). SimLC simulates a low-complexity community dominated by a single, near-clonal population that is flanked by low abundance species. SimMC was designed to mimic a moderately complex community like in the acid mine drainage biofilm (Tyson et al., 2004) or the Olavius algarvensis symbionts' metagenome (Woyke et al., 2006), wherein multiple dominant populations are flanked by low abundant ones. SimHC simulates a highly complex community with no dominant populations, like that present in agricultural soils (Tringe et al., 2005). On all of these data sets, the three different assembly programs Arachne (Jaffe et al., 2003), Phrap and JAZZ, have been used, resulting in a total of nine published test data sets. We excluded the JAZZ assemblies from our analysis, because they yielded a much lower number of correct taxonomic classifications than Phrap and Arachne assemblies. This is hence an effect of the JAZZ assembler (or its parameter settings) that would distort the subsequent taxonomic classification.
Data sets from known organisms for evaluation
A test data set comprising 1401 chromosomes and plasmids was constructed from all completely sequenced bacterial and archaeal genomes within GenBank. One-fifth was randomly cut from each sequence and retained for later classification, while the remaining 80% were used as training sequences for BLSOM and GSOM construction. A total of 10 SOMs were constructed, by splitting the training sequences into 10 or 50 kb fragments and using either di- tri- and tetranucleotide raw counts or z-scores as input data. The sequences remaining for classification were used to construct eight data sets of 0.5, 1, 2.5, 5, 10, 25, 30 and 50 kb lengths, which were subsequently classified by the SOMs (Supplementary Table 1).
Real-world data set
This study is part of the Microbial Interactions in Marine Systems project (MIMAS; http://www.mimas-project.de), which provided the real-world metagenome data (Table 1). The data consisted of pyrosequenced bacterial DNA from the coast off the North Sea Island Helgoland in the German bight (54° 11′ 3′' N; 7° 54′ E) that was sampled at five different dates in 2009 (11 February, 31 March, 7 April, 14 April and 16 June).
At each of these points in time, 500 l of subsurface water (1 m depth) were sampled with the small research vessel Diker, immediately taken to the lab, and pre-filtered with 10 μm polycarbonate filters (TCTP, Millipore, Billerica, MA, USA) and 3 μm polycarbonate filters (TSTP, Millipore). The bacterial fraction was subsequently retained on 0.22 μm polyethersulfone filters (GPWP, Millipore). All filters were 142 mm in diameter and six membrane filtration units were operated in parallel to keep filtration times as low as possible. From the filters, bulk environmental DNA was extracted by a modified standard protocol (Zhou et al., 1996). The DNA was then pyrosequenced directly on the GS FLX Ti platform with one (16 June) or two picotiter plates per sample (454 Life Sciences, Branford, CT, USA) by LGC Genomics (LGC Genomics GmbH, Berlin, Germany), and subsequently assembled with Newbler version 2.0.00.22 (Roche, 454 Life Sciences, Branford, CT, USA). From the assemblies, all sequences at least 2.5 kb long were taken for classification.
The bacterial community composition of the samples was assessed by catalyzed reporter deposition-fluorescence in situ hybridization (CARD-FISH) as follows: samples were fixed with 1% formaldehyde and 10 ml was filtered onto polycarbonate membrane filters (type GTTP, pore size 0.2 μm, Sartorius, Göttingen, Germany). CARD-FISH was performed according to previously published protocols (Pernthaler et al., 2002). All hybridizations were counterstained with 4′,6-diamidino-2-phenylindole (1 μg ml–1) and manually inspected and quantified.
Data sets for SOM construction
The SOMs for the evaluation of the simulated data sets were constructed from all bacterial and archaeal DNA sequences exceeding 485 kb (roughly the size of Nanoarchaeum equitans) in the NCBI GenBank database as of October 2008 (release no. 167). These sequences were extracted using a self-written C++ library termed phyloprint (Waldmann, 2008) that allows any type of sequence selection based on the complete NCBI taxonomy (phyloprint currently includes 462 019 nodes). This resulted in 1521 sequences comprising 3.43 Gb of DNA. All sequences were split into 50 kb fragments and subsequently used for the construction of GSOMs and BLSOMs with two types of inputs: oligonucleotide frequency raw counts (di-, tri- or tetranucleotide counts normalized on values between 0 and 1), or raw counts z-transformed based on a maximal order Markov model (Teeling et al., 2004).
The real-world metagenome data sets were classified on a GSOM with tetranucleotide z-scores as input. The GSOM was trained in a habitat-specific manner using 340 bacterial and archaeal DNA genomic sequences from aquatic habitats, such as open ocean water, hot springs, hydrothermal vents or marine sediments. The respective habitat information was obtained from the EnvO-lite classifications present in the Marine Ecological Genomics (MEGX, http://www.megx.net/) database (Kottmann et al., 2010), and the corresponding sequences were extracted from NCBI GenBank using phyloprint.
Protein-level taxonomic classification of real-world data sets for cross-evaluation
Protein-level taxonomic classification of the assembled 454-sequenced bulk environmental DNA was achieved as follows. First, the sequences were subjected to an open reading frame prediction with MetaGene (Noguchi et al., 2006). Afterward, open reading frames exceeding 150 bp were compared with BLASTP (Altschul et al., 1990) against the non-redundant NCBI database (as of 28 October 2008) and with hmmpfam (Eddy, 1996, 1998) against the Pfam database (release 22) (Sonnhammer et al., 1997, 1998). Hits with good E-values (BLASTP: E ⩽E–15, hmmpfam: E ⩽E–5) were subsequently analyzed.
BLASTP hits were processed with an adaptation of the DarkHorse algorithm (Podell and Gaasterland, 2007). In brief, DarkHorse performs rank-based reasoning on the taxonomic terms from BLASTP hits, calculates for each hit a so-called lineage probability index and assigns the open reading frame to the hit with the highest lineage probability index.
Pfam hits were post-processed with CARMA, an algorithm proposed by Krause et al. (2008) that infers taxonomic affiliations from the alignments underlying Pfam Hidden Markov models. Here, we used a rewritten and improved version of the original algorithm.
A weighted consensus of all three tools was used to derive final taxonomic assignments for reads carrying single and contigs carrying multiple genes. The self-written phyloprint C++ library was used to map the taxonomic terms and their NCBI identifiers during the whole analysis.
Algorithm
The SOM is an unsupervised neural network algorithm that implements a non-linear mapping of high-dimensional input data onto a two-dimensional array of weight vectors (Kohonen, 1982, 1990; Kohonen et al., 1996). The process of reducing the data's dimensionality can be thought of as a compression of the input information, whereby the most important topological and metric relationships are preserved. In this sense, SOMs produce an abstraction of the primary data (Kohonen et al., 2001). The topology of the resulting two-dimensional map can be rectangular or hexagonal, and is easy to visualize (Figure 1). Details about input-data variants and a detailed description of the SOM algorithm variants as implemented in TaxSOM are summarized in the Supplementary Methods.
Example of a GSOM showing phylum-level separation. TaxSOM output of a GSOM constructed from all DNA sequences exceeding 485 kb of all Bacteria and Archaea present in GenBank as of October 2008 (1521 sequences; 3.43 Gb). The figure demonstrates the clustering of sequence fragments of 50 kb with each hexagon representing a single node in the grid. The GSOM was calculated using z-transformed tetranucleotide counts for every fragment. Each color denotes 1 of 23 different phyla, if a node is colored in black it contains fragments of more than one phylum. Nodes displayed in any other color contain only fragments of one particular phylum.
Results
Taxonomic classification of simulated metagenomes
For a close to real-world evaluation, TaxSOM was applied to three published simulated data sets mimicking metagenomes of low, medium and high complexities (simLC, simMC and simHC; see Materials and methods section).
For simLC and simMC, high classification specificities were achieved on both the BLSOM and the GSOM, with almost identical results with either Phrap or Arachne assemblies of at least 8 kb—the length used by Mavromatis et al. (2007) in the publication of the simulated metagenome data sets. For GSOM classification of the simLC data set, specificities and sensitivities of 100% were achieved on the superkingdom level and from there on above 97% down to the genus level. For simMC, the classification specificity of the GSOM dropped slightly but stayed above 95% from superkingdom to the genus level, while the sensitivity stayed above 90% (Figure 2a; Supplementary Figure 1). BLSOM classifications yielded almost identical specificities with slightly decreased sensitivities (Supplementary Figure 2a). SimHC was devoid of assemblies exceeding 8 kb and hence was omitted.
GSOM-based classification specificities of simulated data sets. Taxonomic classification accuracy of TaxSOM for the simulated metagenome data sets mimicking habitats of low (simLC) and medium (simMC) complexities using contigs of 8 kb or larger (a) and all contigs (b). Plot (i) depicts specificities (%) and plot (ii) sensitivities (%), respectively. From left to right: specificity of classifications of the simLC data sets assembled with PHRAP and Arachne; classifications of the simMC data sets assembled by PHRAP and Arachne. The different taxonomic levels are represented by different colors. All classifications were achieved on a GSOM trained with z-transformed tetranucleotide counts.
When Phrap or Arachne assemblies were used without constraints on sequence size, GSOM classification specificities exceeded 94% (Figure 2b) and those of BLSOM exceeded 96% (Supplementary Figure 2b) on the superkingdom level for all three data sets (simLC, simMC and simHC). Both SOMS were still able to correctly classify >67% of the sequences on the phylum level, while classification accuracy deteriorated notably on deeper taxonomic levels.
Taxonomic classification of data sets from known microorganisms
In order to evaluate the taxonomic classification accuracy of SOMs as a function of DNA fragment lengths, a test data set was constructed from DNA sequences of complete bacterial genome sequences. Parts of the sequences were used to construct di-, tri- and tetranucleotide-based SOMs and the remainder was split into fragments of different lengths and subsequently classified using the SOMs (see Materials and methods section).
Classification specificity improved with increasing motif and fragment lengths (Supplementary Table 1). It was mostly above 80% for sequences of at least 5 kb and even above 90% for longer fragments on low-resolution taxonomic levels. Below 5 kb classification specificities quickly dropped to values of mostly below 50%, especially for high-resolution genus and species assignments. One interesting observation was that for fragments of 5 kb or more, z-scores provided better assignments while below 5 kb, raw scores provided more accurate results. Also, GSOMs performed better than BLSOMs. Generally speaking, high-resolution assignments required longer sequences (that is, higher information content) than broad-level assignments. For instance, in order to have a 70% accuracy with dinucleotide-based GSOMs, sequences <0.5 kb were sufficient on the superkingdom level, 2.5 kb on the phylum level, 5 kb on the class level, 25 kb on the family level and >50 kb on the genus level. Similar patterns were observed with longer motif lengths, although longer motifs increased classification accuracy. For example, based on tetranucleotides, 70% classification accuracy on the genus level was possible with <10 kb (Supplementary Table 1).
Taxonomic classification of real-world metagenome data sets
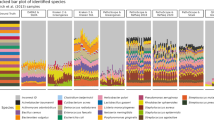
TaxSOM's ability to classify real-world metagenome data was assessed with five North Sea metagenome data sets comprising a total of nine full 454 FLX Ti pyrosequencing runs amounting to almost 8 million reads (Table 1). These could be assembled into 340 143 contigs, of which those of 2.5 kb or more were used for classification. The sample taken in February was highest in biodiversity and yielded only 227 contigs of sufficient length (0.8 Mb). In contrast, the samples taken in March and April had lower biodiversities since they covered a Bacteroidetes bloom, and thus could be assembled into 2321–3229 contigs of sufficient lengths (9.8–16.2 Mb), while the last sample taken in June yielded 1137 such contigs (5 Mb).
In order to assess the plausibility of TaxSOM's taxonomic classifications, we compared the classifications with corresponding CARD-FISH counts of water samples for all five data sets (Figure 3). In the February post-winter situation, the water was low in temperature (∼4 °C) and cell densities (4 E6 cells ml–1). Only in this diverse sample TaxSOM detected Deltaproteobacteria and Epsilonproteobacteria, which were likely dispersed from the sediment by winter storm perturbations.
Biodiversity assessments of the North Sea metagenomes over time. (a) Taxonomic classification of assemblies exceeding 2.5 kb with TaxSOM. (b) Relative CARD-FISH counts of corresponding water samples. Dotted lines indicate congruence in the abundances of Bacteroidetes (green) and the majority of Alphaproteobacteria consisting of the orders Rickettsiales (cyan) and Rhodobacterales (magenta) as assessed by both methods. They do not indicate a smooth transition of the respective abundances, because the community composition fluctuated considerably in between sample time points (data not shown).
The spring situation from end of March to mid-April was characterized by a slight increase in water temperature (∼6 °C) and cell densities (∼1 E6 cells ml–1). TaxSOM and the CARD-FISH data both detected a spring bloom in Bacteroidetes that reached a maximum in mid-April and was accompanied by a decrease in Alphaproteobacteria from the SAR11 and Roseobacter clades. Hence, much of the original biodiversity patterns were retained in the sequence assemblies. Of course, absolute numbers differed. For example, after the bloom maximum in mid-April, equal levels of Gammaproteobacteria (25%) and Alphaproteobacteria (27%, SAR11 and Roseobacter combined), and much higher abundances of Bacteroidetes (43%) were detected in situ with CARD-FISH, while in the assemblies TaxSOM detected more Gammaproteobacteria (36%) than Alphaproteobacteria (13%) and Bacteroidetes (26%).
In addition to CARD-FISH, we compared TaxSOM's classifications with protein-based classifications for the data sets of mid-April (Supplementary Figure 3). Both allow a complete taxonomic breakdown from the superkingdom to the species level. Again, while absolute numbers are different from those of FISH in situ measurements, the overall biodiversity pattern was retained. In comparison with PBC, only TaxSOM was able to resolve the high abundances of Bacteroidetes (TaxSOM: 28%; PBC: 13%). Both tools were able to resolve the key players down to the genus level. Most of the Bacteroidetes were resolved as Polaribacter-like Flavobacteria by both tools. Similarly, a large proportion of the Alphaproteobacteria was resolved as SAR11 and Roseobacter clade species, as indicated by hits to Pelagibacter and Roseobacter on the genus level. This is in line with the CARD-FISH results as well as with reported high abundance of SAR11 species in the oceans by previous metagenome studies (Temperton et al., 2009).
Complete taxonomic breakdowns of all five metagenome data sets are included in the Supplementary Material of this study.
It is noteworthy that as a signature-based method, TaxSOM could classify the contigs without suitable BLAST and HMMer hits that could not be classified on the level of proteins. Especially on deeper taxonomic levels, sequences could oftentimes not be classify based on protein information but could be classified by TaxSOM, which thus provided a much more detailed taxonomic breakdown.
Discussion
In this study, we demonstrate that DNA composition-based SOMs as implemented in TaxSOM are a valuable and useful tool for the taxonomic classification of microbial metagenomes and their subsequent ecological interpretation. Most suitable in this respect are NGS-based deeply sequenced metagenomes of habitats with a low to medium biodiversity, as for example in pelagic ocean waters.
Simulated metagenomes
When applied to simulated metagenome data sets, TaxSOM achieved high classification specificities down to the genus level for the data sets mimicking low- and medium-diversity habitats with fragments of at least 8 kb. These results were obtained even though the corresponding SOMs were constructed from all available fully sequenced prokaryote genomes, and thus comprised a wealth of nodes representing species lacking from the simulated data sets, leading to a high statistical chance of misclassification. This implies that with real-world data from habitats of comparable complexities, respective fragments can be classified with specificities that are sufficient to deduce biologically meaningful results down to the family or even genus level. As in most real-world applications a priori knowledge about the studied habitat is available, more specific SOMs can be constructed from dedicated training sequences, which will further improve classification specificity. Using the simulated data sets without constraining fragment lengths lead to a notable decrease in classification specificities. One reason for this is of course that without length restrictions, large quantities of very short sequences were included whose information content is insufficient for accurate classification. Interestingly, this effect was almost independent of the complexity of the simulated data set, suggesting that at least these data sets were not saturating the resolution of the SOM, that is, the complexity of the analyzed data was not limiting the analysis. An additional reason for the drop in classification specificities, as stated by Mavromatis et al., (2007), is that a high proportion of chimeras among shorter contigs result in low quality classifications. If the number of such misassemblies can be reduced, the minimum required sequence length will drop as well (Chan et al., 2008b). Still, even with inclusion of the short fragments TaxSOM provided respectable results in all simulated data sets at least down to the phylum level, which might be the current limit for reasonable biological conclusions based on mostly short and unassembled sequences (Figure 2; Supplementary Figures 1 and 2).
Data sets from known microorganisms
The results from the artificial data sets of fully sequenced microorganisms show that classification specificity is a function of information content, and hence increases with motif and with sequence length. Longer oligonucleotides provide better specificities than shorter oligonucleotides, and longer sequences can be classified more accurately than shorter ones. Good classification specificities can be obtained for sequences down to 5 kb; below that, information content starts to become limiting (Supplementary Table 1). This is also supported by the fact that below 5 kb SOMs constructed from raw oligonucleotide counts outperformed those constructed from z-transformed counts, while it was the opposite above 5 kb. The z-transformation statistically corrects counts of oligonucleotides of a given length for asymmetries introduced by skews in shorter oligonucleotide frequencies. For example, it is expected that within high GC genomes higher frequencies of GC-rich tetranucleotides (for example, GGCC) are observed than an AT-rich genomes, and thus high frequencies of GGCC in an GC-rich genome convey less information as when they occur in an AT-rich genome. However, the z-transformation compensating this is itself based on a statistical assessment, and hence also limited by the sequence's information content. As the latter deteriorates from about 5 kb on, z-transformation can only enhance results for sequences with sufficient information content and even introduces additional noise when the sequences get too short for proper statistics. Nonetheless, classification accuracies for sequences below 5 kb are still sufficient to conduct NGS-based statistical ecological habitat studies. Here, the ability to discriminate a biological signal from the data's noise is more important than an almost perfect classification, such as when monitoring overall community composition changes or linking abundances of functional genes to taxonomic groups.
Real-world metagenomes
As our results with pyrosequenced bulk DNA show, such studies are possible with sequences of 2.5 kb at least down to the class if not to the order level, especially with suitable habitat-specific SOMs. Although classifications were not perfect with respect to providing a high-resolution quantitative taxonomic breakdown of the analyzed samples, they provide a good description of overall biodiversity and abundances of a given habitat (Supplementary Figure 3) and allow detection of major community composition changes (Figure 3). These data can serve as a guideline for the selection of specific CARD-FISH probes for more detailed biodiversity studies, and furthermore allows mining the taxonomically classified sequences (taxobins) for functions. Such a linkage of taxonomy and function will allow us to gain insights into the ecological functioning of habitats and even to select frequent but as yet unknown genes within dedicated taxa as targets for further studies.
It is our experience from more than a dozen direct pyrosequencing experiments on moderately diverse coastal and deep sea ocean waters (data not shown) that well-run 454 FLX Ti picotiter plates can yield more than a million reads comprising up to 400 Mb of raw sequence that typically can be assembled into 30–70 Mb of non-redundant DNA, equivalent to 7–16 bacterial genomes. In all cases, the longest assemblies were well within the range of typical fosmids (that is, up to 35 kb), and larger proportions of the assemblies were above 2.5 kb and thus suitable for SOM analysis (Table 1).
Biodiversity information from direct DNA sequencing cannot rival in situ measurements like FISH in terms of quantitativeness because of inherent biases, such as lineage-specific DNA extraction efficacies, sequence-dependent differences during the bead-mediated amplification in the 454 library creation step, skews introduced by the assembly diminishing quantities of the most abundant species, and taxa without suitable reference sequences for taxonomic classification, like those without any representation in public sequence databases. FISH on the other hand has to cope with its own inherent limits, like issues with permeabilization, target accessibility or probe sensitivity and specificity. Hence, both methods shed a slightly different light on biodiversity. It is therefore understandable that the biodiversity data obtained by FISH and by direct sequencing of bulk DNA show differences, although they are in broad agreement with respect to major community composition shifts. It is noteworthy that the TaxSOM assignments were well supported by PBC tools. This indicates that the TaxSOM assignments reflect a realistic assessment of the biodiversity within the sampled sequences, which does not necessarily reflect the situation in situ in a perfect manner. FISH does provide only information for the applied probes, whereas in silico taxonomic classifications of directly sequenced DNA do not require a priori assumptions about the community composition, provide a deeper taxonomic resolution in shorter time and enable formation of taxobins that can be mined for gene functions in order to address ecological questions.
TaxSOM website
In order to make such applications accessible for a broader audience in microbial molecular ecology, we implemented TaxSOM as a freely accessible website that allows the usage of GSOMs and BLSOMs for taxonomic classification of microbial DNA sequences. TaxSOM provides either pre-computed SOMs for general taxonomic classification purposes, or the option to compute custom-tailored SOMs. For the latter, TaxSOM provides the ability to upload sequences for SOM construction (for example, with habitat-specific sequences) as well as a dynamic taxonomy tree selection tool that allows for an easy visual as well as textual selection of all sequences of the NCBI nt database with sufficient length. A rich set of features is available for controlling the behavior of SOMs, and the resulting SOMs can be inspected visually. For experts, we provide a rich set of parameters for controlling the SOMs behavior. Unique to TaxSOM is the capability of pre-processing frequencies using a maximal-order Markov model as input data, which improves classification accuracy for sequences exceeding 5 kb. After a SOM is constructed, sequences can be uploaded for classification, whereby a SOM persists and can be used for the classification of multiple data sets. Classification results can be inspected either visually (Figure 1) or downloaded as tables in text files for further use. This will enable a broader audience to use taxonomic classifications in microbial community studies. The TaxSOM web service is available at http://www.megx.net/toolbox/taxsom.
Conclusions
One advantage of SOMs is that taxonomic classification once a SOM is trained takes only minutes, even for large amounts of sequences, while gene-based classification tools rely on time-consuming and computationally intensive BLAST or HMMER searches, and FISH requires labor-intensive laboratory work. For example, the current TaxSOM implementation can classify 100 000 sequences on a SOM of 10 000 nodes within 20 min on moderate hardware (single 2.2 MHz Opteron core). Similarly, a million sequences can be classified within a couple of hours conveniently over night (see Supplementary Tables 4, 5 and 6 for more elaborate data on classification speed). This makes SOM-based taxonomic classifications ideal for processing vast amounts of sequences as they are produced by current NGS platforms.
With promising new sequencing technologies on the horizon that will not need amplification and will deliver more and longer reads at lower prices, like ZMW-based sequencing by Pacific Biosciences (Menlo Park, CA, USA; Eid et al., 2009) or various variants of nanopore sequencing as developed by Oxford Nanopore Technologies (Kidlington, Oxfordshire, UK; Clarke et al., 2009), IBM Deutschland Research & Development GmbH (Böblingen, Germany)/Roche (DNA transistor) and others, there is a need for high-throughput tools to convert the wealth of sequence data into knowledge. The recently introduced Pacific Biosciences single molecule sequencing platform has an average read length of >1 kb and a maximum read length of 5 kb. Hence, technologies that focus on short reads will be mostly obsolete in the not too distant future. As a consequence, microbial biodiversity studies soon will target full-length 16S ribosomal RNA sequences instead of only small hypervariable regions, and metagenomic studies will produce longer assemblies that can be taxonomically classified with high accuracy. Tools like TaxSOM will enable the fast classification of large proportions of metagenomes into taxobins, and thus provide a link between biodiversity and function.
Even with current 454 FLX Titanium pyrosequencing, good results can be expected for SOM-based taxonomic classifications, in particular for habitats with limited diversity, few dominating species or with species that discriminate well in terms of their genomic signatures. For complex habitats leading to metagenomes without longer assemblies, DNA composition-based methods should be combined with if not substituted by PBC methods. These, however, are restricted to sequences harboring well-characterized genes or domains and thus can classify fewer sequences.
Until long read technologies are available, we suggest clustering metagenomes into taxobins by a combination of nucleotide and protein-based taxonomic classification tools. This enables the application of large-scale NGS DNA sequencing as a screening tool for biodiversity and paves the way for insights into the functional ecology of complex microbial communities. For habitats with low-to-medium biodiversity, sufficiently reliable classifications can be achieved down to the genus level, but the amount of sequence that is obtained in praxis with current techniques will be often too small for a sound statistical analysis of gene functions on this level. However, functional analyses of our real-world data have shown that such studies can be done down to the class and for abundant taxa even down to the order level with two to four full picotiter plates of pyrosequencing per sample (data not shown). We anticipate that progress in sequencing with respect to read length and throughput will soon eliminate this bottleneck and thus will enable to study microbial communities in a holistic fashion on a much finer level.
References
Abe T, Sugawara H, Kinouchi M, Kanaya S, Ikemura T . (2005). Novel phylogenetic studies of genomic sequence fragments derived from uncultured microbe mixtures in environmental and clinical samples. DNA Res 12: 281–290.
Abe T, Kanaya S, Kinouchi M, Ichiba Y, Kozuki T, Ikemura T . (2003). Informatics for unveiling hidden genome signatures. Genome Res 13: 693–702.
Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ . (1990). Basic local alignment search tool. J Mol Biol 215: 403–410.
Amann RI, Ludwig W, Schleifer KH . (1995). Phylogenetic identification and in situ detection of individual microbial cells without cultivation. Microbiol Rev 59: 143–169.
Brady A, Salzberg SL . (2009). Phymm and PhymmBL: metagenomic phylogenetic classification with interpolated Markov models. Nat Methods 6: 673 .
Burge C, Campbell AM, Karlin S . (1992). Over- and under-representation of short oligonucleotides in DNA sequences. Proc Natl Acad Sci USA 89: 1358–1362.
Chan C-KK, Hsu AL, Tang S-L, Halgamuge SK . (2008a). Using growing self-organising maps to improve the binning process in environmental whole-genome shotgun sequencing. J Biomed Biotechnol 2008, doi:10.1155/2008/513701.
Chan CK, Hsu AL, Halgamuge SK, Tang SL . (2008b). Binning sequences using very sparse labels within a metagenome. BMC Bioinformat 9: 215.
Clarke J, Wu HC, Jayasinghe L, Patel A, Reid S, Bayley H . (2009). Continuous base identification for single-molecule nanopore DNA sequencing. Nat Nanotechnol 4: 265–270.
Deschavanne PJ, Giron A, Vilain J, Fagot G, Fertil B . (1999). Genomic signature: characterization and classification of species assessed by chaos game representation of sequences. Mol Biol Evol 16: 1391–1399.
Diaz NN, Krause L, Goesmann A, Niehaus K, Nattkemper TW . (2009). TACOA: taxonomic classification of environmental genomic fragments using a kernelized nearest neighbor approach. BMC Bioinformat 10: 56.
Dick GJ, Andersson AF, Baker BJ, Simmons SL, Thomas BC, Yelton AP et al. (2009). Community-wide analysis of microbial genome sequence signatures. Genome Biol 10: R85.
Eddy SR . (1996). Hidden Markov models. Curr Opin Struct Biol 6: 361–365.
Eddy SR . (1998). Profile Hidden Markov Models. Bioinformatics 14: 755–763.
Eid J, Fehr A, Gray J, Luong K, Lyle J, Otto G et al. (2009). Real-time DNA sequencing from single polymerase molecules. Science 323: 133–138.
Fleischmann RD, Adams MD, White O, Clayton RA, Kirkness EF, Kerlavage AR et al. (1995). Whole-genome random sequencing and assembly of Haemophilus influenzae Rd. Science 269: 496–512.
Gupta PK . (2008). Single-molecule DNA sequencing technologies for future genomics research. Trends Biotechnol 26: 602–611.
Hanekamp K, Bohnebeck U, Beszteri B, Valentin K . (2007). PhyloGena—a user-friendly system for automated phylogenetic annotation of unknown sequences. Bioinformatics 23: 793–801.
Huber JA, Mark Welch DB, Morrison HG, Huse SM, Neal PR, Butterfield DA et al. (2007). Microbial population structures in the deep marine biosphere. Science 318: 97–100.
Huse SM, Dethlefsen L, Huber JA, Welch DM, Relman DA, Sogin ML . (2008). Exploring microbial diversity and taxonomy using SSU rRNA hypervariable tag sequencing. PLoS Genet 4: e1000255.
Huson DH, Auch AF, Qi J, Schuster SC . (2007). MEGAN analysis of metagenomic data. Genome Res 17: 377–386.
Jaffe DB, Butler J, Gnerre S, Mauceli E, Lindblad-Toh K, Mesirov JP et al. (2003). Whole-genome sequence assembly for mammalian genomes: Arachne 2. Genome Res 13: 91–96.
Karlin S, Burge C . (1995). Dinucleotide relative abundance extremes: a genomic signature. Trends Genet 11: 283–290.
Karlin S . (1998). Global dinucleotide signatures and analysis of genomic heterogeneity. Curr Opin Microbiol 1: 598–610.
Karlin S, Campbell AM, Mrazek J . (1998). Comparative DNA analysis across diverse genomes. Annu Rev Genet 32: 185–225.
Karlin S, Ladunga I . (1994). Comparisons of eukaryotic genomic sequences. Proc Natl Acad Sci USA 91: 12832–12836.
Karlin S, Ladunga I, Blaisdell BE . (1994). Heterogeneity of genomes: measures and values. Proc Natl Acad Sci USA 91: 12837–12841.
Kohonen T . (1982). Self-organized formation of topologically correct feature maps. Biol Cybernet 43: 59–69.
Kohonen T . (1990). Self-organization maps. Proc IEEE 78: 1464–1480.
Kohonen T, Kohonen T, Schroeder MR, Huang TS, Maps SO . (2001). Springer-Verlag New York Inc.: Secaucus, NJ.
Kohonen T, Oja E, Simula O, Visa A, Kangas J . (1996). Engineering applications of the self-organizing map. Proc IEEE 84: 1358–1384.
Kottmann R, Kostadinov I, Duhaime MB, Buttigieg PL, Yilmaz P, Hankeln W et al. (2010). Megx net: integrated database resource for marine ecological genomics. Nucleic Acids Res 38: D391–D395.
Krause L, Diaz NN, Goesmann A, Kelley S, Nattkemper TW, Rohwer F et al. (2008). Phylogenetic classification of short environmental DNA fragments. Nucleic Acids Res 36: 2230 .
Martin C, Diaz NN, Ontrup J, Nattkemper TW . (2008). Hyperbolic SOM-based clustering of DNA fragment features for taxonomic visualization and classification. Bioinformatics 24: 1568–1574.
Mavromatis K, Ivanova N, Barry K, Shapiro H, Goltsman E, McHardy AC et al. (2007). Use of simulated data sets to evaluate the fidelity of metagenomic processing methods. Nat Methods 4: 495–500.
McHardy AC, Martin HG, Tsirigos A, Hugenholtz P, Rigoutsos I . (2007). Accurate phylogenetic classification of variable-length DNA fragments. Nat Methods S 4: 63–72.
Noguchi H, Park J, Takagi T . (2006). MetaGene: prokaryotic gene finding from environmental genome shotgun sequences. Nucleic Acids Res 34: 5623–5630.
Ochman H . (2007). Single-cell genomics. Environ Microbiol 9: 7.
Pernthaler A, Pernthaler J, Amann R . (2002). Fluorescence in situ hybridization and catalyzed reporter deposition for the identification of marine bacteria. Appl Environ Microbiol 68: 3094–3101.
Peterson J, Garges S, Giovanni M, McInnes P, Wang L, Schloss JA et al. (2009). The NIH Human Microbiome Project. Genome Res 19: 2317–2323.
Podell S, Gaasterland T . (2007). DarkHorse: a method for genome-wide prediction of horizontal gene transfer. Genome Biol S 8: R16.
Pride DT, Meinersmann RJ, Wassenaar TM, Blaser MJ . (2003). Evolutionary implications of microbial genome tetranucleotide frequency biases. Genome Res 13: 145–158.
Reva ON, Tümmler B . (2004). Global features of sequences of bacterial chromosomes, plasmids and phages revealed by analysis of oligonucleotide usage patterns. BMC Bioinformat 5: 90.
Rocha EP, Viari A, Danchin A . (1998). Oligonucleotide bias in Bacillus subtilis: general trends and taxonomic comparisons. Nucleic Acids Res 26: 2971–2980.
Sandberg R, Winberg G, Branden CI, Kaske A, Ernberg I, Coster J . (2001). Capturing whole-genome characteristics in short sequences using a naive Bayesian classifier. Genome Res 11: 1404–1409.
Schloss PD, Handelsman J . (2003). Biotechnological prospects from metagenomics. Curr Opin Biotechnol 14: 303–310.
Seshadri R, Kravitz SA, Smarr L, Gilna P, Frazier M . (2007). CAMERA: a community resource for metagenomics. PLoS Biol 5: e75.
Sogin ML, Morrison HG, Huber JA, Mark Welch D, Huse SM, Neal PR et al. (2006). Microbial diversity in the deep sea and the underexplored ‘rare biosphere’. Proc Natl Acad Sci USA 103: 12115–12120.
Sonnhammer EL, Eddy SR, Durbin R . (1997). Pfam: a comprehensive database of protein domain families based on seed alignments. Proteins 28: 405–420.
Sonnhammer EL, Eddy SR, Birney E, Bateman A, Durbin R . (1998). Pfam: multiple sequence alignments and HMM-profiles of protein domains. Nucleic Acids Res 26: 320–322.
Teeling H, Meyerdierks A, Bauer M, Amann R, Glöckner FO . (2004). Application of tetranucleotide frequencies for the assignment of genomic fragments. Environ Microbiol 6: 938–947.
Temperton B, Field D, Oliver A, Tiwari B, Muhling M, Joint I et al. (2009). Bias in assessments of marine microbial biodiversity in fosmid libraries as evaluated by pyrosequencing. ISME J 3: 792–796.
Tringe SG, von Mering C, Kobayashi A, Salamov AA, Chen K, Chang HW et al. (2005). Comparative metagenomics of microbial communities. Science S 308: 554–557.
Tyson GW, Chapman J, Hugenholtz P, Allen EE, Ram RJ, Richardson PM et al. (2004). Community structure and metabolism through reconstruction of microbial genomes from the environment. Nature 428: 37–43.
Waldmann J . (2008). Phyloprint—Entwicklung und Anwendung eines Frameworks zur taxonomischen Klassifikation. Westfälische Wilhelms-Universität Münster, Department of Mathematics and Computer Science, Diploma Thesis, http://cs.uni-muenster.de/Professoren/Lippe/diplomarbeiten/html/Waldmann/.
Woyke T, Teeling H, Ivanova NN, Huntemann M, Richter M, Gloeckner FO, et al. (2006). Symbiosis insights through metagenomic analysis of a microbial consortium. Nature 443: 950–955.
Wu D, Hugenholtz P, Mavromatis K, Pukall R, Dalin E, Ivanova NN et al. (2009). A phylogeny-driven genomic encyclopaedia of Bacteria and Archaea. Nature 462: 1056–1060.
Zhou J, Bruns MA, Tiedje JM . (1996). DNA recovery from soils of diverse composition. Appl Environ Microbiol S 62: 316–322.
Acknowledgements
We thank Tobin J Hammer for fruitful discussions and proof reading of the paper. This study was supported by the Max Planck society and the MIMAS project (project no. 03F0480A) funded by the German Federal Ministry of Education and Research (BMBF).
Author information
Authors and Affiliations
Corresponding author
Additional information
Supplementary Information accompanies the paper on The ISME Journal website
Supplementary information
Rights and permissions
About this article
Cite this article
Weber, M., Teeling, H., Huang, S. et al. Practical application of self-organizing maps to interrelate biodiversity and functional data in NGS-based metagenomics. ISME J 5, 918–928 (2011). https://doi.org/10.1038/ismej.2010.180
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/ismej.2010.180
Keywords
This article is cited by
-
Glaciers as microbial habitats: current knowledge and implication
Journal of Microbiology (2022)
-
Vulnerability of Zostera noltei to Sea Level Rise: the Use of Clustering Techniques in Climate Change Studies
Estuaries and Coasts (2020)
-
A clinician's guide to microbiome analysis
Nature Reviews Gastroenterology & Hepatology (2017)
-
AKE - the Accelerated k-mer Exploration web-tool for rapid taxonomic classification and visualization
BMC Bioinformatics (2014)
-
A novel approach, based on BLSOMs (Batch Learning Self-Organizing Maps), to the microbiome analysis of ticks
The ISME Journal (2013)