Abstract
Purpose: To evaluate the sensitivity, specificity, advantages, and limitations of multiplex ligation-dependent probe amplification compared with conventional karyotype analysis in the investigation of contributing factors to recurrent pregnancy loss.
Method: A cohort of 284 patients underwent side-by-side analysis of products of conception by both conventional karyotyping and multiplex ligation-dependent probe amplification with direct comparison of results.
Results: Multiplex ligation-dependent probe amplification was shown to enable a diagnosis for an additional 47 (16.5%) patients compared with conventional karyotype analysis. However, this advantage was offset by some disadvantages of the method, including a high false-positive rate (8/104; 7.7%), as demonstrated by single-arm probe abnormalities of uncertain clinical significance, as well as the inability to characterize structural rearrangements, such as Robertsonian translocations, which comprised 2.46% of samples (99% confidence interval = 0.09–4.83), and ploidy changes. The calculated performance characteristics of multiplex ligation-dependent probe amplification in this cohort yielded a sensitivity of 86.9% and specificity of 92.4%.
Conclusions: The advantages of now widely accepted molecular methodologies, such as lower failure rates, faster turnaround times, and lower cost, must be complemented by adequate counseling, family follow-up, and specific diagnostic reporting practices. It is particularly important to specifically address the important limitations of the methodology, including the inability to characterize balanced structural rearrangements and ploidy changes, especially if multiplex ligation-dependent probe amplification is to be performed alone.
Similar content being viewed by others
Main
Aproximately 15–20% of recognized pregnancies end in a spontaneous abortion before 20 weeks gestation. The burden of pregnancy loss is, however, not uniformly distributed. As a result of the increase in the risk of trisomic pregnancies with advancing maternal age, >40% of pregnancies in women older than 40 years end in spontaneous abortion. Recurrent pregnancy loss (RPL) is also frequent, affecting approximately 5% of couples. The major factors contributing to pregnancy loss are chromosomal in origin with approximately 50% of products of conception (POC) demonstrating either aneuploid or polyploid karyotypes.1,2
RPL can be classified into two major groups, those where couples experience RPL due to consecutive sporadic segregation errors that increase with advancing maternal age3 and those where one or more risk factors contribute to the clinical history. Given the high frequency of aneuploid pregnancies, the first group is numerically the largest group.
Chromosomal analysis of POC is an important laboratory investigation for couples who have experienced consecutive miscarriages. Given the known major contributors to RPL, the main practical utility of POC analysis is to differentiate the less commonly occurring fetal loss due to chromosomal structural variants where the recurrence risk may be substantial, from the more frequent whole chromosome aneuploidy where the recurrence risks are lower. Traditionally, POC testing has been performed by microscopic examination of banded chromosomal preparations. Standard cytogenetic analysis is a labor-intensive, comparatively costly, low-throughput technique reliant on highly trained technical staff and is associated with a culture failure rate of 10–40%.4 There are in addition concerns that the process of in vitro culturing may select for the detection of chromosomal abnormalities, which are more viable under these conditions.
Alternative molecular cytogenetic techniques such as fluorescence in situ hybridization (FISH) or comparative genomic hybridization array have other limitations. Although FISH offers rapid diagnosis, quality-assured probes are expensive, and the technique is still labor intensive. The cost of microarray-based diagnosis currently remains prohibitive for the routine detection of chromosomal aneuploidy in many laboratories, but this technique has a major advantage of being able to document and characterize unbalanced chromosomal copy number variants that fall below the resolution of light microscopy. Molecular methods such as multiplex ligation-dependent probe amplification (MLPA) offer an alternative approach for high throughput, cost-efficient chromosomal aneuploidy screening, as sufficient probes can be used to enable a quantitative survey of the entire chromosomal complement and identify all but the most telomeric unbalanced chromosomal translocations. The application and assessment of MLPA as a diagnostic method in this setting has previously been reported in two publications—in 2007 by Diego-Alvarez et al.5 and in 2006 by Bruno et al.6 In this study, we describe the use of MLPA as a screening method for aneuploidy in POC in the largest cohort described to date and provide comparisons with respect to the sensitivity, specificity, and clinical utility of the technique in comparison with classical cytogenetic analysis.
MATERIALS AND METHODS
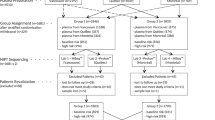
A total of 284 POC samples were referred to the Genetics laboratories of South Eastern Area Laboratory Services, Sydney, between March 2008 and January 2009 for cytogenetic analysis. Cytogenetic analysis of POC for chromosomal aneuploidy is provided as a standard diagnostic tool in the clinical management of patients experiencing pregnancy loss in this hospital. A subgroup of these patients will be referred with RPL, defined as three or more consecutive miscarriages. Additional testing of all POC samples by MLPA was performed as a quality improvement activity of the laboratory. Data analysis was performed on deidentified patient data in a blinded fashion to preserve patient confidentiality.
Cytogenetic analysis
Sample types collected included chorionic villi, umbilical cord, fetal skin, and cartilage. Only those samples where tissue of fetal origin could be identified by microscopic examination were accepted for analysis, and samples showing macroscopic signs of putrefaction were not processed. Isolated fetal tissue was finely chopped using a scalpel blade under sterile conditions. A maximum of 10 mg of chopped tissue was cultured, and genomic DNA was extracted from approximately 5 mg of this material. Samples were then cultured in AmnioMax-C100 (Gibco 17001 and 12556) and harvested using standard cytogenetics conditions. A total of 15 metaphases were analyzed by G-banding by Giemsa and Trypsin staining by an experienced cytogenetics technologist.
MLPA analysis
Coextraction of DNA was performed using the Tissue Kit for EZ1 BioRobot (QIAGEN, Hilden, Germany). The finely chopped tissue was digested in QIAGEN digest buffer with proteinase K at 50°C between 24 and 48 hours following the manufacturer's protocols. MLPA with SALSA P070 probe mixes (MRC-Holland) was performed in a Corbett PalmCycler following the manufacturer's protocol. Polymerase chain reaction products were electrophoresed in an ABI Prism 3130xl Genetic Analyser and analyzed with the GeneMapper 3.5 software package (Applied Biosystems). The probe mix contains probes designed to recognize a well-characterized gene near each subtelomeric region, with quantitation allowing detection of copy number changes at these regions. Normalization of peak area values was achieved according to MRC-Holland recommendations. Probe deletion or duplication was considered where a >25% variation of the relative peak area of the amplification product of the respective probe was obtained in comparison with control DNA from POC samples shown to have a normal karyotype by cytogenetics G-banding by Giemsa and Trypsin staining. Unless otherwise indicated, percentages are expressed relative to the full cohort of 284 enrolled cases.
RESULTS
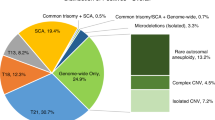
A total of 284 samples were included in the study. Table 1 provides a complete summary of all the findings. A result was obtained from neither the cytogenetics nor MLPA testing in three instances, giving a combined success rate of 98.9%. Structural or numerical chromosomal changes were detected in 148 (52%) cases by either MLPA or cytogenetics, the vast majority of which were clearly pathogenic. The results of MLPA and cytogenetic analyses were compared and categorized in the following manner. Fully concordant cases were defined as agreement between the MLPA and cytogenetics results. Partial results were those cases where data were lacking for one analysis. Incompletely concordant cases were those where consistent abnormalities were detected by MLPA and cytogenetics testing but where one test contained significantly more clinically useful information than the other. Discordant cases were cases where the genetic changes detected by two methodologies were different. The findings for each of these categories are described later. The analytical performance of each method is compared in Table 2.
Fully concordant results
A comparison of the results for samples with both MLPA and cytogenetic results showed that in 189 cases (66%) both techniques yielded the same findings. Among these, 94 samples (33%) showed a concordant result with a normal chromosomal complement, whereas in 95 samples (33%), a concordant chromosomal aneuploidy was identified.
Partial results
A partial result was available from 48 (17%) samples. Of these, 47 (16.5%) samples had no cytogenetic results due to culture failures where MLPA results were available, and one sample (0.5%) failed MLPA, but a cytogenetics result was available. Three samples failed both MLPA and karyotyping. The overall success rate for MLPA was, therefore, 280/284 cases (98.6%) versus that of cytogenetics of 234/284 cases (82%). In the four cases where MLPA failed to produce a result, this was attributed to poor tissue quality in three samples and a low mass of DNA in a single case. The 50 samples that failed to yield a cytogenetics result did so as the result of failure of sample growth in tissue culture in 41 samples, and in the remaining nine samples tissue, culture was not attempted due to putrefaction.
Among the 47 cases where results were only available from MLPA, there were 36 normal results and 11 abnormal cases. The difference in the robustness of the MLPA technique compared with cell culture-based cytogenetics is highly significant and is a useful property of the test. All the abnormalities were single or double aneuploidies, with the exception of one sample that showed two single-probe duplications at 4q and 13q. There was a single instance where a karyotype was obtained where MLPA failed to yield a result. In that case, the karyotype identified a 47, XX,+13 abnormality. Although the reason for the failure of the MLPA in this instance is not known, technical considerations such as low mass of DNA, degradation of the sample during processing, or nuclease contamination cannot be excluded.
Discordant results
There were 42 cases (14.8%) which showed discordant results between cytogenetic and MLPA testing as a result of a variety of different causes.
Discordant results due to undetected structural changes (n = 8)
There were eight cases where testing gave concordant abnormal results but where less clinically useful clinical information was obtained from one testing modality compared with the other. In seven of the eight such events, the MLPA result gave less information than the matched cytogenetics test. The largest group of incompletely concordant results (n = 5) was due to the presence of aneuploidy on the background of a Robertsonian translocation. In each instance, the aneuploid chromosome was correctly identified by MLPA, but the balanced Robertsonian translocation was not. The inability of MLPA to detect and characterize balanced changes is an important limitation of this technology.
The smaller subgroup (n = 3) involved unbalanced autosomal translocations. The first case was of an unbalanced interchange trisomy 47, XY,+4, t(1,4) (q21;q31) where only the chromosome 4 trisomy was identified by MLPA. The second case occurred in a sample where MLPA detected a raised signal intensity for the single probe for the long arm of chromosome 10 but where karyotype analysis showed 46,XY,der(3) t. (3,10). For the purposes of this study where parental samples were not available, this result was classified as an adjacent 2 segregant rather than a de novo unbalanced event as the most cautious interpretation. The third case was the sole instance where MLPA detected a chromosomal anomaly, which evaded full cytogenetic characterization due to poor chromosome morphology. MLPA detected increased signal for 2q and reduced signal for 9q where the cytogenetic analysis showed only the additional material on 9q.
Discordant results due to maternal cell contamination (n = 9)
Maternal cell contamination (MCC) was clearly identified in five cases where the MLPA test identified a male sex constitution but where a female karyotype was detected by cytogenetics. For three of these samples, there was MLPA evidence of aneuploidy on a male genetic background but where karyotype analysis showed a normal female. These cases were a male with a +8; a male with a double aneuploidy +16 and +18; and a male with +17 and +Xp.
MCC also seems to be the most reasonable explanation for the findings in a further three cases. The first case was of chromosome 4 aneuploidy detected by both MLPA probes but where the cytogenetic result was normal female. In the remaining two cases, the MLPA result was consistent with a nonmosaic aneuploidy, but the cytogenetic result identified a coexisting normal female line, consistent with MCC or mosaicism for the aneuploid chromosome. The two cases in this subgroup involved mosaicism for 45,X(80%)/46,XX(20%), where MLPA solely detected the 45,X clone. The second case involved mosaicism for 47,XX,+4(2)/46,XX (25), where MLPA detected the +4 abnormal cell line only. Other possible explanations for these findings include a selective growth advantage in vitro for a normal cell line or placental mosaicism where tissue selected for MLPA contained the abnormal cell line only.
In one case, the uncultured MLPA sample identified a normal female constitution where the karyotype was of a normal male, possibly due to sample contamination or arising as a result of placental chimerism. This sample was not able to be resolved due to lack of additional clinical material.
Discordant results due to polyploidies or balanced structural variants (n = 14)
As MLPA identifies probe copy number changes relative to the haploid set of chromosomes, it is unable to detect certain abnormalities, such as polyploidy or balanced structural variations. In this study, a total of 10 samples with pure polyploidy (3.5%) were detected by karyotype analysis and were predictably identified as normal by MLPA. Similarly, the single balanced chromosomal anomaly detected by cytogenetics (a pericentric inversion of the Y chromosome) was not detected.
There were in addition three cases of aneuploidy occurring on a polyploid background. In two instances, the aneuploidy was detected but the baseline polyploidy karyotype not recognized by MLPA. There was a single case where aneuploidy on a tetraploid background was not detected by MLPA where the fold change in chromosome 13 signals fell below the detection threshold for this technique.
Single-probe discordance (n = 10)
There were two cases where a single-probe abnormality was present in addition to the aneuploid MLPA result and eight samples with single chromosome arm changes in MLPA signal strength where cytogenetics identified a normal karyotype. These findings could represent true copy number variations undetectable at the resolution of karyotyping, such as microduplications. The significance of these abnormalities in this setting and their contribution to pregnancy loss is as yet unknown and has, therefore, been classified in this study as false-positive test results. Future testing may permit revision of this classification.
False-negative MLPA results (n = 3)
There were three samples (1%) in which false-negative MLPA results were identified. In two cases, there was complete failure of MLPA testing to detect a chromosomal aneuploidy clearly present in the cytogenetic sample in nonmosaic form. These cytogenetic findings were of 47,XY,+21 and 47,XX,+6. In the third case, MLPA detected only one of the two aneuploid chromosomes present in a sample where cytogenetics analysis demonstrated 48,XX,+8,+21.
DISCUSSION
Cytogenetic testing in this cohort of 284 samples was shown to be capable of detecting and characterizing 122 genetic changes, 103 normal karyotypes, and gave eight incorrect or incomplete diagnoses. It was in addition associated with 51 culture failures for which a result could not be obtained. In contrast, MLPA testing detected fewer abnormal POC samples (n = 112) and was associated with a higher rate of incorrect or incomplete diagnoses (n = 36) but with a much lower failure rate and better normal result rate. The overall benefit from MLPA testing was that it was able to provide an additional 38 correct diagnoses (13.3%) compared with cytogenetic testing. The cost of this increased success rate was the incomplete characterization of seven unbalanced translocations, which have a moderate recurrence risk and the identification of polyploidy karyotypes with a low recurrence risk.
It is clear that MLPA is capable of producing many more results than cytogenetics as it is possible to generate results from uncultured samples. More detailed analysis of the data shows, however, that MLPA produced the most likely correct diagnosis in 244 cases overall and in 112 with a genetic abnormality. In contrast, cytogenetics produced the likely correct result in 225 but importantly in 123 with a genetic abnormality. The ability of MLPA to characterize a normal genetic constitution outstrips that of cytogenetics, with MLPA calling 132 normal cases compared with cytogenetics call of only 103 due to limitations of cell growth in tissue culture. The errors present in the cytogenetic dataset were predominantly false-negative results due to culture failure, MCC, or as the result of poor chromosome morphology. In contrast, the errors in the MLPA dataset were primarily due to methodological limitations of the technique in the context of balanced karyotypes, polyploidy, and to the production of a larger set of uncertain results.
It is apparent that the two techniques have different strengths and weaknesses, and the optimal situation would be to have both techniques available in the laboratory for the parallel examination of POC samples. The implementation of such an analytical protocol would provide an extremely high success rate for results and would allow the disambiguation of discordant results.
In the publication by Diego-Alvarez et al.,5 who performed MLPA on 221 miscarriage samples, only 102 cases had a karyotype performed. Their results are summarized in Table 3.
Bruno et al.6 also compared the use of MLPA with karyotype analysis but included FISH for the detection of polyploidy, with evaluation of the use of this combination of MLPA and FISH compared with conventional cytogenetics. To enable a direct comparison with our results, we have excluded the results from FISH data in the summary later. The results of their 67 samples where results were available by MLPA and karyotype are also summarized in Table 3.
In addition, however, Diego-Alvarez et al.5 also proved, by microsatellite analysis, that eight normal female karyotypes detected in their study were in fact erroneous and caused by MCC of the culture, demonstrating a maternal contamination rate of ∼12.9%, thus documenting and quantifying a disadvantage of karyotype analysis, long considered to be a “gold standard” in the examination of POC. Although we did not undertake microsatellite analysis, we identified five (2.1%) definite cases of MCC of the karyotype, and one probable case of MCC of MLPA analysis, as identified by sex discrepancies between the two methodologies.
In conclusion, MLPA has been shown to be a robust, semiautomated methodology with acceptable test characteristics for the detection of aneuploidy in POC. Combining the results of this study with previous studies by Diego-Alvarez et al.5 and Bruno et al.,6 the sensitivity of MLPA is estimated to be 87.9%, with a specificity of 90.2%. In our study, we identified seven structural variants (2.46% with 99% confidence interval of 0.09–4.83), highlighting an important need for adequate counseling or diagnostic reporting practices if MLPA is performed alone. It is interesting to also note that the comparative genomic hybridization array technology is subject to some of the same limitations in detecting balanced structural rearrangements as MLPA in the context of RPL. As an example, our MLPA reports now include a request for follow-up parental karyotyping when the result indicates trisomy of an acrocentric chromosome, which is commonly involved in Robertsonian translocations (e.g., 13, 14, and 21). In our study, MLPA was shown to enable a diagnosis in an extra 38 (13%) of patients. Despite the known limitations of the assay, for example, in the detection of polyploidy and structural variants, it still enabled a diagnosis for a significant additional cohort of patients. Given the low rate of polyploidy in POC samples, and their even lower recurrence rate, we believe that MLPA provides a significant advance in automated and multiplexed low-cost detection methodologies for the detection of aneuploidy; however, this study highlights the importance of choosing the right methodology for the appropriate clinical setting. Where there is a history of recurrent miscarriage, a karyotype may be the more appropriate test. If the aim is to make a diagnosis as to whether a genetic cause can be identified to account for a fetal demise, then MLPA may be the more appropriate method.
REFERENCES
Rai R, Regan L . Recurrent miscarriage. Lancet 2006; 368: 601–611.
Sierra S, Stephenson M . Genetics of recurrent pregnancy loss. Semin Reprod Med 2006; 24: 17–24.
Robinson WP, McFadden DE, Stephenson MD . The origin of abnormalities in recurrent aneuploidy/polyploidy. Am J Hum Genet 2001; 69: 1245–1254.
Lomax B, Tang S, Separovic E, et al. Comparative genomic hybridization in combination with flow cytometry improves results of cytogenetic analysis of spontaneous abortions. Am J Hum Genet 2000; 66: 1516–1521.
Diego-Alvarez D, Rodriguez de Alba M, Cardero-Merlo R, et al. MLPA as a screening method of aneuploidy and unbalanced chromosomal rearrangements in spontaneous miscarriages. Prenat Diagn 2007; 27: 765–771.
Bruno DL, Burgess T, Ren H, et al. High-throughput analysis of chromosome abnormality in spontaneous miscarriage using an MLPA subtelomere assay with an ancillary FISH test for polyploidy. Am J Med Genet A 2006; 140: 2786–2793.
Acknowledgements
M.F.B. is a recipient of a Marie Curie fellowship from the European Science Directorate.
Author information
Authors and Affiliations
Corresponding author
Additional information
Disclosure: The authors declare no conflict of interest.
Rights and permissions
About this article
Cite this article
Caramins, M., Saville, T., Shakeshaft, R. et al. A comparison of molecular and cytogenetic techniques for the diagnosis of pregnancy loss. Genet Med 13, 46–51 (2011). https://doi.org/10.1097/GIM.0b013e3181faa0d2
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1097/GIM.0b013e3181faa0d2
Keywords
This article is cited by
-
Detection rates of abnormalities in over 10,000 amniotic fluid samples at a single laboratory
BMC Pregnancy and Childbirth (2023)
-
Aneusomy detection with Karyolite-Bac on Beads® is a cost-efficient and high throughput strategy in the molecular analyses of the early pregnancy conception losses
Molecular Cytogenetics (2015)
-
Molecular analysis of products of conception obtained by hysteroembryoscopy from infertile couples
Journal of Assisted Reproduction and Genetics (2015)