Abstract
A multidisciplinary approach has identified sigmoidal genetic clines on the east and west coasts in central New Zealand where low-density ecological interactions occur between northern and southern lineages of the endemic greenshell mussel, Perna canaliculus. The sigmoidal clines indicate the existence of a mussel hybrid zone in a region of genetic discontinuities for many continuously distributed coastal taxa, in particular marine invertebrates. Examination of the genetic architecture of the hybrid zone revealed the differential contribution of individual microsatellite loci and/or alleles to defining the zone of interaction and no evidence of increased allelic richness or heterozygosity inside versus outside the hybrid zone. Genomics cline analysis identified one locus in particular (Pcan1–27) as being different from neutral expectations, thereby contributing to lineage differentiation. Estimates of contemporary gene flow revealed very high levels of within-lineage self-recruitment and a hybrid zone composed mostly (~85%) of northern immigrants. Broad scale interpretation of these results is consistent with a zone of genetic interaction that was generated between 0.3 and 1.3 million years before present at a time of pronounced global sea-level change. At that time, the continuous distribution of the greenshell mussel was split into northern and southern groups, which differentiated to become distinct lineages, and which have subsequently been reunited (secondary contact) resulting in the generation of the hybrid zone at ~42°S.
Similar content being viewed by others
Introduction
Hybrid zones are of interest to biologists for insights that they provide into interactions between genetically differentiated groups such as species or lineages (Harrison, 1990; Gardner, 1997; Abbott et al., 2013). Natural hybridisation, on land and in the sea, is reasonably common and widespread across plant and animal taxa, and has been occurring for millions of years (Harrison, 1990; Arnold, 1997; Gardner, 1997). The geographic location of a hybrid zone may reflect the evolutionary histories of the two taxa (for example, Hewitt, 2000), or it may reflect differences in the environmental preferences of the species, for example, as a mosaic or an environmental gradient (Harrison, 1990; Gardner, 1994; Bierne et al., 2002, 2003; Luttikhuizen et al., 2012). In addition, hybrid zones may be important sources of genetic novelties that may ultimately spread through the zone and into one or both parental populations and such new genetic combinations may have a role in the formation of new species (Hird and Sullivan, 2009; Abbott et al., 2013).
The architecture of a hybrid zone is dependent on the genetic interactions of the genomes, the extent of which will largely depend on the degree of similarity between them (Arnold, 1997; Hewitt, 2000; Gompert and Buerkle, 2009). Genomes with extensive genetic differentiation may be limited in their interactions, whereas lineages with small genetic differences may give rise to extensive hybridisation (that is, a secondary contact zone). Interbreeding between the two distinct source populations may break-up the coadapted gene complexes of parental types to reveal fitness differences among the admixed multilocus genotypes (Harrison, 1990; Gardner, 1997; Gompert and Buerkle, 2011). The presence/absence of backcrosses (admixture or hybrid swarm) is indicative of the relative fitness of individuals within the zone (Gardner, 1994; Bierne et al., 2003; Gompert and Buerkle, 2009).
Hybrid zones are barriers to gene flow between parental types and as such can be sigmoidal in shape, describing the transition from one genetic group to the other through a transition zone (Harrison, 1990; Arnold, 1997; Sotka and Palumbi, 2006). One of the key features of hybrid zones is the rate of change of gene frequency across the zone, and the associated strength of the barrier to gene flow. Gene flow through the hybrid zone may be unidirectional (asymmetric) or bidirectional (symmetric), and may be short or long in distance depending on the circumstances (Arnold, 1997; Abbott et al., 2013). Genes associated with reduced fitness in hybrids or those that contribute to assortative mating will not move far into the hybrid zone or be able to introgress into the genome of the other parental type, whereas genes that promote hybrid fitness are likely to introgress rapidly (Fitzpatrick et al., 2009). Whilst hybridisation may be extensive, introgression does not have to occur at high rates, if at all (Brannock et al., 2009) and rates of introgression are genome-dependent (Abbott et al., 2013).
In recent years, the concept of speciation in the face of gene flow has received considerable attention, although the role of partial reproductive isolation in the speciation process is still not fully understood (Hird and Sullivan, 2009; Luttikhuizen et al., 2012). Recent advances have increased the ability to detect loci that contribute to partial reproductive isolation and adaptive evolution, and the generation of recombinant individuals provides an opportunity to better understand the genetic architecture of reproductive isolation (Gompert and Buerkle, 2009, 2010). For example, genomic clines analysis can be used to quantify the change in frequency of marker genotypes along a genome-wide admixture gradient (Luttikhuizen et al., 2012; Fraïsse et al., 2014). Such analysis is different from the more usual geographic cline analysis that is traditionally applied to geographic gradients. In individuals of mixed ancestry, appropriate contrasts among loci facilitate the identification of how many and which loci decrease fitness of hybrids and thereby contribute to reproductive isolation, or that increase fitness and promote adaptive introgression (Gompert and Buerkle, 2010).
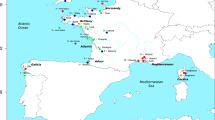
Marine hybrid zones have been recorded from many different taxonomic groups and in many different geographic locations (Gardner, 1997). This study focusses on a marine mollusc, the greenshell mussel, Perna canaliculus, that is endemic to New Zealand (NZ). This mussel is widely distributed with population sizes into the millions, has external fertilisation, produces larvae with a pelagic duration of ~4 weeks and has the potential to produce millions of offspring per female with considerable dispersal potential (Wei et al., 2013a, 2013b). A pronounced genetic discontinuity between northern and southern lineages occurs in central NZ (Figure 1) where genetic discontinuities exist for many coastal taxa (Ross et al., 2009; Gardner et al., 2010; Wei et al., 2013a). Dating places the time of the genetic divergence at ~0.3 to 1.3 million years before present (M ybp) in the greenshell mussel, P. canaliculus, and to ~0.2 to 0.3 M ybp in limpets, Cellana spp. (Apte and Gardner, 2002; Goldstien et al., 2006). This timing is coincident with global fluctuations of sea levels during the Pleistocene and is consistent with the dynamic and complex hydrographic conditions that ultimately gave rise to Cook Strait, the body of water between NZ’s North and South Islands (Knox, 1980; Apte and Gardner, 2002). The patterns of genetic structure that are reported for this region are generally consistent with ancestral single populations of taxa having been split into two (a northern and a southern lineage) with some degree of subsequent isolation during a period of global sea-level upheaval, before being reunited. Thus, interpreting the present situation requires an understanding of the historical events that led to the creation of genetically differentiated lineages within taxa, and contemporary events that may now be eroding or maintaining such genetic differences. Recent investigations have focussed on the population genetics, phylogeography and seascape genetics of multiple taxa (Ross et al., 2009; Gardner et al., 2010; Wei et al., 2013a, 2013b), but no previous study has looked at the genetic architecture of the hybrid zone of any taxon in this region. In this study, we use a variety of different analytical approaches to examine the structure of the greenshell mussel hybrid zone to better understand its architecture and how it is maintained in the face of extensive potential gene flow.
Collection sites of P. canaliculus and hydrographic patterns around NZ. Opononi (OPO); Maunganui (MAU); Castlepoint (CAP); Westhaven (WEST); Fletchers Beach (FLE); Tasman Bay (TAS); Cape Campbell (CAM); Little Wanganui River (LWR); Kaikoura (KAI); Gore Bay (GOB); Timaru (TIM); Fiordland (FIO); Horseshoe Bay (HSB); Big Glory Bay (BGB). ●—northern lineage; ▴—southern lineage; …—areas inside the dashed lines on the east and west coasts contain the centres of the hybrid zone (but not necessarily the full spatial extent of the hybrid zone) and are also regions of pronounced seasonal coastal upwelling. Populations to the north and south of the dashed lines are the northern and southern lineage mussels, respectively; c, cultured mussels; WCC, Wairarapa Coastal Current; WE, Wairarapa Eddy. Reproduced from Wei et al. (2013a).
Materials and methods
Sample collection
Mussels were collected from the low intertidal and shallow subtidal (1–20 m depth) zones at 14 sites from North, South and Stewart Islands, from 1996 to 2008 (Figure 1), covering the distributional range of the mussel in NZ. One sample (Big Glory Bay (BGB)) is from an aquaculture population that is derived from northern mussels transported to Stewart Island, and another (Horseshoe Bay (HSB)) is a wild/native Stewart Island population that shows evidence of introgression of northern genes (Apte et al., 2003; Star et al., 2003; Wei et al., 2013a). These samples are the same mussels as those used previously in studies of population and seascape genetics (Wei et al., 2013a, 2013b).
Microsatellite markers
In total, 311 mussels (mean of 22 mussels per population) were genotyped at 10 polymorphic microsatellite loci (Table 1) with two multiplex reactions using different fluorescent labels or widely separated allelic size ranges (Wei et al., 2013a, 2013b). DNA fragment sizes were identified by comparison with an internal size standard (GeneScan-500) using GENEMAPPER version 3.5 and PEAK SCANNER version 1.0 (Applied Biosystems, Foster City, CA, USA).
Rationale for analytical approach
A variety of different molecular markers have shown that the greenshell mussel can be divided into northern and southern lineages (Apte and Gardner, 2002; Star et al., 2003; Wei et al., 2013a, 2013b). The northern lineage includes all populations north of 42°S on the east coast and 41°S on the west coast (seven populations—Opononi (OPO), Maunganui (MAU), Castlepoint (CAP), Fletchers Beach (FLE), Westhaven (WEST), Tasman Bay (TAS) and Cape Campbell (CAM)), whereas the southern lineage consists of all populations below 42°S on the east coast and 41°S on the west coast, including the BGB aquaculture population (seven populations—Little Wanganui River (LWR), Kaikoura (KAI), Gore Bay (GOB), Fiordland (FIO), Timaru (TIM), BGB and HSB). As a result of the influence of anthropogenic activities on BGB and HSB, these populations were not included in all analyses.
Statistical analysis of the mussel hybrid zone is challenging for several different reasons. First, most marine bivalve populations are characterised by heterozygote deficiencies at many loci (Wei et al., 2013a). Not only is this ‘universal’ phenomenon a potential problem in its own right in terms of violating assumptions of many analyses, it may also contribute to difficulties in the detection of linkage disequilibrium (Sotka and Palumbi, 2006). Second, there are no fixed allelic differences that define the two lineages (Wei et al., 2013a). Third, mussel occurrence is very sparse in the region of interaction (Apte and Gardner, 2002; Wei et al., 2013a, 2013b). In recognition of these challenges and limitations, we have used a variety of different approaches to better understand the architecture of the hybrid zone that exists between northern and southern lineages of the greenshell mussel.
Hardy–Weinberg equilibrium, linkage disequilibrium, allelic richness, null alleles and F-statistics
Estimates of observed (HO) and expected (HE) heterozygosity, conformation to Hardy–Weinberg equilibrium, genotypic linkage equilibrium, allelic richness per locus (A) and Wright’s F-statistics were calculated using FSTAT v 2.9.3 (Lausanne, Switzerland), POPGENE v1.32 (Edmonton, AB, Canada), and ARLEQUIN v3.11 (Bern, Switzerland). MICROCHECKER v 2.2.3 (Hull, UK) was used to check for stutters and allele drop-out, and both MICROCHECKER and GENEPOP (Montpellier, France) to check for null alleles (Wei et al., 2013a). We corrected for multiple testing using the false discovery rate procedure (Verhoeven et al., 2005). Locus-specific values of HO, HE and A were tested using permutational analysis of variance using unrestricted permutation of the raw data and 999 permutations (PERMANOVA in PRIMER6—Anderson et al., 2008) to test for differences in mean values for (a) northern (reference populations=OPO, MAU and CAP) and southern (reference populations=GOB, TIM and FIO) lineage and for populations in the central region (putative admixed populations—FLE, WEST, TAS, CAM, KAI and LWR) where the hybrid zone exists, and (b) outside (OPO, MAU, CAP, GOB, TIM and FIO) versus inside the hybrid zone (FLE, WEST, TAS, CAM, KAI and LWR).
Cline shape, width and location
A curve fitting function (SLIDEWRITE PLUS v3, Advanced Graphics, Sunnyvale, CA, USA) was used to fit a sigmoidal plot [y=a0+a1/(1+exp(–(x–a2)/a3))] that defines hybrid zones, where a0 and a1 are the start and end frequencies of the gene or locus or hybrid index score (that is, values at the northern and southern ends of the cline), and a2 and a3 are the mid-point and width of the cline, respectively. When the curve fitting routine had difficulty finding the minimum coefficients such values were provided in the general area of the solution to locate the global minimum (a true least squares fit). We plotted as a function of latitude the change in allele proportion for all alleles where the difference in the allele proportion at the ends of the cline was >0.05 (that is, 10 of 202 alleles). Values for all 14 populations were plotted, but the BGB and HSB populations were not included in the calculations of cline parameters.
Admixture mapping
The R-language genomic clines analysis programme INTROGRESS, a form of admixture mapping that quantifies the behaviour of individual loci within the context of genome-wide introgression, was used to examine the genetic architecture of the hybrid zone (Gompert and Buerkle, 2009, 2010). Parental allele frequencies are calculated from populations on either side of the hybrid zone using a bi-allelic system (for example, P1 and P2) so that alleles of individuals in the admixture are coded as 0, 1 or 2. A hybrid index is then calculated using a maximum likelihood approach to provide genome-wide estimates of admixture for each individual in the hybrid zone. In this case, low hybrid index scores (values <0.5) were typical of the northern lineage (OPO, MAU and CAP) and high hybrid index scores (values >0.5) were typical of the southern lineage (FIO, GOB and TIM). Logistic regression was used to fit genomic clines for individuals in the region of the hybrid zone (that is, the admixed population). The regressions use the observed data to estimate probabilities of observing homozygous and heterozygous genotypes as a function of the hybrid index values. Genomic clines are estimated for the observed data and then significance testing for departures from neutral expectations is carried out for each genomic cline. A parametric approach was used to generate neutral expectations because the northern and southern lineages do not exhibit fixed allele frequency differences. This involves the simulation of individual genotypes from the parental and admixed population data (that is, the allele frequency differentials between parental populations, hybrid index estimates and deviations from expected heterozygosity). A plot of patterns of introgression was produced for all markers and individuals in the region of admixture. This analysis takes an overview of all markers and all individuals at the same time, and produces a plot where individual mussel affiliation (north P1/P1, hybrid P1/P2 and south P2/P2) is represented for the 10 loci. Each rectangle within the plot denotes an individual’s genotype at a given locus. In addition, a plot of the fraction of the genome that was inherited from population 2 (the southern lineage) reveals the nature of the transition from the P1/P1 via the P1/P2 to the P2/P2 state. In an ideal theoretical data set, this curve will follow a trajectory with lowest rates of change at both ends (where proportions of P2 are 0 and 1, respectively) and the greatest rate of change in the centre (where proportion of P2 is 0.5). In other words, if a hybrid zone is present the plot represents a sigmoidal change from the southern to the northern lineage that captures the multi-locus and multi-individual nature of the full data set. In addition to the above, but at the level of each individual marker or locus, Gompert and Buerkle (2010) have shown for a simulated data set that locus-specific genomic cline plots can represent a hybrid zone particularly well. Such plots aid in the identification of individual loci that are (or are not) under selection. Each locus-specific plot shows a response that is representative of the P1/P1 state (the northern lineage in dark green) and also a response that is representative of the P1/P2 state (the hybrid mussels in light green). Each response is also represented by a 95% confidence interval (solid line for P1/P1, dashed line for P1/P2). For an individual locus under selection, the P1/P1 response will start at the top left of the plot (where hybrid index values are 0, or nearly so) and will decrease rapidly as hybrid index values approach 0.5, and will be 0, or nearly so, as hybrid index values approach 1.0. The P1/P2 response is expected to be 0 when hybrid index values are low, to increase as hybrid index values approach 0.5, and then to decrease as hybrid index values approach 1.0. That is, the P1/P2 response describes a tightly defined peak around hybrid index values of ~0.5. In contrast, for an individual locus that is not under such pronounced selection (or not under any selection), the graphical form of the response will be relaxed (that is, the gradient of change of P1/P1 will be less steep and the shape of the P1/P2 curvilinear response will be flatter and more widely spread). Different forms of selection (underdominance, overdominance and epistasis, directional with incomplete dominance) modify the clines in predictable ways away from expectations under neutrality, and can therefore be tested for (Gompert and Buerkle, 2009). The data represented in each locus-specific plot are tested (as described above) for fit to neutral expectations and the associated significance level is presented with each plot so that the possible contribution of each locus to the structure of the hybrid zone can be assessed. Inevitably, the results of these analyses reflect the information content of the markers being used. As we do not know to what extent the microsatellite markers represent the mussel genome as a whole (that is, we do not know their chromosomal locations), the interpretation of the extent of genomic differentiation between the lineages may change when additional markers are included in the analysis. Gompert and Buerkle (2009, 2010) provide a comprehensive explanation of the genomic clines approach, including examples of genomic admixture plots and marker-specific introgression plots, and their interpretations. We ran the analysis with the admixed population containing the four populations closest to the hybrid zone (that is, CAM, FLE, TAS and WEST) and then with the admixed population containing the six populations closest to the hybrid zone (that is, CAM, FLE, TAS, WEST, KAI and LWR).
Recent migration rates
BAYESASS v3 software (Wilson and Rannala, 2003) was used to estimate recent migration rates among/within the northern (OPO, MAU and CAP) and southern (FIO, GOB and TIM) lineages and the populations nearest to the hybrid zone (WEST, FLE, TAS, CAM, KAI and LWR), and to determine individual posterior probabilities of migrant ancestry (populations BGB and HSB were excluded from this analysis). BAYESASS detects migrants within the past few generations, using a Markov chain Monte Carlo approach to calculate posterior probabilities for migrant ancestries (non-migrant, first- or second-generation migrant). As it does not assume Hardy–Weinberg equilibrium for these assignments, it is appropriate for the present data set. Evaluation of BAYESASS indicates that it works well when FST⩾0.05 (Faubet et al., 2007), a condition that is met some, but not all of the time for the two mussel lineages. Following preliminary testing to ascertain mixing parameters, we used 106 iterations, a burn-in run of 16 and interval sampling of 100. Multiple runs with different seed numbers were used to check for consistency of results and trace plots were examined using TRACER software (v1.5.0) to ensure adequate mixing (http://beast.bio.ed.ac.uk/). We tested two different scenarios: first, with the four populations in closest proximity to the hybrid zone (TAS, FLE, WEST and CAM), and second with the six populations in closest proximity to the hybrid zone (TAS, FLE, WEST, CAM, KAI and LWR).
Results
Hardy-Weinberg equilibrium, linkage disequilibrium, allelic richness, null alleles and F-statistics
Of the 139 tests, 104 revealed heterozygote deficiencies, 27 revealed heterozygote excesses and 9 revealed no difference between HO and HE (Table 1). After correction for multiple testing, 25 tests revealed significant heterozygote deficiencies and 1 test revealed a significant heterozygote excess, with two loci, Pcan6–17 and Pcan1–29, accounting for most instances. PERMANOVA testing for differences in mean HO or mean HE revealed no significant differences among the mussels of the northern, central and southern regions (P=0.523 and P=0.527, respectively) or between the mussels outside and inside the hybrid zone (P=0.811 and P=0.696, respectively).
There was no evidence of significant linkage disequilibrium between pairs of loci within each population or between pairs of loci within the northern and southern lineages (P>0.05 in all cases). Only Pcan2–17 × Pcan1–44 exhibited statistically significant linkage disequilibrium (P<0.0001) when data for all 14 populations were pooled. As only 1 of 765 tests was statistically significant after correction for multiple testing, and then only when data were pooled for all populations, we retained all loci in the analyses.
PERMANOVA testing for differences in mean allelic richness (A) per locus (Table 1) revealed no significant differences among the north, centre and south regions (P=0.212) or between inside versus outside the hybrid zone (P=0.129).
Estimates of null allele frequency per population ranged from 0.036 to 0.077, whereas null allele frequency per locus ranged from 0.021 to 0.136. There was no pattern in null allele frequency with respect to population or locus.
Estimates of FIS were significantly different from 0 at 1 locus, FIT was significantly different from 0 at 6 of 10 loci and for ‘all loci’ (Weir and Cockerham, 1984; Nei, 1987), and FST was significantly different from 0 at 2 loci (Weir and Cockerham, 1984) and significantly different from 0 at 7 loci and over ‘all loci’ (Nei, 1987) (refer to Supplementary Table S1 of Supplementary Information).
Cline shape, width and location
Plots of allele proportion as a function of latitude revealed sigmoidal shapes, although the plot of Pcan10–369 had a mid-point outside the hybrid zone (Figure 2). For the other nine plots, the mid-point value (a2) of the hybrid zone was in the range 40.8648 to 42.5147 (°S), with a mean±s.d. of 41.686±0.625, indicating that the centre location of the hybrid zone is geospatially consistent regardless of allele or locus being considered. In some cases, the cline was a steep sigmoidal curve (narrow hybrid zone, limited penetration of allele across the zone), whereas in others it was relatively wide (for example, Pcan1–2712 versus Pcan10–3611). Change in allele proportion as a function of latitude revealed variable allelic responses within a locus (Table 2). Estimates of cline width ranged from 0.35 km (Pcan10–3611) to 121 km (Pcan1–2712), with a mean±s.d. of 35.57 km±0.23, indicative of pronounced allele-specific differences of penetration across the hybrid zone (Table 2).
Sigmoidal cline plots for the 10 alleles showing greatest change in allele frequency as a function of latitude. In each case, the cline fit is based on data from 12 populations; the HSB population (circle ○) and the BGB population (black triangle ▴) shown for comparison. The hybrid zone is located in the areas of steepest gradient of change of allelic frequency. Note the different y axis ranges that are necessary to show detail of the sigmoidal curves.
Admixture mapping
INTROGRESS analyses confirmed the geographic location of the hybrid zone and identified those loci that contributed to significant differences between the northern and southern lineages. Population-specific hybrid index mean scores for mussels from the four populations closest to the hybrid zone were <0.5 and indicated greater affinity with northern than southern lineages (mean±s.d.; all 0.407±0.349; CAM 0.400±0.372; FLE 0.441±0.444; TAS 0.284±0.233; WEST 0.151±0.291). The addition of the next two geographically proximate populations emphasised these northern affinities (mean±s.d.; all 0.341±0.359; CAM 0.326±0.330; FLE 0.437±0.441; TAS 0.303±0.298; WEST 0.186±0.290; KAI 0.630±0.284; LWR 0.576±0.309) and highlighted the southern affinities of the KAI and LWR populations, which had mean hybrid index scores >0.5. In the four population analysis, 3 of the 10 loci were significantly different from neutral expectations (Pcan6–17, P=0.012; Pcan2–17, P=0.041; Pcan1–27, P=0.008), whereas in the six population analysis 2 loci were significantly different from neutral expectations (Pcan1–29, P=0.023; Pcan1–27, P=0.022). Differences between the northern and southern lineages were most pronounced for the four population analysis (Figure 3). However, none of the genome plots fitted the theoretical distributions expected of admixed populations, although in both cases locus Pcan1–27 came closest (see Gompert and Buerkle, 2009, 2010), and as a consequence it was not possible to identify the model of selection (underdominance, overdominance, epistasis, directional with incomplete dominance). Plots of the proportions of lineage ancestry for the four population test supported the genome clines analysis and revealed the change from northern (dark green) to southern (light green) genetic identity across the region of the hybrid zone (Figure 4a). Although this transition from northern to southern identity was not as pronounced as in a theoretical example (Gompert and Buerkle, 2010), the summary proportion plot clearly indicates the change in hybrid index scores for all mussels across all loci in this region of central NZ. The addition of the KAI and LWR (southern) populations to the admixture group substantially changed the balance of north versus south ancestry within the analysis, with more of the plot being light green, that is, exhibiting southern affinities (Figure 4b). Nonetheless, the summary proportion plot showed a change in hybrid index scores for all mussels across all loci in this region of central NZ, but with a more shallow gradient of change than the four population test.
Genomic cline plots from the INTROGRESS analysis for 10 locus-specific microsatellite markers. (a) Four population analysis, (b) six population analysis. Locus designation and P-values per locus are given in each plot (P-values are after false discovery rate (FDR) correction). Solid coloured regions represent the 95% confidence intervals for the northern (dark green and solid line) and hybrid (light green and dashed line) genotypes. Circles indicate the raw genotype data (northern on the top line; hybrid in the middle; southern on the bottom line), with counts of individuals on the right vertical axis. A full colour version of this figure is available at the Heredity journal online.
Plot of patterns of introgression for all 10 microsatellite loci and individuals in an admixed population (the hybrid zone) composed of (a) four populations and (b) six populations in central NZ. Plot on the right shows the fraction of the genome inherited from population 2 (that is, the northern lineage).
Recent migration rates
BAYESASS analyses supported the scenario of four hybrid zone populations (TAS, FLE, WEST and CAM), rather than six (KAI, LWR, TAS, FLE, WEST and CAM). For the four hybrid zone populations, the posterior probabilities of migrant ancestry indicated that most mussels in the northern (OPO, MAU and CAP) and the southern (FIO, GOB and TIM) regions were directly derived from those regions (98.8% and 85.7%, respectively). One individual in the northern region was a second-generation southern migrant, whereas 16 individuals in the southern region originated from the north. The hybrid zone populations were composed of first-generation northern (84.5%) and southern (15.5%) individuals (Table 3a). The scenario with six hybrid zone populations completely changed the interpretation. All of the southern mussels were identified as first-generation hybrid zone migrants, whilst 53 and 47% of the northern mussels were identified as first- or second-generation hybrid zone immigrants, respectively. All but one of the hybrid zone mussels (99%) were identified as hybrid zone non-migrants (Table 3a).
Detection of migrants over recent generations provided evidence of moderate to high levels of self-recruitment within each of the three regions. For the four population scenario, the northern and southern regions both had self-recruitment levels of >90%, with correspondingly low levels of recruitment from other regions (fractionally more northern individuals contributed to the southern region than vice versa). Populations within the hybrid zone received more migrants from the north than the south. For the six population scenario, the hybrid zone had the highest level of self-recruitment, with the northern and southern regions having very similar moderate levels of self-recruitment. The hybrid zone region contributed far more migrants to other regions than did either the northern or southern regions (Table 3b).
Discussion
A multilocus microsatellite data set from the NZ endemic greenshell mussel, P. canaliculus, has been used to examine the genetic architecture of a natural zone of admixture in central NZ. The data set is challenging to analyse and interpret for several reasons, including its spatial nature (non-linear coastal distances between sites), heterozygote deficiencies, low level of genetic differentiation and absence of fixed differences between lineages, and the sparsely populated region of the hybrid zone itself. In addition, we do not have information about sex-linked markers and nor do we have even a rudimentary genetic linkage map for the greenshell mussel. We do, however, have evidence of the existence of two lineages that interact at a geographic location that is coincident with the existence of contact zones for several other marine coastal taxa (Ross et al., 2009; Gardner et al., 2010). Despite the existence of the challenges noted above, the combined analyses provided a consistent set of results, which has permitted us to develop new insights into the origin and maintenance of this multitaxon hybrid zone.
The origin and structure of the hybrid zone
The region just south of Cook Strait is an area of genetic discontinuities for many continuously distributed coastal marine taxa (Ross et al., 2009; Gardner et al., 2010). Mussels of the genus Perna are subtropical in origin and the fossil record suggests that they arrived in NZ ~25 M ybp (Fleming, 1979; Wood et al., 2007). The distribution of the greenshell mussel includes all three major islands (North, South and Stewart), but excludes the many offshore islands. Analyses reported here support the contention that the greenshell mussel genetic discontinuity is a hybrid zone, formed via secondary contact. That is, ~0.3 to 1.3 M ybp (Apte and Gardner, 2002; Goldstien et al., 2006) the northern and southern populations of the mussel were partially or fully isolated, at a time of global sea-level fluctuations and more recently contact has been re-established between mussels of the two regions. Consistent with many such examples, both terrestrial and marine (Harrison, 1990; Gardner, 1997; Abbott et al., 2013), the mussel hybrid zone is a sigmoidal cline of gene frequency changes over a short geographic distance. All analyses support the existence of northern and southern lineages and the existence of populations and/or individuals of mixed ancestry geographically located between the two parental lineages. Cline shape analysis has confirmed the locations of the centres of the hybrid zone on either coast and has identified loci and alleles that describe a wide or a narrow cline (that is, genes that do or do not travel through the hybrid zone, respectively—for example, Nolte et al., 2009; Abbott et al., 2013; Sá-Pinto et al., 2013).
Analyses reported here indicate that the shape and width of the hybrid zone is similar on the east and west coasts, even if the geographic centre of the zone is slightly different (42.20°S on the east, 41.36°S on the west coast). Overall, the findings indicate that the underlying genetic architecture of the mussel hybrid zones on NZ’s east and west coasts is similar and that pronounced environmental factors such as localised currents and/or upwelling are coincident with the present day geographic location of the mussel hybrid zone on either coast. The similarity of the genetic architecture reported here for east and west coasts is in contrast to the situation reported for other aquatic taxa. For example, Nolte et al. (2009) noted that despite the similarity in genomic composition of two independent fish hybrid zones (the sculpin, Cottus spp.), patterns of microsatellite locus introgression were different in the two zones, reflecting different genotypic models of fitness. Such a difference in the extent of genomic isolation (at least as defined by the number and type of markers used for the analyses) between hybrid zones highlights the uncertain role played by different ecological factors and the need to better understand how long-term environmental variability influences the structure of hybrid zones. In addition, the role of life-history characteristics may prove to be important in explaining the location of patterns of genetic structure among coastal taxa. For example, in NZ many sessile invertebrates but no fish species exhibit genetic discontinuities at the same locations (that is, at ~42°S on the east coast and ~41°S on the west coast) as the greenshell mussel hybrid zones (Gardner et al., 2010), suggesting that adult mobility may be an important factor explaining population genetic differentiation in the central region of the country.
Non-neutral loci and reproductive isolation
The approach of admixture mapping of isolation across the genomes of hybrids can provide new and powerful insights into the genetic architecture of hybrid zones (Gompert and Buerkle, 2009). Although many analytical methods require selectively neutral loci (hence widespread use of outlier detection to identify and discard markers under selection), markers under selection (directly or indirectly) may be informative on ecological rather than evolutionary timescales for assigning individuals to populations of origin, and for defining lineage-specific differences (Waples and Gaggiotti, 2006; Wei et al., 2013a). Loci that are different from neutral (genome-wide background) expectations are believed to be linked to genomic regions involved in reproductive isolation (Gompert and Buerkle, 2009; Luttikhuizen et al., 2012) or may be able to introgress at different levels from other neutral loci because of selection effects within or across the hybrid zone (Bierne et al., 2011). As noted by Gompert and Buerkle (2011) it is unknown from genome scans what fraction of the genome has experienced selection and contributes to reproductive isolation.
Within the greenshell mussel hybrid zone, most loci showed evidence of introgression consistent with neutral expectations. The main exception was Pcan1–27, a locus already identified as an outlier (Wei et al., 2013a, 2013b). Although we do not have knowledge of male/female sex determination mechanisms as used in analysis of other hybrid zones (for example, Gompert and Buerkle, 2009), and nor do we have information about chromosomal locations of the 10 microsatellite loci nor linkage relationships, our results indicate that there is little evidence of intralineage variation in the genetic architecture of reproductive isolation between northern and southern mussels. As noted by Wei et al. (2013a, 2013b) the Pcan1–27 locus is a candidate locus for further evaluation for its possible role as a marker of environmental variation (for example, temperature variation, perhaps via heat-shock proteins) or perhaps for a role in reproductive isolation (for example, gamete recognition proteins) across the mussel hybrid zone.
Population genetic structural complexity and gene connectivity
There is now increasing evidence of strong and complex genomic differentiation between groups that at the same time exhibit evidence of a hybrid swarm structure (for example, the marine bivalve Macoma balthica (Luttikhuizen et al., 2012) and the malarial mosquito species Anopheles gambiae (Lawniczak et al., 2010)). Such findings suggest that genomes may be ‘leaky’ and subject to introgression despite the extent of differentiation and the fitness differences between them (Luttikhuizen et al., 2012). In another context, hybridisation may generate novel combinations of genotypes, and if selectively beneficial these may travel beyond the region of the hybrid zone (Hird and Sullivan, 2009; Abbott et al., 2013). Results for the greenshell mussel hybrid zone provide no strong evidence for extensive introgression across the hybrid zone and also no evidence for increased genetic diversity (allelic richness or heterozygosity) in the region of the zone of interaction. These findings most probably reflect the limited genetic differentiation between the two lineages.
Estimates of contemporary gene flow reveal very high levels of self-recruitment within the northern, hybrid zone and southern regions. Although interpretation of the BAYESASS results requires care (not all FST values were >0.05), these estimates of contemporary gene flow and high levels of self-recruitment are consistent with estimates of historical gene flow that also point to very high levels of self-recruitment (Wei et al., 2013a). Where microsatellite marker exchange exists between/among the northern, hybrid zone and southern regions, the results all point to the direction of flow as being predominantly from north to south. This is consistent with earlier work based on mitochondrial markers that reported a unique single strand conformational polymorphism mtDNA haplotype at ~20% frequency in the southern region, with this haplotype being absent from the larger northern region (Apte and Gardner, 2002). These results are consistent with the known patterns of coastal flow, with the West Auckland Current and the East Cape Current flowing from north to south down the west and east coasts, respectively, and with the intermittent flow of the Westland Current and the Southland Current being deflected offshore as they flow south to north, respectively (Apte and Gardner, 2002, and references therein). Thus, historical processes such as global sea-level change may have generated the genetic differences between the two lineages, whereas contemporary processes such as coastal upwelling in the region of the hybrid zone that have only existed much more recently may be acting as a semi-permeable barrier to modern gene flow. Overall, these findings lend support to a growing body of literature that points to the influence of oceanic or coastal features on genetic differentiation and the low effective dispersal rates for many marine species, often independent of early life-history traits and despite a high dispersal potential (Galarza et al., 2009; Luttikhuizen et al., 2012).
Human influence on mussel hybridisation
The status of the BGB aquaculture population and the neighbouring native HSB population in the far south (Stewart Island) is informative in the context of hybridisation. Mussel spat from the far north are collected and trucked to aquaculture sites, including BGB. This northern genetic signal in the far south is clearly identifiable using a variety of genetic markers and analytical approaches (Apte et al., 2003; Star et al., 2003; this study). Thus, human-mediated transfer of mussels over the last ~40 years has created a new geographic area of hybridisation, >750 km south of the natural zone of interbreeding.
Southern and northern mussels are genetically distinct and have not been able to interbreed for many generations because of allopatric separation. Despite this, the recent introduction of northern mussels into Stewart Island has resulted in extensive interbreeding over only a few generations, indicating that there is, at best, limited reproductive isolation between the two lineages despite ~0.3 to 1 M years of isolation. This indicates that the genetic differences that have evolved between the northern and southern lineages affect genes that are not related to reproductive isolation (this may explain why the genomic clines analysis could not produce easy to interpret results), which in turn suggests that it may be environmental factors at the region of natural hybridisation at ~42°S that influence the location and structure of the hybrid zone, rather than pre- or post-zygotic genetic differences that dictate the nature of the interaction. Although this is speculative it deserves further examination and the existence of both the east and west coast natural hybrid zones plus the man-made hybrid region in Stewart Island provide excellent opportunities to further investigate the interactive effects of genetic and environmental factors on hybridisation in the sea.
Data archiving
The data used in this study are available from the Dryad Digital Repository: http://doi.org/10.5061/dryad.gt223.
References
Abbott R, Albach D, Ansell S, Arntzen JW, Baird SJE, Bierne N et al. (2013). Hybridization and speciation. J Evol Biol 26: 229–246.
Anderson MJ, Gorley RN, Clark M . (2008). PERMANOVA+ for PRIMER: Guide to Software and Statistical Methods. PRIMER-E Ltd: Plymouth, UK, 214 p.
Apte S, Gardner JPA . (2002). Population genetic variation in the New Zealand greenshell mussel, Perna canaliculus: SSCP and RFLP analyses of mitochondrial DNA. Mol Ecol 11: 1617–1628.
Apte S, Star B, Gardner JPA . (2003). A comparison of genetic diversity between cultured and wild populations, and a test of genetic introgression in the New Zealand greenshell mussel, Perna canaliculus (Gmelin 1791). Aquaculture 219: 193–220.
Arnold ML . (1997). Natural Hybridization and Evolution. Oxford University Press: New York.
Bierne N, Borsa P, Daguin C, Jollivet D, Viard F, Bonhomme F et al. (2003). Introgression patterns in the mosaic hybrid zone between Mytilus edulis and M. galloprovincialis. Mol Ecol 12: 447–461.
Bierne N, David P, Langlade A, Bonhomme F . (2002). Can habitat specialisation maintain a mosaic hybrid zone in marine bivalves? Mar Ecol Prog Ser 245: 157–170.
Bierne N, Welch J, Loire E, Bonhomme F, David P . (2011). The coupling hypothesis: why genome scans may fail to map local adaptation genes. Mol Ecol 20: 2044–2072.
Brannock PM, Wethey DS, Hilbish TJ . (2009). Extensive hybridisation with minimal introgression in Mytilus galloprovincialis and M. trossulus in Hokkaido, Japan. Mar Ecol Prog Ser 383: 161–171.
Faubet P, Waples RS, Gaggiotti OE . (2007). Evaluating the performance of a multilocus Bayesian method for the estimation of migration rates. Mol Ecol 16: 1149–1166.
Fitzpatrick BM, Johnson JR, Kump DK, Shaffer HB, Smith JJ, Voss SR . (2009). Rapid fixation of non-native alleles revealed by genome-wide SNP analysis of hybrid tiger salamanders. BMC Evol Biol 9: 176.
Fleming CA . (1979). The Geological History of New Zealand and its Life. Auckland University Press: New Zealand, p 141.
Fraïsse C, Roux C, Welch JJ, Bierne N . (2014). Gene-flow in a mosaic hybrid zone: Is local introgression adaptive? Genetics 197: 939–951.
Galarza JA, Carreras-Carbonell J, Macpherson E, Pascual M, Roques S, Turner GF et al. (2009). The influence of oceanographic fronts and early-life-history traits on connectivity among littoral fish species. Proc Natl Acad Sci USA 106: 1473–1478.
Gardner JPA . (1994). The structure and dynamics of naturally occurring hybrid Mytilus edulis Linnaeus, 1758 and Mytilus galloprovincialis Lamarck, 1819 (Bivalvia, Mollusca) populations: review and interpretation. Archiv für Hydrobiologie, Monographische Beitrage 99: 37–71.
Gardner JPA . (1997). Hybridization in the sea. Adv Mar Biol 31: 1–78.
Gardner JPA, Bell JJ, Constable HB, Hannan D, Ritchie PA, Zuccarello GC . (2010). Multi-species coastal marine connectivity: a literature review with recommendations for further research. N Z Aquat Environ Biodiversity Rep 58: 1–47. ISSN 1176-9440.
Goldstien SJ, Schiel DR, Gemmell NJ . (2006). Comparative phylogeography of coastal limpets across a marine disjunction in New Zealand. Mol Ecol 15: 3259–3268.
Gompert Z, Buerkle CA . (2009). A powerful regression-based method for admixture mapping of isolation across the genome of hybrids. Mol Ecol 18: 1207–1224.
Gompert Z, Buerkle CA . (2010). INTROGRESS: a software package for mapping components of isolation in hybrids. Mol Ecol Resour 10: 378–384.
Gompert Z, Buerkle CA . (2011). Bayesian estimation of genomic clines. Mol Ecol 20: 2111–2127.
Harrison RG . (1990). Hybrid zones: windows on evolutionary process. Oxf Surv Evol Biol 7: 69–128.
Hewitt GM . (2000). The genetic legacy of the quaternary ice ages. Nature 405: 907–913.
Hird S, Sullivan J . (2009). Assessment of gene flow across a hybrid zone in red-tailed chipmunks (Tamias ruficaudus). Mol Ecol 18: 3097–3109.
Knox GA . (1980). Plate tectonics and the evolution of intertidal and shallow-water benthic biotic distribution patterns of the southwest Pacific. Palaeogeogr Palaeoclimatol Palaeoecol 31: 267–297.
Lawniczak MKN, Emrich SJ, Holloway AK, Regier AP, Olson M, White B et al. (2010). Widespread divergence between incipient Anopheles gambiae species revealed by whole genome sequences. Science 330: 512–514.
Luttikhuizen PC, Drent J, Peijnenburg KTCA, van der Veer HW, Johannesson K . (2012). Genetic architecture in a marine hybrid zone: comparing outlier detection and genomic clines analysis in the bivalve Macoma balthica. Mol Ecol 21: 3048–3061.
Nei M . (1987). Molecular Evolutionary Genetics. Columbia University Press: New York.
Nolte AW, Gompert Z, Buerkle CA . (2009). Variable patterns of introgression in two sculpin hybrid zones suggest that genomic isolation differs among populations. Mol Ecol 18: 2615–2627.
Ross PM, Hogg ID, Pilditch C, Lundquist CJ . (2009). Phylogeography of New Zealand’s coastal benthos. NZ J Mar Freshwater Res 43: 1009–1027.
Sá-Pinto A, Branco MS, Alexandrino PB, Fontaine MC, Baird SJE . (2013). Barriers to gene flow in the marine environment: insights from two common intertidal limpet species of the Atlantic and Mediterranean. PLoS One 7: e50330.
Sotka EE, Palumbi SR . (2006). The use of genetic clines to estimate dispersal distances of marine larvae. Ecology 87: 1094–1103.
Star B, Apte S, Gardner JPA . (2003). Genetic structuring among populations of the greenshell mussel Perna canaliculus (Gmelin 1791) revealed by analysis of randomly amplified polymorphic DNA. Mar Ecol Prog Ser 249: 171–182.
Verhoeven KJF, Simonsen KL, McIntyre LM . (2005). Implementing false discovery rate control: increasing your power. Oikos 108: 643–647.
Waples RS, Gaggiotti O . (2006). What is a population? An empirical evaluation of some genetic methods for identifying the number of gene pools and their degree of connectivity. Mol Ecol 15: 1419–1439.
Wei K, Wood AR, Gardner JPA . (2013a). Population genetic variation in the New Zealand greenshell mussel: Locus-dependent conflicting signals of weak structure and high gene flow balanced against pronounced structure and high self-recruitment. Mar Biol 160: 931–949.
Wei K, Wood AR, Gardner JPA . (2013b). Seascape genetics of the New Zealand greenshell mussel: sea surface temperature explains macrogeographic scale genetic variation. Mar Ecol Prog Ser 477: 107–121.
Weir BS, Cockerham CC . (1984). Estimating F-statistics for the analysis of population structure. Evolution 38: 1358–1370.
Wilson GA, Rannala B . (2003). Bayesian inference of recent migration rates using multilocus genotypes. Genetics 163: 1177–1191.
Wood AR, Apte S, MacAvoy ES, Gardner JPA . (2007). A molecular phylogeny of the marine mussel genus Perna (Bivalvia: Mytilida) based on nuclear (ITS 1 & 2) and mitochondrial (COI) DNA sequences. Mol Phylogenet Evol 44: 685–698.
Acknowledgements
We thank Smita Apte for sample collection and Smita Apte, Liz MacAvoy, Rose McColl and Ann Wood for help in the lab. This research was supported by funding from the New Zealand Foundation for Research, Science and Technology (contract CAWX0303 to JPAG) and by the China Research Council (to K-JW). We thank three anonymous reviewers and the editor for helpful comments that have improved the manuscript.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Additional information
Supplementary Information accompanies this paper on Heredity website
Supplementary information
Rights and permissions
About this article
Cite this article
Gardner, J., Wei, KJ. The genetic architecture of hybridisation between two lineages of greenshell mussels. Heredity 114, 344–355 (2015). https://doi.org/10.1038/hdy.2014.108
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/hdy.2014.108