Abstract
OBJECTIVE:
Obesity is a growing health concern in the Oceanic populations. To investigate the genetic factors associated with adult obesity in the Oceanic populations, the association of single nucleotide polymorphisms (SNPs) of the beta-2 adrenergic receptor (ADRB2) gene with obesity was examined in 694 adults living in Tonga and Solomon Islands.
RESULTS:
A screening for variation in 16 Oceanic subjects detected 17 SNPs in the entire region of ADRB2, of which nine SNPs including two non-synonymous ones, rs1042713 (Arg16Gly) and rs1042714 (Gln27Glu), were further genotyped for all subjects. The rs34623097-A allele, at a SNP located upstream of ADRB2, showed the strongest association with risk for obesity in a logistic regression analysis adjusted for age, sex, and population (P=5.6 × 10−4, odds ratio [OR]=2.5, 95% confidence interval [CI]=1.5–4.2). The 27Glu was also significantly associated with obesity in the single-point association analysis (P=0.013, OR=2.0, 95%CI=1.2–3.4); however, this association was no longer significant after adjustment for rs34623097 since these SNPs were in linkage disequilibrium with each other. A copy of the obesity-risk allele, rs34623097-A, led to a 1.6 kg/m2 increase in body mass index (BMI; defined as weight in kilograms divided by height in meters squared) (P=0.0019). A luciferase reporter assay indicated that rs34623097-A reduced the transcriptional activity of the luciferase reporter gene by approximately 10% compared with rs34623097-G. An electrophoretic mobility shift assay demonstrated that rs34623097 modulated the binding affinity with nuclear factors. An evolutionary analysis implies that a G>A mutation at rs34623097 occurred in the Neandertal genome and then the rs34623097-A allele flowed into the ancestors of present-day humans.
CONCLUSION:
The present results suggest that rs34623097-A, which would lead to lower expression of ADRB2, contributes to the onset of obesity in the Oceanic populations.
Similar content being viewed by others
Introduction
The overall prevalence of adult obesity is increasing in Oceanic populations. Although the change from a traditional to a modern lifestyle largely contributed to the rapid increase in the prevalence of obesity, environmental factors cannot fully explain individual differences in the predisposition to obesity in Oceanic populations as well as other populations. Along with environmental factors, genetic factors also play a key role in the development of obesity.1
Genome-wide association studies have shown that common variants in the FTO gene are significantly associated with body mass index (BMI) or obesity in European populations,2, 3, 4 and a number of studies have confirmed the association of FTO polymorphisms with obesity in European populations. However, our previous study did not replicate the association of FTO polymorphism with BMI in Oceanic populations, even though the possibility of false-negative results due to the small sample size (320 subjects) was not excluded.5 A recent study with a larger sample size (465 American Samoans and 624 Samoans) also revealed no significant association after corrections for multiple testing.6 These results suggest differences in genetic factors between Oceanic and European populations that predispose them to obesity.
Total energy consumption in excess of total energy expenditure leads to obesity. It has been suggested that polymorphisms of genes involved in the regulation of catecholamine function are associated with obesity, since catecholamines play a key role in energy expenditure and lipolysis. The beta-2 adrenergic receptor (ADRB2), a member of the G protein coupled receptors, mediates the catecholamine-induced activation of adenylate cyclase through the action of G proteins. Considering the biological function of ADRB2, polymorphisms of the ADRB2 gene (OMIM 109690) are plausible candidates that are associated with obesity. Previous association studies have especially focused on two common nonsynonymous SNPs of the ADRB2 gene, Arg16Gly (rs1042713) and Gln27Glu (rs1042714). In some studies, Arg16Glu and/or Gln27Glu were associated with obesity and/or BMI,7, 8, 9 but these associations were not found in other studies.10, 11 A meta-analysis for Arg16Gly and Gln27Glu showed that the 27Glu allele was a significant risk factor for obesity in Asians, Pacific Islanders, and Native Americans.12 However, in the original study of the Tongan population (ie, Pacific Islanders) referred by the meta-analysis, 27Glu tended to be associated with obesity, but the association was not statistically significant,13 implying questionable association in Oceanic populations. It is also the fact that few Oceanic populations have been investigated. Therefore, in this study, we examined the association of ADRB2 polymorphisms with obesity in Oceanic populations and performed functional analyses of the SNP associated with risk for obesity.
Subjects and methods
Subjects
A total of 694 adult subjects (18 years old or older) were recruited from five Oceanic populations: (1) Austronesian (AN)-speaking Polynesians in Ha’ano and Fakakakai villages of Ha’apai Island, Tonga (n=32); (2) AN-speaking Polynesians in Nuku’alofa, Tonga (n=139); (3) AN-speaking Melanesians in Munda, Solomon Islands (n=170); (4) AN-speaking Melanesians in Kusaghe (referred to as Paradise in our previous papers), Solomon Islands (n=202); and (5) AN-speaking Micronesians in Rawaki, Solomon Islands (n=151). Rawaki village is located on New Georgia Island, Solomon Islands, but Rawaki villagers are AN-speaking Micronesians who migrated from the Gilbert Islands, Kiribati, about 40 years ago. Thus, in this paper, Rawaki villagers are regarded as Micronesians.
To investigate the allele frequencies of rs34623097 and rs1042714 (Gln27Glu) in other Oceanic populations, 32 non-Austronesian (NAN)-speaking Melanesians (Gidra) living in Western Province, Papua New Guinea, and 32 AN-speaking Melanesians in Balopa, Manus Province, Papua New Guinea, were also analyzed. In addition, the allele frequencies of rs34623097 and rs1042714 (Gln27Glu) were also investigated in 894 Mongolian individuals. The following genomic DNA samples used in the International HapMap Project14, 15 were obtained from the Coriell Cell Repository and subjected to rs34623097 genotyping: 57 subjects from YRI (Yoruba in Ibadan, Nigeria), 58 CEU (CEPH Utah residents with ancestry from northern and western Europe), 43 JPT (Japanese in Tokyo, Japan), and 45 CHB (Han Chinese in Beijing, China). The features of the Oceanic populations have been previously described in detail.16, 17 The locations of the Oceanic populations studied are shown in Supplementary Material, Supplementary Figure S1.
Blood samples were collected after obtaining informed consent from each participant. This study was approved by the Health Ethics Committee, Ministry of Health, Solomon Islands; the Medical Research Advisory Committee of Papua New Guinea; National Health Ethics & Research Committee of Tonga; the Ethics Committee of the Health Science University of Mongolia; the Ethics Committee of Jichi Medical University; and the Research Ethics Committee of the Graduate School of Comprehensive Human Sciences, University of Tsukuba.
Anthropometric measurement
Anthropomorphic phenotypes were directly measured in field settings. Measurements were taken of subjects dressed in light clothing. Body height was measured to the nearest 1 mm by using a field anthropometer (GPM, Zurich, Switzerland), and weight was recorded to the nearest 0.1 kg by using a portable digital scale (Tanita model BC-518, Tokyo, Japan). Subjects with BMI ⩾27 kg/m2 were classified as obese regardless of the population, ethnicity, and sex in this study. The reason why obesity was not defined as BMI ⩾30 kg/m2 is because there were no obese male subjects in Kusaghe if defined so.
Screening for variation of ADRB2
Genomic DNA was extracted from peripheral blood by using a QIAamp Blood Kit (Qiagen, Hilden, Germany). Variations in the ∼3.8 kb region containing the entire ADRB2 gene were screened for 16 subjects: two from Nuku’alofa, two from Ha’apai, four from Kusaghe, four from Munda, and four from Rawaki. For the putative regulatory region, the genomic region ∼2 kb upstream of the initiator codon ATG was analyzed. Primers were designed within the overlapping region on the basis of the genomic sequence obtained from the National Center for Biotechnology Information database (GRCh37/hg19) (Supplementary Material, Supplementary Figure S2a). Primer sequences are listed in Supplementary Material, Supplementary Figures S2b and S2c. PCR amplification was performed with the GeneAmp PCR System 9700 (Applied Biosystems, Foster City, CA, USA) by using a FastStart Taq DNA Polymerase kit (Roche, Molecular Biochemicals, Mannheim, Germany). PCR products were sequenced with an ABI Prism 3100 Genetic Analyzer (Applied Biosystems, Foster City, CA, USA).
Genotyping
From 17 SNPs detected by the screening, eight tag SNPs were selected as candidates to be genotyped. Of eight candidate SNPs, seven SNPs, excluding rs2400706, were successfully genotyped. Here, rs2400706 could not be genotyped, since the TaqMan probe for rs2400706 was not designed. Two SNPs, rs1042711 and rs1042714, were additionally selected, since the significant associations of these SNPs with obesity had been previously reported.7, 8, 9, 18 Accordingly, a total of nine SNPs were genotyped (Table 2 and Figure 1). The remaining seven SNPs were in LD (r2⩾0.8) with at least one of nine SNPs genotyped (Supplementary Material, Supplementary Figure S3). Two SNPs, rs1042713 (Arg16Gly) and rs1042714 (Gln27Glu), were genotyped by using the DigiTag2 assay.19 The other seven SNPs were genotyped using the TaqMan SNP genotyping assays. The assays were run on an ABI 7300 machine (Applied Biosystems, Foster City, CA, USA).
Significant association of rs34623097-A with obesity. (a) Location of SNPs. A screening for variation detected 17 SNPs in the entire region of ADRB2 gene in 16 subjects. (b) Association plot of nine SNPs. The association of each ADRB2 SNP with obesity was examined by multiple regression analysis adjusted for age, sex, and population. Each SNP is plotted as a diamond along the chromosomal position, and r2 between the SNP and rs34623097 is colored as a scale from low (blue) to high (red).
Cell culture
HEK293T (human embryonic kidney cell line) cells were cultured in D-MEM (Wako, Osaka, Japan) supplemented with 10% heat-inactivated FBS in 5% CO2 at 37 °C.
Luciferase reporter assay
The DNA samples from subjects with homozygotes of rs34623097-A and -G were amplified with a forward primer (5′-TTTTTTGGATCCAGCTCCAAGTTTGGTGAATCTGGAAC-3′) and a reverse primer (5′-TTTTTTGCTAGCTGGGTGACAGAACAAGACTCCATCTC-3′). Accordingly, two different PCR products with 619 bp, including the rs34623097-A and rs34623097-G alleles, were separately obtained. The PCR products had no sequence difference at any sites other than rs34623097. The PCR products for each allele were gel purified by using NucleoSpin Extract II (Macherey Nagel, Düren, Germany) and then cut with BamHI and NheI restriction enzymes. These products were ligated into the BglII and NheI sites in the promoter region located 5’ to the Renilla luciferase gene in the psiCHECK2 vector (Promega, Madison, WI, USA) by using T4 DNA ligase (Thermo Fisher Scientific, Waltham, MA, USA). Accordingly, SV40 early enhancer/promoter in the psiCHECK2 vector was completely replaced by the PCR products. The inserts containing rs34623097-A or rs34623097-G were confirmed by PCR-direct sequencing. Plasmid with rs34623097-A or rs34623097-G was transfected into HEK293T cells using Lipofectamine 2000 (Life Technologies, Carlsbad, CA, USA) and incubated for 24 h. The luciferase activities were determined by using the Dual-Glo Luciferase Assay system (Promega, Madison, WI, USA) with a luminometer. In this system, transcriptional activity was measured by Renilla luciferase activity and was normalized by Firefly luciferase activity in each assay (ie, in the same well). The experiment was replicated five times, with quintuplicate assays performed within each replicate.
Electrophoretic mobility shift assay
Nuclear extracts were extracted from the cultured HEK293T cells by using NE-PER Nuclear and Cytoplasmic Extraction Reagents (Thermo scientific, Rockford, IL, USA) according to the manufacturer’s instruction. The oligonucleotide probes (5′-TAATTTCACTTTAGCAATAAAGTCACATGC-3′ for rs34623097-A, 5′-TAATTTCACTTTAGCAGTAAAGTCACATGC-3′ for rs34623097-G, and both complementary sequences) were labeled with a Biotin 3’ End DNA Labeling Kit (Thermo scientific, Rockford, IL, USA). EMSA was performed using a Light Shift Chemiluminescent EMSA Kit (Thermo scientific, Rockford, IL, USA) according to the manufacturer’s instructions. Briefly, the reaction mixtures (20 μl) containing 6 μg of nuclear extract, 1 × binding buffer, 50 ng/μl of poly (dI-dC), 5 mM MgCl2, 0.05% NP-40 and 2.5% glycerol were pre-incubated on ice for 10 min and then incubated with 1 nM of the biotin-labeled probe for 20 min at room temperature The reaction mixtures containing no nuclear extract were also incubated with the labeled probes as negative control samples. Each sample was loaded onto the 5% polyacrylamide gel (ATTO, Tokyo, Japan) in 0.5 × TBE. Samples were electrophoresed until the bromophenol blue dye had migrated approximately 3/4 down the length of the gel. The gel and nylon membrane were sandwiched in a clean electrophoretic transfer unit, and the DNA was transferred at 380 mA (∼100 V) for 30 min. Then DNA and membrane were crosslinked by using a UV-light cross-linker instrument. Biotin-labeled DNA was detected by chemiluminescence provided by the LightShift Chemiluminescent EMSA Kit (Thermo scientific, Rockford, IL, USA). For competition EMSA, to compete specifically with labeled oligonucleotide probes, a 200-fold excess of unlabeled oligonucleotide probes for rs34623097-A or rs34623097-G were added to reaction mixtures containing 6 μg of nuclear extract.
Statistical analyses
Deviation of genotype frequencies from Hardy-Weinberg equilibrium was examined by chi-square test. Haplotype frequency and pairwise LD parameters, r2 and D’, were estimated by using Haploview software.20 In association analyses, only subjects who had been 18 years old or older at the time of anthropometric survey were analyzed. A logistic regression analysis that was adjusted for age, sex, and population was performed to assess the association of each ADRB2 SNP of ADRB2 with obesity (BMI ⩾27 kg/m2). To assess the independent effect of each SNP on obesity, a logistic regression analysis with rs34623097, rs11959427, rs1042711, rs1042714, and rs1042720 as independent variables adjusted for age, sex, and population was also performed. A multiple regression analysis adjusted for age, sex, and population was performed to assess the association of rs34623097 with BMI. In both regression analyses, the number of copies of a derived allele at each ADRB2 SNP was used as an independent variable (ie, homozygotes of an ancestral allele, those of a derived allele, and heterozygotes were coded as 0, 2, and 1, respectively). The association of rs34623097 genotypes with BMI was further assessed by analysis of covariance (ANCOVA) adjusted for age, sex, and population. The genotypes of 626 SNPs with minor allele frequency of more than 0 in JPT and CHB were retrieved from the HapMap database,14, 15 and then the r2 values between rs34623097 and each of 626 SNPs around the ADRB2 were calculated.
In the luciferase assay, to adjust for the variation of experimental conditions among five replications (ie, transfections) each with quintuplicate assays, the difference in transcriptional activity between rs34623097-A and rs34623097-G was assessed by ANCOVA adjusted for replication.
A SNP upstream of ADRB2, rs34623097, was analyzed for a putative transcription factor-binding site by using the web-based software Transcription Factor Binding Sites Search (TFSEARCH; http://www.cbrc.jp/research/db/TFSEARCH.html) (version 1.3),21 with a threshold score of 90.0.
The genotype data of rs34623097 and rs1042714 for CHB, JPT, CHS, FIN, GBR, IBS, TSI, YRI, LWK, PUR, MXL, and CLM were obtained from the 1000 Genomes Project database (http://www.1000genomes.org).22 The genotype data of SNPs around rs34623097 in CHB (Han Chinese in Beijing, China), JPT (Japanese in Tokyo, Japan), CHS (Southern Han Chinese), and FIN (Finnish in Finland) populations were also retried from the 1000 Genomes Project database. The Neandertal genome sequences (Vi33.16, Vi33.25, and Vi33.26) at the corresponding sites were obtained from the UCSC Genome Browser (http://genome.ucsc.edu/).23
In this study, the significance level was set at 0.05 (ie, P-values of less than 0.05 were regarded as statistically significant), unless otherwise stated. The Bonferroni correction was used when appropriate.
Results
Association test
To assess the association between ADRB2 polymorphisms and obesity, 694 adults between 18 and 65 years of age were recruited from five Oceanic populations (Table 1 and Supplementary Material, Supplementary Figure S1). Although the mean BMI varied among the five populations, subjects with BMI ⩾27 kg/m2 were classified as obese regardless of the population, ethnicity, and sex. In this study, obesity was not defined as BMI ⩾30 kg/m2, since no male subjects were obese in Kusaghe if defined so. The prevalence of obesity was the highest in the Polynesians (87.8% for Nuku’alofa and 78.1% for Ha’apai), followed by the Micronesians (48.3% for Ravaki) and the Melanesians (42.9% for Munda and 13.4% for Kusaghe) (Table 1).
A screening for variation in the entire region of ADRB2 detected 17 SNPs in the Oceanic subjects (Figure 1a and Supplementary Material, Supplementary Figure S3a). Seven SNPs were selected from the 17 SNPs based on the linkage disequilibrium (LD) between SNPs (Supplementary Material, Supplementary Figure S3b). Two SNPs, rs1042711 and rs1042714, were additionally selected, since the significant associations of these SNPs with obesity had been previously reported.7, 8, 9, 18 These nine SNPs were genotyped for 694 Oceanic subjects (Supplementary Material, Supplementary Table S1). Although a significant deviation from the Hardy-Weinberg equilibrium was observed at rs1042714 in the Kusaghe subjects (Supplementary Material, Supplementary Table S1), their genotype data were included in the following statistical analyses since the Hardy-Weinberg P-value of 0.04 for rs1042714 in Kusaghe was not markedly small.
A multiple logistic regression analysis adjusted for age, sex, and population was performed for each of the nine SNPs (Table 2 and Figure 1b). The results revealed that five SNPs, rs34623097, rs11959427, rs1042711, rs1042714 (Gln27Glu), and rs1042720, were significantly associated with obesity. Among these five SNPs, rs34623097-A was the strongest risk factor for obesity (P=5.6 × 10−4, odds ratio [OR]=2.5, 95% confidence interval [CI]=1.5–4.2). After the significance level α was corrected by the number of SNPs tested with the Bonferroni correction (ie, α=0.0055=0.05/9), rs1042714 and rs1042720 were not significantly associated with obesity. To evaluate the independent effect of each SNP on risk for obesity, a logistic regression analysis with rs34623097, rs11959427, rs1042711, rs1042714, and rs1042720 as independent variables adjusted for age, sex, and population was also performed (Supplementary Material, Supplementary Table S2). Since only rs34623097 showed a statistically significant P-value (P=0.033), the other four SNPs were judged to have non-significant independent effects. In other words, the significant association between each of rs11959427, rs1042711, rs1042714, and rs1042720 and obesity may have resulted from the LD with rs34623097 (Supplementary Material, Supplementary Figure S4). The association of rs34623097-A with obesity was also evaluated in each population (Supplementary Material, Supplementary Table S3). Although the association was statistically significant only in Rawaki (P=0.0263, OR=2.4, 95% CI=1.1–5.0), the ORs were more than 1 in all the populations. Lack of significant association in each of the other four populations would have come from low statistical power due to the low allele frequency of rs34623097-A and the small sample size of each population.
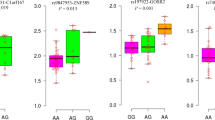
Next, to assess the effect of rs34623097 on BMI, mean BMI was compared among rs34623097 genotypes by using an analysis of covariance (ANCOVA) model. Since age, sex, and population were also detected as significant factors, the mean and standard error of BMI adjusted for age, sex, and population were calculated for each genotype (Figure 2). There was a significant difference in BMI among rs34623097 genotypes (P=0.0048), and BMI was higher in subjects with rs34623097-A. A multiple regression analysis adjusted for age, sex, and population indicated that a copy of the obesity-risk allele, rs34623097-A, was associated with a 1.6-kg/m2 increase in BMI (P=0.0019).
The means and standard errors of BMI, stratified by rs34623097 genotypes. Mean BMI was compared among AA, AG, and GG genotypes at rs34623097 using the analysis of covariance (ANCOVA) by adjusting for age, gender, and population. The significant difference in BMI among three genotypes was detected (P=0.0048).
To further examine the possibility that the significant association of rs34623097 with obesity came from LD between rs34623097 and a SNP outside the ADRB2 gene, the values of r2 between rs34623097 and each of 626 SNPs spanning the 500 kb genomic region were calculated in the HapMap samples,14, 15 JPT (Japanese in Tokyo, Japan) and CHB (Han Chinese in Beijing, China) (Supplementary Material, Supplementary Figure S5). The reason why JPT and CHB were used rather than YRI or CEU was because Oceanic populations are genetically closer to JPT and CHB (East Asians) than to YRI (African) and CEU (European) populations.24 No genes are located closely to ADRB2 and no SNPs outside the ADRB2 gene showed the strong LD with rs34623097. From this result, we conclude that rs34623097 is the primary polymorphism, in this genomic region, associated with obesity in the Oceanic populations.
Luciferase assay
Since rs34623097 is located ∼1.8 kb upstream of the translation start codon, rs34623097 may influence the transcriptional activity of ADRB2 if it has a functional significance. We performed a luciferase reporter assay to evaluate the impact of rs34623097 on the transcriptional activity in HEK293T cells. We detected a significant difference in transcriptional activity between the rs34623097-A and -G alleles (Figure 3a, P=7.5 × 10−6). The rs34623097-A allele showed an approximately 10% reduction in the transcriptional activity compared to the rs34623097-G allele.
Functional analysis of rs34623097. (a) Luciferase reporter assay of ADRB2 rs34623097 SNP. The reporter gene construct with rs34623097-A or rs34623097-G was transfected in HEK293T cells. Transcriptional activity was measured using Renilla luciferase as a reporter and was normalized using Firefly luciferase activity. The experiment was replicated five times, with quintuplicate assays performed within each replicate. The results of a total of 25 assays of each allele are divided by the mean of rs34623097-G. The values presented are mean±s.e. after the division (ie, relative values against rs34623097-G). The P-value was obtained by ANCOVA adjusted for replication. The significant difference in transcriptional activity between rs34623097-A and rs34623097-G was detected (P=7.5 × 10−6). (b) Electrophoretic mobility shift assay (EMSA) of ADRB2 rs34623097 SNP. The biotin-labeled probes carrying rs34623097-A and rs34623097-G were incubated with reaction mixtures containing no nuclear extracts (lanes 1 and 4) and with those containing 6 μg of nuclear extracts from HEK293T cells (lanes 2 and 5); lanes 3 and 6: as lanes 2 and 5 plus 200-fold molar excess of unlabeled probes carrying rs34623097-A and rs34623097-G.
Electrophoretic mobility shift assay
The significant difference in transcriptional activity between rs34623097-A and -G may have resulted from a change in the binding affinity with nuclear factors. To elucidate the allelic difference of the nuclear protein binding, we performed an electrophoretic mobility shift assay (EMSA) with HEK293T cell nuclear extract. The DNA-protein complex was detected for the rs34623097-A probe, but not for rs34623097-G, and the rs34623097-A allele-specific binding protein was inhibited by unlabeled oligonucleotide probes (Figure 3b). We further conducted an in-silico search with the TFSEARCH software21 to examine whether rs34623097 is located in any putative mammalian transcription factor-binding sites. The results indicated no putative transcription factor whose binding affinity is affected by rs34623097.
Implications for the origin of rs34623097-A
The rs34623097-A allele is a derived allele since the homolog sites in other mammalian genomes, including the chimpanzee genome, have G. The rs34623097-A allele is found among Oceanic Islanders, Asians, and the Finnish, but it is not found in Africans (Supplementary Material, Supplementary Figure S6). Of particular interest, the Neandertal genome has A at the corresponding site (Supplementary Material, Supplementary Figure S6).23 These observations imply that a G>A mutation at rs34623097 occurred in the Neandertal genome, and then the rs34623097-A allele flowed into the ancestors of present-day humans. To examine this possibility, based on the genotype data retrieved from the 1000 Genomes Project database,22 we analyzed the LD structure around rs34623097 in Asian and Finnish people (Supplementary Material, Supplementary Figure S7a). Next, we estimated the frequencies of haplotypes bearing SNPs that had a minor allele frequency of more than 0.1 and were in LD (D’>0.9) with rs34623097 (Supplementary Material, Supplementary Figure S7b). In this analysis, we considered only SNPs whose sequences of the Neandertal genome were available in the UCSC database (http://genome.ucsc.edu/). A comparison of the haplotypes in human populations with the Neandertal genome revealed that the haplotype bearing rs34623097-A in present-day humans had the same allele (ie, sequence) at each SNP as the Neandertal haplotype (Supplementary Material, Supplementary Figure S7b). In other words, Neandertals and present-day humans share rs34623097-A on the same 21-kb genomic segment. Although only genomes from three Neandertals were analyzed, and the read depth obtained by using next generation sequencing was very low,23 all Neandertal individuals seem to have the same segment or haplotype bearing rs34623097-A, and the population frequency is estimated to have been high in Neandertals. These results suggest that a G>A mutation at rs34623097 occurred in Neandertals and then the rs34623097-A allele flowed into the common ancestors of the Oceanic Islanders, Asians, and Finnish.
Discussion
To our knowledge, this is the first study in which a significant association of rs34623097 with obesity or BMI was detected. Functional analyses indicated that rs34623097-A reduced the transcriptional activity of the luciferase reporter gene compared to rs34623097-G. Our results suggest that the lower expression of ADRB2 caused by rs34623097-A increases the risk for obesity. It has been reported that ADRB2 expression is decreased in omental adipose tissues obtained from obese subjects as compared to those from lean subjects.25 Taken together, these observations indicate that decreased expression of ADRB2 in adipose tissue is one of the underlying causes of obesity. ADRB2 would be a potential therapeutic target for the development of anti-obesity drugs.
Since the gene flow between Neandertals and present-day humans is considered to have occurred before the divergence of non-African populations,23 rs34623097-A would have existed for at least 50 000 years in ancestors of Asians, Oceanic Islanders, and the Finnish. This period could be long enough for rs34623097-A to increase in frequency due to positive selection, if it operates. However, the frequency of rs34623097-A is still low in present-day human populations, and no extended LD is observed for rs34623097-A (Supplementary Material, Supplementary Figure S7a). Thus, it would be reasonable to postulate that rs34623097-A has not been subjected to strong positive selection in present-day human populations, although we do not exclude the possibility that rs34623097-A was beneficial for Neandertals.
It has been considered that obesity in Oceanic populations may be explained by Neel’s ‘thrifty genotype’ model26 in which genotypes responsible for improved energy storage conferred a selective advantage during their voyage to Oceania; however, in modern societies with a constant abundance of food, these genotypes increase the risk for obesity. In this model, the frequencies of alleles associated with the risk for obesity are expected to be higher in populations that have experienced long distance voyages, such as the Polynesians. The allele frequency of rs34623097-A was not increased in the Polynesians compared to the other Oceanic populations (Supplementary Material, Supplementary Figure S6). The present observation does not seem to support Neel’s ‘thrifty genotype’ model for Oceanic populations. To evaluate ‘thrifty genotype’ model for the Oceanic populations and obtain conclusive evidence, more polymorphisms associated with obesity are required to be identified.
In this study, we showed that rs34623097-A upstream of ADRB2 was significantly associated with an increased risk for obesity in the Oceanic populations. The functional analysis revealed that rs34623097-A leads to lower expression of the luciferase reporter gene, suggesting lower expression of ADRB2. The lower expression of ADRB2 may be a risk for obesity in the Oceanic populations. A previous meta-analysis reported that the 27Glu allele significantly increased the risk for obesity in Asians, Pacific Islanders, and Native Americans, but not in Europeans.12 The 27Glu allele is commonly observed in worldwide populations (Supplementary Material, Supplementary Figure S8), whereas rs34623097-A is observed in Asian and Oceanic populations, but not in Europeans except for the Finnish (Supplementary Material, Supplementary Figure S5). Since rs34623097 is in LD with Gln27Glu (rs1042714) in CHB, CHS, JPT, and FIN populations (Supplementary Material, Supplementary Figure S7) as well as in the Oceanic populations (Supplementary Material, Supplementary Figure S4), the association of 27Glu with obesity that has been found in populations of Asian ancestry12 may have come from the positive LD between rs34623097-A and 27Glu. Since Europeans do not have rs34623097-A, an apparent association of 27Glue with obesity, as found in Asian populations, may have not been detected in populations of European ancestry in a meta-analysis.12 This should be investigated in future studies.
References
Friedman JM. Obesity: Causes and control of excess body fat. Nature 2009; 459: 340–342.
Frayling TM, Timpson NJ, Weedon MN, Zeggini E, Freathy RM, Lindgren CM et al. A common variant in the FTO gene is associated with body mass index and predisposes to childhood and adult obesity. Science 2007; 316: 889–894.
Dina C, Meyre D, Gallina S, Durand E, Korner A, Jacobson P et al. Variation in FTO contributes to childhood obesity and severe adult obesity. Nature genetics 2007; 39: 724–726.
Scuteri A, Sanna S, Chen WM, Uda M, Albai G, Strait J et al. Genome-wide association scan shows genetic variants in the FTO gene are associated with obesity-related traits. Plos Genet 2007; 3: 1200–1210.
Ohashi J, Naka I, Kimura R, Natsuhara K, Yamauchi T, Furusawa T et al. FTO polymorphisms in oceanic populations. J Hum Genet 2007; 52: 1031–1035.
Karns R, Viali S, Tuitele J, Sun G, Cheng H, Weeks DE et al. Common variants in FTO are not significantly associated with obesity-related phenotypes among Samoans of Polynesia. Annals of human genetics 2012; 76: 17–24.
Gjesing AP, Andersen G, Burgdorf KS, Borch-Johnsen K, Jorgensen T, Hansen T et al. Studies of the associations between functional beta2-adrenergic receptor variants and obesity, hypertension and type 2 diabetes in 7808 white subjects. Diabetologia 2007; 50: 563–568.
Gjesing AP, Sparso T, Borch-Johnsen K, Jorgensen T, Pedersen O, Hansen T et al. No consistent effect of ADRB2 haplotypes on obesity, hypertension and quantitative traits of body fatness and blood pressure among 6514 adult Danes. PloS one 2009; 4: e7206.
Hayakawa T, Nagai Y, Kahara T, Yamashita H, Takamura T, Abe T et al. Gln27Glu and Arg16Gly polymorphisms of the beta2-adrenergic receptor gene are not associated with obesity in Japanese men. Metabolism: clinical and experimental 2000; 49: 1215–1218.
Pereira AC, Floriano MS, Mota GF, Cunha RS, Herkenhoff FL, Mill JG et al. Beta2 adrenoceptor functional gene variants, obesity, and blood pressure level interactions in the general population. Hypertension 2003; 42: 685–692.
Bengtsson K, Orho-Melander M, Melander O, Lindblad U, Ranstam J, Rastam L et al. Beta(2)-adrenergic receptor gene variation and hypertension in subjects with type 2 diabetes. Hypertension 2001; 37: 1303–1308.
Jalba MS, Rhoads GG, Demissie K. . Association of codon 16 and codon 27 beta 2-adrenergic receptor gene polymorphisms with obesity: a meta-analysis. Obesity (Silver Spring) 2008; 16: 2096–2106.
Duarte NL, Colagiuri S, Palu T, Wang XL, Wilcken DE. . Obesity, Type II diabetes and the beta 2 adrenoceptor gene Gln27Glu polymorphism in the Tongan population. Clin Sci (Lond) 2003; 104: 211–215.
The International HapMap Consortium. A haplotype map of the human genome. Nature 2005; 437: 1299–1320.
The International HapMap Consortium. A second generation human haplotype map of over 3.1 million SNPs. Nature 2007; 449: 851–861.
Ohashi J, Naka I, Tokunaga K, Inaoka T, Ataka Y, Nakazawa M et al. Brief communication: mitochondrial DNA variation suggests extensive gene flow from Polynesian ancestors to indigenous Melanesians in the northwestern Bismarck Archipelago. American journal of physical anthropology 2006; 130: 551–556.
Ohashi J, Yoshida M, Ohtsuka R, Nakazawa M, Juji T, Tokunaga K. . Analysis of HLA-DRB1 polymorphism in the Gidra of Papua New Guinea. Human biology 2000; 72: 337–347.
Yamada K, Ishiyama-Shigemoto S, Ichikawa F, Yuan X, Koyanagi A, Koyama W et al. Polymorphism in the 5'-leader cistron of the beta2-adrenergic receptor gene associated with obesity and type 2 diabetes. The Journal of clinical endocrinology and metabolism 1999; 84: 1754–1757.
Nishida N, Tanabe T, Takasu M, Suyama A, Tokunaga K. . Further development of multiplex single nucleotide polymorphism typing method, the DigiTag2 assay. Analytical biochemistry 2007; 364: 78–85.
Barrett JC, Fry B, Maller J, Daly MJ. . Haploview: analysis and visualization of LD and haplotype maps. Bioinformatics 2005; 21: 263–265.
Heinemeyer T, Wingender E, Reuter I, Hermjakob H, Kel AE, Kel OV et al. Databases on transcriptional regulation: TRANSFAC, TRRD and COMPEL. Nucleic acids research 1998; 26: 362–367.
1000 Genomes Project Consortium. A map of human genome variation from population-scale sequencing. Nature 2010; 467: 1061–1073.
Green RE, Krause J, Briggs AW, Maricic T, Stenzel U, Kircher M et al. A draft sequence of the Neandertal genome. Science 2010; 328: 710–722.
Ohashi J, Naka I, Kimura R, Tokunaga K, Yamauchi T, Natsuhara K et al. Polymorphisms in the ABO blood group gene in three populations in the New Georgia group of the Solomon Islands. Journal of human genetics 2006; 51: 407–411.
Gomez-Ambrosi J, Catalan V, Diez-Caballero A, Martinez-Cruz LA, Gil MJ, Garcia-Foncillas J et al. Gene expression profile of omental adipose tissue in human obesity. FASEB journal: official publication of the Federation of American Societies for Experimental Biology 2004; 18: 215–217.
Neel JV. Diabetes mellitus: a ‘thrifty’ genotype rendered detrimental by ‘progress’? Am J Hum Genet 1962; 14: 353–362.
Acknowledgements
We would like to sincerely thank the people of the Solomon Islands, Tonga, Papua New Guinea, and Mongol for their kind approval and support of our research. We are grateful to Dr Taniela Palu (Diabetes Clinic, Kingdom of Tonga), Dr Viliami Tangi (Minister of Health, Kingdom of Tonga), and Prof. Kazumichi Katayama (Kyoto University, Japan) for their cooperation in the study of the Tongan populations. We also thank the chiefs, elders, and church leaders, especially Sir. Ikan Rove of the Christian Fellowship Church of the Solomon Islands and the staff of the National Gizo Hospital and Helena Goldie Hospital, especially Mr. Ricky Eddie, for their help with the surveys in the Solomon Islands. We would like to express our gratitude to Dr Lkhagvasuren Munkhtulga (Health Science University of Mongolia, Mongolia) for his cooperation in the blood sampling from the Mongol subjects. We thank two anonymous reviewers for their helpful comments and suggestions. This work was partly supported by a KAKENHI (22370084) Grant-in-Aid for Scientific Research (B) and a Grant-in-Aid for JSPS Fellows (24.5) from the Ministry of Education, Culture, Sports, Science, and Technology of Japan.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Additional information
Supplementary Information accompanies the paper on International Journal of Obesity website
Supplementary information
Rights and permissions
This work is licensed under the Creative Commons Attribution-NonCommercial-No Derivative Works 3.0 Unported License. To view a copy of this license, visit http://creativecommons.org/licenses/by-nc-nd/3.0/
About this article
Cite this article
Naka, I., Hikami, K., Nakayama, K. et al. A functional SNP upstream of the beta-2 adrenergic receptor gene (ADRB2) is associated with obesity in Oceanic populations. Int J Obes 37, 1204–1210 (2013). https://doi.org/10.1038/ijo.2012.206
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/ijo.2012.206
Keywords
This article is cited by
-
Workplace bullying increases the risk of anxiety through a stress-induced β2-adrenergic receptor mechanism: a multisource study employing an animal model, cell culture experiments and human data
International Archives of Occupational and Environmental Health (2021)