Abstract
Association mapping has become a widely applied genomic approach to identify quantitative trait loci (QTL) and dissect the genetic architecture of complex traits. However, approaches to assess the quality of the obtained QTL results are lacking. We therefore evaluated the potential of cross-validation in association mapping based on a large sugar beet data set. Our results show that the proportion of the population that should be used as estimation and validation sets, respectively, depends on the size of the mapping population. Generally, a fivefold cross-validation, that is, 20% of the lines as independent validation set, appears appropriate for commonly used population sizes. The predictive power for the proportion of genotypic variance explained by QTL was overestimated by on average 38% indicating a strong bias in the estimated QTL effects. The cross-validated predictive power ranged between 4 and 50%, which are more realistic estimates of this parameter for complex traits. In addition, QTL frequency distributions can be used to assess the precision of QTL position estimates and the robustness of the detected QTL. In summary, cross-validation can be a valuable tool to assess the quality of QTL parameters in association mapping.
Similar content being viewed by others
Introduction
Association mapping was developed by human geneticists but in recent years has become a widely applied approach in plant genetics to dissect the genetic architecture underlying complex traits. In contrast to linkage mapping, which is based on a biparental family, association mapping uses a diversity panel of lines with different degrees of relatedness (Würschum, 2012). This population structure is a major issue for association mapping as it increases the risk of detecting false-positive quantitative trait loci (QTL) and must therefore be accounted for in the biometrical model (Sillanpää, 2011). Model comparison studies revealed that a model incorporating a kinship matrix performs well, as it adequately controls the false-positive rate without overly reducing the QTL detection power (Yu et al., 2006; Würschum et al., 2011a). Association mapping has been applied to detect QTL for agronomically important traits in different crop species, for example, maize (Buckler et al., 2009; Liu et al., 2011), wheat (Reif et al., 2011a, 2011b; Kulwal et al., 2012; Kollers et al., 2013), barley (Wang et al., 2012a; Berger et al., 2013; Zhou and Steffenson 2013), sugar beet (Stich et al., 2008; Würschum et al., 2011b) and rapeseed (Snowdon et al., 2010; Wang et al., 2012b; Würschum et al., 2012a).
The primary intention of all QTL mapping approaches in crops is the identification of QTL underlying the traits of interest and estimation of their chromosomal positions, their effects, as well as the proportion of genotypic variance explained by them. In plant breeding, the identified QTL can then be used for marker-assisted selection programs that promise to enhance selection gain per time unit. However, for marker-assisted selection to be superior to classical phenotypic selection based on field evaluation, the following criteria must be met: (i) the QTL positions must be estimated accurately to minimize the number of recombinations between the QTL and the markers used for selection, (ii) QTL effects must be estimated with high precision and (iii) the genotypic variance explained by the detected QTL must be high and estimated without bias. For linkage mapping, it has been shown through simulation studies (Beavis 1998) and experimental data analysis (Utz et al., 2000; Schön et al., 2004), that both the QTL effects and the proportion of explained genotypic variance are often overestimated. This results in a wrong, oftentimes excessively optimistic assessment of the prospects of marker-assisted selection. Reasons for this overestimation of the genotypic variance explained by QTL include epistatic and genotype-by-environment interactions, but also that estimation of QTL effects and assessment of their predictive power are done with the same data. To obtain unbiased estimates of QTL effects and to determine the magnitude of bias of the predictive power, different approaches have been suggested. Of these, cross-validation has been shown to perform well for linkage mapping (Utz et al., 2000) but also for association mapping in multiple families (Liu et al., 2013). For cross-validation, QTL detection and effect estimation are done in a subset of the lines (estimation set, ES) while the remaining lines of the population are set aside to serve as an independent validation set (test set, TS). Although this resampling can potentially alter the existing population structure in the subsets as compared with the full data set, it also offers the advantage to evaluate the robustness of the obtained results. QTL which are only detected given a specific composition of the mapping population have a higher probability of being false positives and are also of little interest for use in marker-assisted selection. In plant breeding, the detected QTL will not be used in the population in which they have been detected but in different, oftentimes related germplasm, a situation that is mimicked by cross-validation.
Despite the routine application of association mapping in crop genetics, little attention is paid to the validation of the estimated QTL-related parameters in association mapping. Here, we used a large sugar beet data set to evaluate the potential of cross-validation to address these questions. In particular, the objectives of this study were to (1) determine the optimum proportion of the independent TS in dependence on the population size, (2) assess the bias in the estimation of the proportion of genotypic variance explained by the detected QTL and (3) investigate the robustness of the detected QTL and the precision of QTL position estimates in association mapping.
Materials and methods
Plant materials, field experiments and molecular markers
This study was based on 924 diploid elite sugar beet (Beta vulgaris L.) inbred lines described by Würschum et al. (2011a, 2011b). In brief, testcross progenies were produced by crossing the genotypes to a single-cross hybrid as tester. All material used in this study was provided by the breeding company Syngenta Seeds AB (Landskrona, Sweden).
The 924 genotypes were evaluated in routine plant breeding trials with two replicates at 1–7 locations in 2008. The evaluated traits were white sugar yield (WSY, t ha−1), sugar content (SC, %), root yield (RY, t ha−1), potassium content (K, mM), sodium content (Na, mM) and α-amino nitrogen content (N, mM). Genotypes were grown in different trials per location that were connected by common checks. Based on these, adjusted entry means (best linear unbiased estimates (BLUEs)) were estimated for each location as: yij=μ+gi+tj+eij, where yij is the phenotypic value of the ith sugar beet line in the jth trial, μ the intercept term, gi the genetic effect of the ith sugar beet line, tj the effect of the jth trial, and eij the residual. Heritability (h2) on an entry-mean basis was estimated as the ratio of genotypic to phenotypic variance according to Melchinger et al. (1998).
The 924 genotypes were fingerprinted following standard protocols with 677 single-nucleotide polymorphism markers. These markers were randomly distributed across the sugar beet genome with an average marker distance of 1 cM and a maximum gap between adjacent markers of 23 cM. Map positions of all markers were based on the linkage map of Syngenta Seeds AB with a total map length of 698 cM.
Association mapping
For association mapping, a mixed model incorporating a kinship matrix was used: yijp=μ+ap+gi+lj+eijp, where yijp is the adjusted entry mean of the ith sugar beet line at the jth location carrying allele p, μ the intercept term, ap the allele substitution effect of allele p, gi the genetic effect of the ith sugar beet line, lj the effect of the jth location and eijp the residual including the genotype times location interaction effect. The allele substitution effect ap was modeled as fixed effect whereas gi and lj were regarded as random effects. The variance of the random genetic effect was assumed to be Var(g)=Kσ2g, where σ2g refers to the genetic variance estimated by REML and K was a 924 × 924 matrix of kinship coefficients that define the degree of genetic covariance between all pairs of entries. We followed the suggestion of Bernardo (1993) and calculated the kinship coefficient Kij between inbreds i and j on the basis of marker data as described by Würschum et al. (2011a, 2011b, 2012b). For the detection of main effect QTL, a genome-wide scan for marker–trait associations was conducted. To control for multiple testing, we followed the suggestion of Kraakman et al. (2004) and tested at a false discovery rate of 0.20 (Benjamini and Hochberg, 1995). All mixed model calculations were performed using the software ASReml 2.0 (Gilmour et al., 2006).
The total proportion of genotypic variance (pG) explained by the detected QTL was calculated by fitting all QTL simultaneously in a linear model in the order of the strength of their association (most significantly associated markers first) to obtain R2adj. The ratio pG=R2adj/h2, where h2 refers to the heritability of the trait, yielded the proportion of genotypic variance (Utz et al., 2000).
Cross-validation
To obtain unbiased estimates of the proportion of genotypic variance explained by the detected QTL, a cross-validation approach was established similar to that used by Utz et al. (2000) in the context of linkage mapping in biparental populations. We used a cross-validation in which a certain percentage of the lines were used as ES in which QTL detection was done. The remaining lines represented the TS, which was used for validation. The following proportions of TS were tested: 5%, 10%, 20%, 30%, 40%, 50%. For validation, the QTL mapping results from the ES were used to predict the genotypic value of line j in TS QTS.ESj according to QTS.ESj=XTSj βES, where XTSj is the vector of marker information of line j at the QTL positions, and βES is the vector of genetic effects of these QTL estimated as partial regression coefficients from a simultaneous fit in the ES (Utz et al., 2000). The proportion of genotypic variance explained by the QTL in the TS (pG−TS) was calculated from the adjusted squared correlation coefficient, R2adj, between the phenotypic values observed for the lines in the TS and the predicted genotypic values QTS.ESj, divided by the heritability of the trait. The bias in the proportion of explained genotypic variance was calculated as the difference in pG between the ES (pG−ES) and the TS (pG−TS) and the relative bias as 1–(pG−TS/pG−ES). The presented results are the median values from 300 cross-validation runs for the different proportions of TS tested and 1000 runs for the fivefold cross-validation (80% ES and 20% TS).
Results
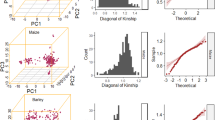
This study is based on a large population of sugar beet with a total of 924 individuals. Details on the population structure present in this population, the linkage disequilibrium and the results from QTL analyses have been described in detail by Würschum et al. (2011a). Briefly, three yield-related (WSY, SC and RY) and three quality-related traits (Na, K and N) have been investigated and the number of detected QTL ranged between 4 for SC and K to 15 for Na. The proportion of genotypic variance explained by these QTL was 73.4% for WSY, 38.1% for SC, 63.1% for RY, 58.7% for Na, 12.8% for K and 10.4% for N. We first assessed the effect of genotypic sampling on population structure and allele frequencies. For fivefold cross-validation, that is, 80% of the lines as ES and 20% as TS, we observed only negligible changes for both population structure and genetic kinship, as well as for the polymorphic information content (Figure 1).
Changes in population structure and allele frequencies in the ES during fivefold cross-validation with the full data set (N=924). (a) Explained variance of the first four principal coordinates, (b) mean kinship among the individuals and (c) mean polymorphic information content (PIC). The dashed lines indicate the mean and the arrowheads the mean of the full data set.
Next, we used a yield-related trait (RY) and a quality-related trait (Na) with comparable heritability of 0.55 and 0.53, respectively, to evaluate the optimum proportion of the TS for cross-validation in dependence on the population size. To this end, we varied the proportion of plants used as TS from 5 to 50% and performed the analyses for the full data set (N=924), as well as reduced data sets with N=500 and N=200 randomly sampled genotypes (Figure 2). As expected, the median number of QTL detected in the ES decreased with decreasing size of the ES, that is, increasing proportion of the TS. This effect was more pronounced for the largest population size. The median cross-validated proportion of genotypic variance explained by detected QTL (pG−TS) was also affected by the percentage of plants in the TS. It decreased with decreasing size of the ES for the largest population, remained on a similar level for N=500, and increased for N=200. Notably, the observed increase in pG−TS for N=200 was mainly for the proportion of the TS of 5–20% while it did not change much between 20 and 50%. The relative bias in the estimation of pG, that is, the difference in pG between estimation and test set, was relatively stable for N=924 and N=500, but decreased for N=200 with increasing size of the TS, especially between 5 and 20%. Taken together, considering the most important parameter, the cross-validated proportion of genotypic variance explained by QTL, the optimum proportion of plants to be used as TS is dependent on the population size. For commonly used population sizes, however, a fivefold cross-validation, that is, 20% of the lines as TS, appears most appropriate. In the following, we therefore further characterized fivefold cross-validation for association mapping.
Applying fivefold cross-validation in the full data set, we observed for the six traits a substantially reduced cross-validated proportion of explained genotypic variance (pG−TS) as compared with that estimated in the full data set (pG−DS) (Figure 3). For all traits the relative bias averaged 38%. The number of detected QTL, the proportion of genotypic variance explained by QTL in the ES (pG−ES) and in the TS (pG−TS), and the relative bias all showed considerable variation across runs (Figure 4). The range in pG−TS was of similar magnitude or even smaller than that in pG−ES.
To assess the precision and the reliability of the identified QTL positions, we used the QTL frequency distributions derived from fivefold cross-validation (Figure 5). With the exception of α-amino nitrogen, most of the QTL detected with the full data set were identified in at least 40% of the runs. The QTL frequency distributions also revealed that some QTL detected with the full data set were only identified as QTL in a small number of runs, whereas a few other positions were identified as QTL in >20% of the runs, but not in the full data set (for example, SC QTL on chromosome 7).
Discussion
Association mapping has become a popular genomic tool to dissect the genetic architecture of complex traits and to identify QTL, which in plant breeding are of interest for their implementation in marker-assisted selection programs. However, a prerequisite is that the QTL parameters are estimated with high accuracy and precision. In this study, we used a large sugar beet data set to evaluate cross-validation as an approach to obtain more robust estimates of QTL parameters in association mapping.
Genetic kinship and population structure
Biparental populations as used for linkage mapping possess balanced allele frequencies and are not burdened with the confounding effects of population structure or genetic relatedness. By contrast, both effects are well known to be present in association mapping populations and must be accounted for in the analysis as they can inflate the false-positive rate (Sillanpää, 2011). In addition, association mapping populations will show the full range of allele frequencies including rare alleles with a frequency below 10%. Thus, in contrast to biparental populations, genotypic subsampling in association mapping populations may affect both the population structure and the allele frequencies and consequently the false-positive rate and the QTL detection power. For fivefold cross-validation, that is, 80% of the lines as subsample for QTL detection, we observed only slight variations in population structure among the cross-validation runs suggesting that the genotypic subsampling does not substantially affect population structure (Figures 1a and b). The same holds true for the allele frequencies as illustrated by the negligible variation in polymorphic information content values (Figure 1c). It must be noted, however, that for rare alleles the frequency in the ES may drop below a threshold that does not permit a significant association any more. Taken together, although genotypic subsampling in association mapping is potentially more critical than in linkage mapping, our results suggest that the effect on population structure and allele frequencies, and consequently on the false discovery rate and the QTL detection power, are small and should not prohibit the use of cross-validation in association mapping. A major advantage of the genotypic subsampling is that it enables an evaluation of the robustness of the mapping results and mirrors the situation in breeding programs where the population in which QTL are detected is different from that in which the QTL are to be used for marker-assisted selection.
Cross-validation approach
The correct statistical model to estimate QTL effects would require their true number and their precise position to be known. Consequently, all approaches for QTL analysis share the problem of model selection. In association mapping, QTL are identified based on their association with the trait given a predefined significance threshold. As shown extensively for linkage mapping, model selection generally results in an overestimation of the QTL effects and the proportion of explained genotypic variance, especially for small sample sizes (Beavis, 1998). For linkage mapping, cross-validation is therefore commonly used to obtain an unbiased assessment of the prospects of marker-assisted selection. Here, we evaluated cross-validation for association mapping and first addressed the question of the proportion of the ES and the TS in dependence on the population size (Figure 2).
For population sizes of N=924 and to a lesser extent also for N=500, a decrease in the size of the ES resulted in fewer QTL being detected, that is, in the selection of models with too few variables, and consequently in a lower accuracy of prediction. In contrast, for the smallest population size (N=200), we observed the opposite trend as the median proportion of cross-validated genotypic variance (pG−TS) increased with decreasing size of the ES, especially up to a proportion of the TS of 20% after which pG−TS reached a plateau. A likely reason for this observation is that the TS must have a certain minimum size to enable a reliable validation of the estimated QTL effects. If for small population sizes, like, for example, N=200, the proportion of the TS is chosen too small, the number of individuals left in the TS will be insufficient for robust validation. Consistently, we observed an extremely high median relative bias for N=200 and proportions of the TS below 20%. For N=924 and 500 the median relative bias was not affected by varying sizes of the TS for RY whereas for Na it slightly increased with increasing size of the TS. This illustrates that the proportion of plants that should be used as independent TS strongly depends on the population size. With population sizes of 1000 individuals or more, 5% of the lines will provide a sufficiently large TS for robust validation while enabling a maximum QTL detection power because of a large ES. Conversely, for population sizes of a few hundred individuals as commonly used for association mapping, a fivefold cross-validation, that is, 20% of the lines as TS, as routinely used in linkage mapping, also appears appropriate for association mapping. In the following, we therefore characterize fivefold cross-validation in more detail.
Predictive power and its bias
The predictive power for the proportion of genotypic variance explained by the detected QTL is an important parameter as it determines whether a marker-assisted selection is justified in a breeding program. Recent work has shown this parameter to be overestimated with a relative bias between 10 and 60% depending on the population and the complexity of the trait (Utz et al., 2000; Schön et al., 2004; Liu et al., 2013). This reduction of pG−TS as compared with pG−ES indicates a large upward bias in predictors of the proportion of explained genotypic variance, that is, QTL effects inferred from the ES. The poor precision of QTL effect estimation is also reflected in the large range of estimated pG−ES and pG−TS values (Figure 4). Our results corroborate previous findings (Schön et al., 2004; Liu et al., 2013) as we observed an average relative bias of 38%. This is higher than what was observed in a previous study using the same data set where a cross-validation was used assuming the QTL detected with the full data set as fixed (Würschum et al., 2011a). This in combination with the results from the QTL frequency distributions illustrates that a robust cross-validation to obtain realistic estimates for the predictive power must include a QTL detection in each cross-validation run.
The bias in QTL effect estimates is introduced when effects are estimated in the same data set that was used to identify the QTL. The degree of overestimation is thereby a function of the power of the study such that QTL effects will be more upwardly biased with low power, that is, small effect sizes (Xu 2003; Zöllner and Pritchard 2007). Ingvarsson et al. (2008) performed a candidate gene association mapping approach in European aspen and identified two single-nucleotide polymorphisms significantly associated with bud set. They used an ad hoc method described by Allison et al. (2002) to obtain less biased estimates of QTL effects. Based on data-perturbation simulations and a methods-of-moments approach they showed the effects of the two single-nucleotide polymorphisms and consequently their contribution to the phenotypic variance to be strongly overestimated. Our findings corroborate this overestimation of QTL effects observed in a candidate gene approach, which emphasizes the need for robust validation of association mapping results.
It must be noted that the relative bias observed here was higher for K and N, the two traits for which the predictive power was low (∼10%). For the other four traits, the relative bias averaged 30%, which thus appears to be the level of overestimation that must be expected in association mapping for complex traits, even with large population sizes close to 1000 as applied here. It must be noted that for smaller population sizes this relative bias will be even higher (Figure 2) and that in addition to the trait will to some extent also depend on the underlying data set.
The cross-validated proportion of genotypic variance was highest for WSY (50%) despite the presumably high complexity of this trait. For SC, RY and Na, it was approximately 30% while for K and N it was only 5% or below. These values are consistent with the quantitative nature of the traits under consideration and illustrate the unbiased potential of marker-assisted selection.
Robustness of QTL and precision of QTL localization
Another important QTL parameter is the precision of QTL position estimates. Only if QTL positions are estimated precisely can closely linked markers be identified for efficient selection of plants carrying the favorable QTL allele. Sillanpää and Arjas (1998) suggested the use of QTL intensity distributions for detailed analysis of putative QTL regions. This concept has been applied in cross-validation to judge the robustness and the reliability of QTL position estimates as the number of runs in which each position is identified as QTL in the different estimation sets (Utz et al., 2000; Liu et al., 2013). In our study, most QTL identified in the full data set were supported by their detection in a high number of runs. However, as mentioned above, a difference to linkage mapping is that minor allele frequency QTL may not be detected in some estimation sets if the allele frequency becomes too low to permit detection. Broad QTL frequency peaks indicate a fairly vague localization of the QTL and consequently a biased estimation of their true genetic effects. In association mapping, the achievable mapping resolution will depend on the linkage disequilibrium in the respective QTL regions (Würschum, 2012). The shape of the QTL frequency peaks is thus affected by the linkage disequilibrium varying among the resampled ESs and, in addition, by the resampling of individuals with variable errors of their phenotypic value estimates. We mainly observed rather narrow, clearly defined peaks, which illustrates the high precision of QTL position estimates (Figure 5). This will, however, also depend on the available marker density and QTL frequency distributions will be most informative applying high marker densities. In summary, QTL frequency distributions can be used to further assess the quality of the obtained mapping results.
Conclusions
In this study, we evaluated the potential of cross-validation for association mapping. Our results suggest that the interpretation of association mapping results can be improved applying cross-validation. The predictive power of the proportion of genotypic variance explained by detected QTL can be assessed more realistically. In addition, the construction of QTL frequency distributions can serve as a measure for the precision of QTL localization and the robustness of the detected QTL. Taken together, cross-validation can be a valuable tool for association mapping to draw realistic conclusions on the prospects of the identified QTL for marker-assisted selection.
Data archiving
The data underlying this study is provided as Supplementary files and is also available from the Dryad Digital Repository: doi:10.5061/dryad.db521.
References
Allison DB, Fernadez JR, Heo M, Zhu S, Etzel C, Beasley TM et al. (2002). Bias in estimates of quantitative-trait-locus effect in genome scans: demonstration of the phenomenon and a method-of-moments procedure for reducing bias. Am J Hum Genet 70: 575–585.
Beavis WD . (1998). QTL analyses: power, precision and accuracy. Paterson AH, edited by Molecular Dissection of Complex Traits. CRC Press: Boca Raton, FL, USA. pp 145–162.
Benjamini Y, Hochberg Y . (1995). Controlling the false discovery rate: a practical and powerful approach to multiple testing. J R Stat Soc 57: 289–300.
Berger GL, Liu S, Hall MD, Brooks WS, Chao S, Muehlbauer GJ et al. (2013). Marker-trait associations in Virginia Tech winter barley identified using genome-wide mapping. Theor Appl Genet 126: 693–710.
Bernardo R . (1993). Estimation of coefficient of coancestry using molecular markers in maize. Theor Appl Genet 85: 1055–1062.
Buckler ES, Holland JB, Bradbury PJ, Acharya CB, Brown PJ, Browne C et al. (2009). The genetic architecture of maize flowering time. Science 325: 714–718.
Gilmour AR, Gogel BJ, Cullis BR, Thompson R . (2006) ASReml User Guide Release 2.0. VSN International Ltd: Hermel Hempstead, UK.
Ingvarsson PK, Garcia MV, Luquez V, Hall D, Jannson S . (2008). Nucleotide polymorphism and phenotypic associations within and around the phtyochrome B2 locus in European aspen (Populus tremula, Salicaceae). Genetics 178: 2217–2226.
Kollers S, Rodemann B, Ling J, Korzun V, Ebmeyer E, Argillier O et al. (2013). Whole genome association mapping of fusarium head blight resistance in European winter wheat (Triticum aestivum L.). PLoS One 8: e57500.
Kraakman ATW, Niks RE, PMMM Van den Berg, Stam P, Van Eeuwijk FA . (2004). Linkage disequilibrium mapping of yield and yield stability in modern spring barley cultivars. Genetics 168: 435–446.
Kulwal P, Ishikawa G, Benscher D, Feng Z, Yu L-X, Jadhav A et al. (2012). Association mapping for pre-harvest sprouting resistance in white winter wheat. Theor Appl Genet 125: 793–805.
Liu W, Gowda M, Steinhoff J, Maurer HP, Würschum T, Longin CF et al. (2011). Association mapping in an elite maize breeding population. Theor Appl Genet 123: 847–858.
Liu W, Maurer HP, Reif JC, Melchinger AE, Utz HF, Tucker MR et al. (2013). Optimum design of family structure and allocation of resources in association mapping with lines from multiple crosses. Heredity 110: 71–79.
Melchinger AE, Utz HF, Schön CC . (1998). Quantitative trait locus (QTL) mapping using different testers and independent population samples in maize reveals low power of QTL detection and larger bias in estimates of QTL effects. Genetics 149: 383–403.
Reif JC, Gowda M, Maurer HP, Longin CFH, Korzun V, Ebmeyer E et al. (2011a). Association mapping for quality traits in soft winter wheat. Theor Appl Genet 122: 961–970.
Reif JC, Maurer HP, Korzun V, Ebmeyer E, Miedaner T, Würschum T . (2011b). Mapping QTLs with main and epistatic effects underlying grain yield and heading time in soft winter wheat. Theor Appl Genet 123: 283–292.
Schön CC, Utz HF, Groh S, Truberg B, Openshaw S, Melchinger AE . (2004). Quantitative trait locus mapping based on resampling in a vast maize testcross experiment and its relevance to quantitative genetics for complex traits. Genetics 167: 485–498.
Sillanpää MJ, Arjas E . (1998). Bayesian mapping of multiple quantitative trait loci from incomplete inbred line cross data. Genetics 148: 1373–1388.
Sillanpää MJ . (2011). Overview of techniques to account for confounding due to population stratification and cryptic relatedness in genomic data association analyses. Heredity 106: 511–519.
Snowdon RJ, Wittkop B, Rezaidad A, Hasan M, Lipsa F, Stein A et al. (2010). Regional association analysis delineates a sequenced chromosome region influencing antinutritive seed meal compounds in oilseed rape. Genome 53: 917–928.
Stich B, Melchinger AE, Heckenberger M, Möhring J, Schechert A, Piepho HP . (2008). Association mapping in multiple segregating populations of sugar beet (Beta vulgaris L.). Theor Appl Genet 117: 1167–1179.
Utz HF, Melchinger AE, Schön CC . (2000). Bias and sampling error of the estimated proportion of genotypic variance explained by quantitative trait loci determined from experimental data in maize using cross validation and validation with independent samples. Genetics 154: 1839–1849.
Wang M, Jiang N, Jia T, Leach L, Cockram J, Comadran J et al. (2012a). Genome-wide association mapping of agronomic and morphologic traits in highly structured populations of barley cultivars. Theor Appl Genet 124: 233–246.
Wang X, Zhang C, Li L, Fritsche S, Endrigkeit J, Zhang W et al. (2012b). Unraveling the genetic basis of seed tocopherol content and composition in rapeseed (Brassica napus L.). PLoS One 7: e50038.
Würschum T, Maurer HP, Kraft T, Janssen G, Nilsson C, Reif JC . (2011a). Genome-wide association mapping of agronomic traits in sugar beet. Theor Appl Genet 123: 1121–1131.
Würschum T, Maurer HP, Schulz B, Möhring J, Reif JC . (2011b). Genome-wide association mapping reveals epistasis and genetic interaction networks in sugar beet. Theor Appl Genet 123: 109–118.
Würschum T . (2012). Mapping QTL for agronomic traits in breeding populations. Theor Appl Genet 125: 201–210.
Würschum T, Liu W, Maurer HP, Abel S, Reif JC . (2012a). Dissecting the genetic architecture of agronomic traits in multiple segregating populations in rapeseed (Brassica napus L.). Theor Appl Genet 124: 153–161.
Würschum T, Liu W, Gowda M, Maurer HP, Fischer S, Schechert A et al. (2012b). Comparison of biometrical models for joint linkage association mapping. Heredity 108: 332–340.
Xu S . (2003). The theoretical basis of the Beavis effect. Genetics 165: 2259–2268.
Yu J, Pressoir G, Briggs WH, Vroh BiI, Yamasaki M, Doebley JF et al. (2006). A unified mixed-model method for association mapping that accounts for multiple levels of relatedness. Nat Genet 38: 203–208.
Zhou H, Steffenson B . (2013). Genome-wide association mapping reveals genetic architecture of durable spot blotch resistance in US barley breeding germplasm. Mol Breeding 32: 139–154.
Zöllner S, Pritchard JK . (2007). Overcoming the winner’s curse: estimating penetrance parameters from case-control data. Am J Hum Genet 80: 605–615.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Additional information
Supplementary Information accompanies this paper on Heredity website
Supplementary information
Rights and permissions
About this article
Cite this article
Würschum, T., Kraft, T. Cross-validation in association mapping and its relevance for the estimation of QTL parameters of complex traits. Heredity 112, 463–468 (2014). https://doi.org/10.1038/hdy.2013.126
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/hdy.2013.126
Keywords
This article is cited by
-
Genome-wide association study for resistances to yellow rust, powdery mildew, and Septoria tritici blotch in cultivated emmer
Euphytica (2024)
-
Do lower nitrogen fertilization levels require breeding of different types of cultivars in triticale?
Theoretical and Applied Genetics (2022)
-
Exploiting genetic diversity in two European maize landraces for improving Gibberella ear rot resistance using genomic tools
Theoretical and Applied Genetics (2021)
-
Improving the baking quality of bread wheat by genomic selection in early generations
Theoretical and Applied Genetics (2018)
-
Deciphering the regulation of porcine genes influencing growth, fatness and yield-related traits through genetical genomics
Mammalian Genome (2017)