Abstract
BubR1 mitotic checkpoint kinase monitors attachment of microtubules to kinetochores and links regulation of the chromosome-spindle attachment to mitotic checkpoint signaling. Defects in BubR1-mediated signaling severely perturb checkpoint control and are linked to diseases such as cancer. Studies using BubR1 mouse models suggest that BubR1 activities prevent premature aging and infertility. In this study, we show that BubR1 depletion in human adipose-derived mesenchymal stem cells (ASCs) precedes loss of the differentiation potential and induction of replicative senescence. These effects occur independently of p16INK4A expression and may involve DNA methylation. Our results reveal a new and unsuspected feature of BubR1 expression in regulation of adult stem cell differentiation.
Similar content being viewed by others
Introduction
BubR1 is a critical component of the mitotic check-point that is localized to kinetochores. BubR1 associates tightly with Cdc20 and/or with another protein that is essential to APC/C E3 ubiquitin ligase inhibition during checkpoint activation. BubR1 also monitors attachment of microtubules to kinetochores and links regulation of chromosome-spindle attachment to mitotic checkpoint signaling (Taylor et al., 1998; Mao et al., 2003; Lampson and Kapoor, 2005). Studies show that BubR1 disruption results in loss of checkpoint control, chromosomal instability (caused by premature anaphase), and/or early onset of malignancy (Cahill et al., 1998; Taylor et al., 1998; Baker et al., 2004; Dai et al., 2004). The regulatory profile of BubR1 exemplifies the tight interrelationship of checkpoint control, genomic integrity, and cell proliferative in diseases such as cancer.
The actions of BubR1, however, are likely to extend beyond checkpoint control. Our findings show, for example, that BubR1 promotes apoptosis in abnormal mitotic cells with chromosomal instability and may thereby influence post-mitotic adaptation (Shin et al., 2003; Ha et al., 2007). Mutant mice with low levels of BubR1 develop features of age-related diseases, such as cachectic dwarfism, sarcopaenia, cataracts, lordokyphosis, and impaired wound healing (Baker et al., 2004; 2008). Consistent with accelerated aging, the mutations significantly shortened life spans of these mutant mice compared to mice with normal BubR1 expression. BubR1-mutant mice showed defective chromosome segregation in meiosis and were infertile (Baker et al., 2004). In addition, gradual reduction of BubR1 expression in mouse embryonic fibroblasts promotes cellular senescence. These observations indicate that normal BubR1 expression prevents early aging and infertility in mice. Whether and how BubR1 influences the life span and senescence in human adult stem cells and primary cells is not yet known.
Results
BubR1 levels decline during replicative senescence in hASCs
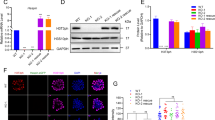
To determine the effect of BubR1 expression in human adult stem cells, we isolated human adipose-derived stem cells (hASCs) from the fatty portion of liposuction aspirates. hASCs, which may differentiate into adipocytes, chondrocytes, osteocytes, and neuronal cells, were positive for known mesenchymal markers (e.g., CD44, CD73 and CD105), but negative for CD45 (data not shown), as reported previously (Noel et al., 2008). Passage 1 hASCs were trypsinized and replated when cells reached approximately 80% culture-confluence. During several passages in vitro, ASCs gradually decline in proliferative potential and multipotentiality. To pursue these typical signs of senescence (Supplementary Figure S1), we measured p16INK4A mRNA expression (as a senescence marker) and GAPDH mRNA (as a control). After passage 9, we observed a marked increase in the level of p16INK4A mRNA, with protein expression increasing in parallel (data not shown). The correlation of p16INK4A induction and protein activity with the process of cellular senescence has been previously established (Krishnamurthy et al., 2004). Under these same experimental conditions, we monitored expression of the BubR1 and senescence-associated β-galactosidase (SA-β-gal) proteins. BubR1 protein levels increased slightly in early passages then, during propagation (passage 5), began to decline and were almost undetectable by passage 9 (Figure 1B). In contrast, the SA-β-gal level markedly increased by passage 9. These results point to an inverse correlation between BubR1 and SA-β-gal expression in ASC proliferation. Although the potential to differentiate varied between adipocyte preparations, we confirmed these results in ASCs from different patients. In early and mid-passage cells (passages 2 and 5), we confirmed the commitment of the cells to adipogenesis, as shown through lipid droplet formation (Figure 1C), as well as to osteogenesis and neurogenesis (data not shown). Late passage ASCs (passage 9), however, showed no lipid droplets and were unable to differentiate into adipocytes, consistent with positive staining for senescence markers (in situ acid-β-galactosidase staining).
Correlation between BubR1 expression, the potential to differentiate, and senescence in adipose-derived mesenchymal stem cells. (A) hASCs were isolated from liposuction aspirates, cultured (passage 1), and propagated for 2-11 passages. Expression of p16INK4A as a senescence marker was determined using quantitative real-time PCR. Triplicate samples were normalized to GAPDH. (B) Protein extracts (100 µg) from each passage of ASCs were processed and immunoblotted with anti-BubR1, anti-SA-β-gal, and anti-actin antibodies. (C) Early (passage 1), middle (passage 5), and late (passage 9) ASCs were cultured in a control medium and an adipogenic medium. Passage 5 ASCs that showed the highest level of BubR1 in (B) formed the lipid droplets typical of the adipogenic phenotype. Passage 9 ASCs had lost their adipogenic potential, but significantly increased the proportion of SA-β-gal positive (blue-green) cells. (D) Differentiation into adipocytes was confirmed using Oil-Red-O staining, as described in Methods.
BubR1 depletion leads to loss of stemness and induces cellular senescence independently of p16INK4A expression
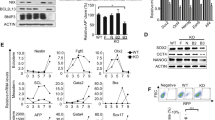
To further explore the direct involvement of BubR1 in ASC differentiation, we generated recombinant adenoviruses that depleted both BubR1 (rAd-sh-BubR1) and luciferase (rAd-shLuc). However, the transduction efficiency of recombinant adenoviruses (rAd) is extremely low in stem cells that express very low levels of the primary rAd receptor (Bergelson et al., 1997). The presence of protein transduction domains (PTDs) enables transduction of rAd into adult stem cells (Youn et al., 2008). As expected, co-treatment with HP4-PTD derived from herring protamine (Youn et al., 2008) and with rAd expressing the green fluorescence protein (rAd-GFP) dramatically enhanced in vitro transduction of rAd-GFP into ASCs (Figure 2A). Under these experimental conditions, we transduced either rAd-shLuc or rAd-shBubR1 into ASCs, and thereby significantly depleted endogenous BubR1 with rAd-shBubR1 transduction, but not with rAd-shLuc transduction (Figures 2B and 2C). rAd-shBubR1 transduction into passage 5 ASCs, which contain optimally active BubR1, significantly induced SA-β-gal production, whereas cells transduced with control rAd-shLuc showed no change in the SA-β-gal level. However, we did not observe changes in p16INK4A expression with depletion of BubR1 in passage 5 ASCs. We confirmed these results using cytochemical staining of SA-β-gal. As expected, ASCs transduced with rAd-shBubR1 significantly increased the number of SA-β-gal-positive cells (Figure 2D, left panels), consistent with failure of the ASCs to differentiate into adipocytes (Figure 2D, right panels). These results indicate that BubR1 probably contributes to the lineage commitment of ASCs, and that loss of BubR1 expression induces cellular senescence.
Targeted inhibition of BubR1 in ASCs blocks the differentiation potential and induces cellular senescence. (A) Introduction of the protein transduction domain HP4 enables transduction of rAd into ASCs. An rAd expressing GFP (rAd-GFP) was introduced into the cells, with or without the HP4 peptide, and GFP signals were visualized using fluorescence microscopy. (B) Passage 5 ASCs were transduced with rAd, in conjugation with the HP4 peptide, to selectively inhibit either BubR1 (rAd-shBubR1) or luciferase (rAd-shLuc) expression. After a 24 h transduction, cell lysates were prepared and immunoblotted with anti-BubR1, anti-SA-β-gal, anti-p16INK4A, and anti-actin antibodies. 'No' indicates non-transduced control ASCs. (C) Subcellular localization of BubR1 in ASCs. ASCs were co-stained with anti-BubR1 and anti-CENP-C (as a positive control for the kinetochore protein) antibodies, and then with either FITC- or rhodamine-conjugated secondary antibodies. DNA was visualized using Hoechst dye staining. BubR1 localizes at kinetochores in the prometaphase and the metaphase, but begins to dissociate from kinetochores at the anaphase. (D) Passage 5 ASCs were transduced with either rAd-shLuc or rAshBubR1, as described in (B), and then assayed using in situ acid-β-galactosidase (SA-β-gal) staining (left panels). Transduced ASCs, as above, were cultured in an adipogenic medium, and then monitored via formation of lipid droplets (right panels).
DNA methylation mediates the decline in the BubR1 level during replicative senescence
Changes in promoter methylation play an important role in BubR1 regulation (Park et al., 2007). To test whether DNA methylation is integral to down-regulation of BubR1 expression in senescent ASCs, cells from both early and late passages were cultured in the absence or presence of the irreversible DNA methyltransferase inhibitor 5-aza-2'-deoxycytidine (5-Aza-2-DC). BubR1 levels were determined using immunoblotting analysis (Figure 3A). Passage 10 (late) ASCs, which contain very low levels of BubR1, markedly restored BubR1 expression following treatment with 5-Aza-2-DC, whereas early passage cells (passage 3) that contain competent levels of BubR1 showed no apparent change in the BubR1 level. To determine the methylation status of the BubR1 promoter, we isolated genomic DNAs from early and late passage ASCs that were cultured in the absence or presence of 5-Aza-2-DC. The BubR1 promoter region was PCR-amplified using unmethylated primers as described in Methods. This procedure resulted in amplification products from the BubR1 promoter region in bisulfite-treated gDNAs from passage 3 cells, but not from passage 10 cells (Figure 3B). 5-Aza-2-DC treatment of the passage 10 ASCs markedly restored amplification of the unmethylated BubR1 promoter. We also performed methylation-specific PCR using both methylated and unmethylated primers for the BubR1 promoter (Figure 3C). Consistent with previous results, the BubR1 promoter was unmethylated in passage 3 (early) ASCs, but highly methylated in passage 10 (late) ASCs. These results suggest that promoter methylation mediates down-regulation of BubR1 and induction of cellular senescence in proliferating ASCs.
DNA methylation down-regulates BubR1 during replicative senescence. (A) ASCs at passages 3 and 10 were cultured in the absence (-) or presence (+) of the DNA methyltransferase inhibitor 5'-aza-2'-deoxycytidine (5-Aza-2-DC). Cell lysates were immunoblotted with anti-BubR1 and anti-actin antibodies. (B) PCR amplification of the unmethylated BubR1 promoter. Genomic DNA (gDNA) was isolated from the ASC samples from (A). The BubR1 promoter was amplified using PCR primers specific for the unmethylated region of bisulfite-treated DNA samples. A PCR template control (1 µg gDNA) is shown. (C) Methylation-specific PCR. Primers were designed to amplify methylated and unmethylated BubR1 promoters. M, methylated. UM, unmethylated. An asterisk indicates the non-specific band. (D) Model of BubR1 regulation in adult stem cell differentiation commitment.
Conversely, our findings support a role for BubR1 in maintenance of the differentiation potential and delay of cellular senescence in proliferating ASCs.
Discussion
During extended propagation, ASCs show a marked decline in BubR1 expression that correlates with loss of the potential to differentiate. Targeted inhibition of BubR1 expression in ASCs significantly reduces the potential to differentiate and leads to cellular senescence. These observations support a role for BubR1 in lineage commitment. Studies in mouse models further link these BubR1 activities to processes related to aging.
The cyclin-dependent kinase inhibitor p16INK4A participates in cellular senescence induction and physiological correlates of aging in a number of mouse models (Zindy et al., 1997; Krishnamurthy et al., 2004). In BubR1-depleted ASCs, however, we did not detect an increase in p16INK4A expression, in contrast to results from a previous report (Baker et al., 2008). Although BubR1 and p16 levels are inversely correlated in mouse tissues and human ASCs, it does not follow that BubR1 directly affects the transcriptional and/or post-translational regulation of p16INK4A. Accordingly, the loss of stemness through BubR1 depletion may not trigger p16INK4A-mediated senescence directly, but rather, loss of the potential to differentiate may induce senescence as a consequence. Depletion of BubR1 in human cancer cells (e.g., HeLa and HCT116) does not induce SA-β-gal production (data not shown). However, we were unable to determine whether introduction of exogenous BubR1 increased stem cell proliferation and the differentiation potential because of its strong apoptotic effect when overexpressed (Shin et al., 2003).
A small proportion of human cancers display mutations in BubR1 (Lengauer et al., 1997; Kops et al., 2005), and some of these mutations may be linked to mosaic variegated aneuploidy (Hanks et al., 2004). It is not known, however, whether the BubR1 mutations actually cause this rare recessive defect. We recently demonstrated that BubR1 expression is significantly regulated by DNA methylation (Park et al., 2007) and we show here that DNA methylation also mediates suppression and inactivation of BubR1 during replicative senescence.
In conclusion, our research has shown that the mitotic checkpoint kinase BubR1 probably plays an essential role in stem cell differentiation, and that promoter methylation is probably the major mechanism for BubR1 inactivation during the natural aging process.
Methods
Isolation of human adipose tissue-derived mesenchymal stem cells (hASCs)
The Institutional Review Board (IRB) at Kangbuk Samsung Hospital, Sungkyunkwan University School of Medicine approved this study, and for all liposuctioned tissues used herein we obtained written informed consent from all donors prior to surgery. hASCs were isolated from adipose tissue using methods previously described (Schaffler and Buchler, 2007), with minor modifications. Briefly, the adipose tissue was washed with PBS and centrifuged at 1,200 g for 5 min to remove red blood cells. Washed aspirates were treated with 0.0075% collagenase (Type XI, Sigma-Aldrich) in PBS for one hour on a shaker at 37℃. Enzymatic digestion was stopped by addition of an equal volume of a growth medium (high glucose DMEM, 10% FBS, 1% Pen/Strep), followed by centrifugation at 1,200 g for 10 min. The pellet was reconstituted using a normal growth medium, plated in a T-75 tissue culture flask, and incubated at 37℃ in 5% CO2. The medium was changed every two to three days.
Differentiation assay
For adipogenesis, cells were plated onto multi-well plates in a growth medium and maintained until they reached 100% confluency (approximately three weeks). Cells were cultured in an adipogenic medium (low glucose DMEM, 10% FBS, 1% Pen/Strep supplemented with 1 µM dexamethasone, 1 µM indomethacin, 10 µg/ml of insulin, and 500 µM IBMX) for three days, then transferred to an adipocyte maintenance medium (low glucose DMEM, 10% FBS, 1% Pen/Strep supplemented with 10 µg/ml of insulin) for one day (Lin et al., 2005).
Histological assays
Adipogenic differentiation was determined using Oil Red O staining to detect lipid droplet formation. Cells were fixed in 10% formalin solution for 30 min, washed with deionized water to remove formalin, and then washed with 60% isopropanol for another 5 min. An Oil Red O working solution was added and the cells were incubated for 30 min. After staining, cells were washed with deionized water until the fluid became clear (Zuk et al., 2001).
Senescence-associated β-galactosidase (SA-β-gal) assay
Cells were fixed with 3% formaldehyde for 10 min and incubated overnight at 37℃ with 1 mg/ml of X-gal (5-bromo-4-chloro-3-indolyl-β-D-galactopyranoside), 5 mM K3Fe(CN)6, 5 mM K4Fe(CN)6, 150 mM NaCl, and 2 mM MgCl2 in 40 mM citric acid/sodium phosphate pH 6.0 (Dimri et al., 1995).
Immunoblotting and immunofluorescence
Cells for immunoblots were lysed in NE buffer (20 mM Hepes, pH 7.6, 250 mM NaCl, 1.5 mM MgCl2, 20% glycerol, 0.1% Triton X-100, 1 mM DTT, 1 mM PMSF, protease inhibitor cocktail), and lysates were clarified by centrifugation at 13,200 rpm for 30 min. After blocking in TBS-T (20 mM Tris, 137 mM NaCl, 0.1% Tween-20, pH 7.4) with 5% skim milk, membranes were incubated with the primary antibodies at appropriate dilutions in 5% skim milk/TBS-T for a minimum of three hours at room temperature or overnight in a cold room. Membranes were then washed three times with TBS-T and incubated with HRP-conjugated secondary antibodies (1:3000) in 5% skim milk/ TBS-T. Membranes were developed for visual analysis using enhanced chemiluminescence. For immunofluorescence staining, hASCs were grown on coverslips (1 × 104 cells/each) for two days. Cells were fixed with 5% formaldehyde and incubated with antibodies in a 3% skim milk solution. DNA was stained with Hoechst dye.
Quantitative real-time RT-PCR
Total RNA was extracted using an RNeasy Mini Kit (Qiagen, Germany) and quantified using UV spectroscopy. To prepare RNA for PCR analysis, 1.2 µg of RNA was converted to cDNA using the ImProm-II Reverse Transcription System (Promega). qRT-PCR reactions were performed with SYBR Green PCR Master Mix in a MicroAmp optical 96-well reaction plate (Applied Biosystems). The mRNA expression of candidate genes was determined using real-time quantitative PCR with an AMI PRISM 7000 SDS v1.1 instrument according to the manufacturer's instructions (Applied Biosystems). The following specific primers were used : GAPDH (5'-GGCATGGACTGTGGTCATGAG-3', 5'-TGCACCACCAACTGCTTAGC-3'), p16INK4A (5'-CATCAGTCACCGAAGGTCCT-3', 5'-AATGGACATTTACGGTAGTGGG-3').
Sodium bisulfite PCR and methylation-specific PCR
Genomic DNAs were isolated using a DNeasy tissue Kit (Qiagen, Germany) and quantified using UV spectroscopy. gDNA was digested with HindIII and the isolated DNA fragments were denatured with 0.3 N sodium hydroxide at 39℃ for 30 min, then treated with 5.5 mM hydroquinone and 2.5 M sodium bisulfite at pH 5.0. Samples were incubated in a thermocycler at 55℃ for 16 h and punctuated every three hours by a 5 min denaturation at 95℃. Converted DNAs were desalted using an Expin Gel SV Kit (GeneAll, Korea), denatured in 0.3 N sodium hydroxide at 37℃ for 15 min, and purified using ethanol precipitation. The bisulfite-modified DNAs were amplified using PCR with BubR1 promoter-specific primers (5'-GTAGTTTGTTTAAGGTTGTATATTTGGTATG-3', 5'-CCATCCTACATTCCTAACTCAAAT-3'). Products from nested PCR amplifications (5'-TAAGGGTGGGGTAGGAAATAGTTAGG-3', 5'-ACTACAAACCTTTAATCTAAACCAC-3') were obtained as described previously (Park et al., 2007). For methylation-specific PCR (MS-PCR), bisulfate-modified DNAs were amplified using PCR with primers for methylated BubR1 (5'-ACGGAGGAGCGGAGGGGC-3', 5'-CACGTACACGACTCCCTAACAAACCG-3') or unmethylated BubR1 (5'-ATGGAGGAGTGGAGGGGT-3', 5-CACATACACAACTCCCTAACAAACCA-3') regions.
Generation of recombinant adenovirus and transduction in vitro
To construct a recombinant adenoviral vector expressing shBubR1, the H1 promoter for the shBubR1-oligo region of the pSuper-puro-shBubR1 vector was subcloned into the pShuttle vector (Quantum Biotech, Canada) to generate pShuttle-shBubR1. The resultant constructs were co-transformed with pAdEasy-1 DNA into E. coli BJ5183 cells for homologous recombination. Recombinant adenoviruses (rAd shBubR1) were recovered from 293A cells that were calcium phosphate precipitate-transfected with the linearized recombinant adenoviral DNA pAd shBubR1 (Shin et al., 2003). Amplified shBubR1 adenoviruses were purified using a Vivapure AdenoPACK 100RT Kit (Sartorius, Germany) according to the manufacturer's instructions. HP4, one kind of protein transduction domain (PTD), was synthesized by PEPTRON Inc. (Daejeon, Korea). A mixture of rAds at a multiplicity of infection (MOI) of 100 plaque-forming units (PFU) per cell, and HP4 (100 nM) was incubated in a serum-free medium for 30 min at room temperature. Cells were washed and incubated with the rAds and HP4 preparation at 37℃ in a CO2 incubator. After two hours, the cells were washed and incubated with a serum-containing medium (Youn et al., 2008).
Abbreviations
- 5-Aza-2-DC:
-
5'-aza-2'-deoxycytidine
- ASCs:
-
adipose-derived stem cells
- PTD:
-
protein transduction domain
- rAd:
-
recombinant adenovirus
- SA-β-gal:
-
senescence-associated β-galactosidase
References
Baker DJ, Jeganathan KB, Cameron JD, Thompson M, Juneja S, Kopecka A, Kumar R, Jenkins RB, de Groen PC, Roche P, van Deursen JM . BubR1 insufficiency causes early onset of aging-associated phenotypes and infertility in mice . Nat Genet 2004 ; 36 : 744 - 749
Baker DJ, Perez-Terzic C, Jin F, Pitel K, Niederlander NJ, Jeganathan K, Yamada S, Reyes S, Rowe L, Hiddinga HJ, Eberhardt NL, Terzic A, van Deursen JM . Opposing roles for p16INK4A and p19Arf in senescence and ageing caused by BubR1 insufficiency . Nat Cell Biol 2008 ; 10 : 825 - 836
Bergelson JM, Cunningham JA, Droguett G, Kurt-Jones EA, Krithivas A, Hong JS, Horwitz MS, Crowell RL, Finberg RW . Isolation of a common receptor for Coxsackie B viruses and adenoviruses 2 and 5 . Science 1997 ; 275 : 1320 - 1323
Cahill DP, Lengauer C, Yu J, Riggins GJ, Willson JK, Markowitz SD, Kinzler KW, Vogelstein B . Mutations of mitotic checkpoint genes in human cancers . Nature 1998 ; 392 : 300 - 303
Dai W, Wang Q, Liu T, Swamy M, Fang Y, Xie S, Mahmood R, Yang YM, Xu M, Rao CV . Slippage of mitotic arrest and enhanced tumor development in mice with BubR1 haploinsufficiency . Cancer Res 2004 ; 64 : 440 - 445
Dimri GP, Lee X, Basile G, Acosta M, Scott G, Roskelley C, Medrano EE, Linskens M, Rubelj I, Pereira-Smith O . A biomarker that identifies senescent human cells in culture and in aging skin in vivo . Proc Natl Acad Sci USA 1995 ; 92 : 9363 - 9367
Gimble J, Guilak F . Adipose-derived adult stem cells: isolation, characterization, and differentiation potential . Cytotherapy 2003 ; 5 : 362 - 369
Ha GH, Baek KH, Kim HS, Jeong SJ, Kim CM, McKeon F, Lee CW . p53 activation in response to mitotic spindle damage requires signaling via BubR1-mediated phosphorylation . Cancer Res 2007 ; 67 : 7155 - 7164
Hanks S, Coleman K, Reid S, Plaja A, Firth H, Fitzpatrick D, Kidd A, Mehes K, Nash R, Robin N, Shannon N, Tolmie J, Swansbury J, Irrthum A, Douglas J, Rahman N . Constitutional aneuploidy and cancer predisposition caused by biallelic mutations in BUB1B . Nat Genet 2004 ; 36 : 1159 - 1161
Kops GJ, Weaver BA, Cleveland DW . On the road to cancer: aneuploidy and the mitotic checkpoint . Nat Rev Cancer 2005 ; 5 : 773 - 785
Krishnamurthy J, Torrice C, Ramsey MR, Kovalev GI, Al-Regaiey K, Su L, Sharpless NE . INK4A/Arf expression is a biomarker of aging . J Clin Invest 2004 ; 114 : 1299 - 1307
Lampson MA, Kapoor TM . The human mitotic checkpoint protein BubR1 regulates chromosome-spindle attachments . Nat Cell Biol 2005 ; 7 : 93 - 98
Lengauer C, Kinzler KW, Vogelstein B . Genetic instability in colorectal cancers . Nature 1997 ; 386 : 623 - 627
Lin TM, Tsai JL, Lin SD, Lai CS, Chang CC . Accelerated growth and prolonged lifespan of adipose tissue-derived human mesenchymal stem cells in a medium using reduced calcium and antioxidants . Stem Cells Dev 2005 ; 14 : 92 - 102
Mao Y, Abrieu A, Cleveland DW . Activating and silencing the mitotic checkpoint through CENP-E-dependent activation/inactivation of BubR1 . Cell 2003 ; 114 : 87 - 98
Noel D, Caton D, Roche S, Bony C, Lehmann S, Casteilla L, Jorgensen C, Cousin B . Cell specific differences between human adipose-derived and mesenchymal-stromal cells despite similar differentiation potentials . Exp Cell Res 2008 ; 314 : 1575 - 1584
Park HY, Jeon YK, Shin HJ, Kim IJ, Kang HC, Jeong SJ, Chung DH, Lee CW . Differential promoter methylation may be a key molecular mechanism in regulating BubR1 expression in cancer cells . Exp Mol Med 2007 ; 39 : 195 - 204
Schaffler A, Buchler C . Concise review: adipose tissue-derived stromal cells--basic and clinical implications for novel cell-based therapies . Stem Cells 2007 ; 25 : 818 - 827
Shin HJ, Baek KH, Jeon AH, Park MT, Lee SJ, Kang CM, Lee HS, Yoo SH, Chung DH, Sung YC, McKeon F, Lee CW . Dual roles of human BubR1, a mitotic checkpoint kinase, in the monitoring of chromosomal instability . Cancer Cell 2003 ; 4 : 483 - 497
Taylor SS, Ha E, McKeon F . The human homologue of Bub3 is required for kinetochore localization of Bub1 and a Mad3/Bub1-related protein kinase . J Cell Biol 1998 ; 142 : 1 - 11
Youn JI, Park SH, Jin HT, Lee CG, Seo SH, Song MY, Lee CW, Sung YC . Enhanced delivery efficiency of recombinant adenovirus into tumor and mesenchymal stem cells by a novel PTD . Cancer Gene Ther 2008 ; 15 : 703 - 712
Zindy F, Quelle DE, Roussel MF, Sherr CJ . Expression of the p16INK4A tumor suppressor versus other INK4 family members during mouse development and aging . Oncogene 1997 ; 15 : 203 - 211
Zuk PA, Zhu M, Ashjian P, De Ugarte DA, Huang JI, Mizuno H, Alfonso ZC, Fraser JK, Benhaim P, Hedrick MH . Human adipose tissue is a source of multipotent stem cells . Mol Biol Cell 2002 ; 13 : 4279 - 4295
Zuk PA, Zhu M, Mizuno H, Huang J, Futrell JW, Katz AJ, Benhaim P, Lorenz HP, Hedrick MH . Multilineage cells from human adipose tissue: implications for cell-based therapies . Tissue Eng 2001 ; 7 : 211 - 228
Acknowledgements
This work was supported by a Korea Science and Engineering Foundation (KOSEF) grant funded by the Korean government (MEST) [R01-2007-000-20007-0 (2008)] and by the Korea Research Foundation (KRF-2005-041-C00452).
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
This is an Open Access article distributed under the terms of the Creative Commons Attribution Non-Commercial License (http://creativecommons.org/licenses/by-nc/3.0/) which permits unrestricted non-commercial use, distribution, and reproduction in any medium, provided the original work is properly cited.
About this article
Cite this article
Lee, J., Lee, C., Lee, KW. et al. Cross-talk between BubR1 expression and the commitment to differentiate in adipose-derived mesenchymal stem cells. Exp Mol Med 41, 873–879 (2009). https://doi.org/10.3858/emm.2009.41.12.093
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.3858/emm.2009.41.12.093
Keywords
This article is cited by
-
Mps1 kinase regulates tumor cell viability via its novel role in mitochondria
Cell Death & Disease (2016)
-
Ablation of Dido3 compromises lineage commitment of stem cells in vitro and during early embryonic development
Cell Death & Differentiation (2012)