Abstract
Background:
Overexpression of Galectin-3 (Gal-3), a β-galactoside binding protein, has been noted in many tumour types but its functional significance and clinical utility in gastric adenocarcinoma (GAC) are not well known.
Methods:
We studied 184 GAC patients characterised by histologic grade, sub-phenotypes (diffuse vs intestinal), and ethnicity (Asians vs North Americans). Immunohistochemistry was performed to assess the expression of Gal-3 in human GACs and we correlated it to the clinical outcomes. Cell proliferation, invasion, co-immunoprecipitation and kinase activity assays were done in genetically stable Gal-3 overexpressing GC cell lines and the parental counterparts to delineate the mechanisms of action and activity of inhibitors.
Results:
Most patients were men, Asian, and had a poorly differentiated GAC. Gal-3 was over-expressed in poorly differentiated (P=0.002) tumours and also in diffuse sub-phenotype (P=0.02). Gal-3 overexpression was associated with shorter overall survival (OS; P=0.026) in all patients. Although, Gal-3 over-expression was not prognostic in the Asian cohort (P=0.337), it was highly prognostic in the North American cohort (P=0.001). In a multivariate analysis, Gal-3 (P=0.001) and N-stage (P=<0.001) were independently prognostic for shorter OS. Mechanistically, Gal-3 induced c-MYC expression through increasing RalA activity and an enhanced YAP1/RalA/RalBP complex to confer an aggressive phenotype. YAP1/BET bromodomain inhibitors reduced Gal-3-mediated aggressive phenotypes in GAC cells.
Conclusions:
Gal-3 is an independent prognostic marker of shorter OS and a novel therapeutic target particularly in diffuse type GAC in North American patients.
Similar content being viewed by others
Main
Gastric adenocarcinoma (GAC) is rampant in many endemic areas of the world with more than 800 000 new cases and it remains the third most common cause of cancer death globally (Jemal et al, 2011; Siegel et al, 2017). In most countries, GAC is diagnosed in the late stages as patients are diagnosed based on symptoms. Localised GAC is difficult to cure but metastatic GAC is invariably incurable (Ajani and Takiuchi, 1999; Ajani, 2007; Wadhwa et al, 2014). Therapeutic options are limited because currently, we have only two lines of therapies available and many patients in good general physical and physiologic condition lack therapeutic options. Only a few have an access to clinical trials (Jiang and Ajani, 2010; Wadhwa et al, 2013, 2014). GAC often exhibits primary resistance to systemic therapy but when responsive to initial therapy, the benefit is transient and tumour becomes resistant. Advanced genetic and molecular studies have yielded a vast quantity of new information for potential exploitation. Unfortunately, only a few target agents are useful to GAC patients. Rational therapeutic approaches are urgently needed.
Galectin-3 (Gal-3) is a β-galactoside binding protein in the lectin family that has multiple physiologic and pathologic functions (Song et al, 2009; Newlaczyl and Yu, 2011; Radosavljevic et al, 2012; Song et al, 2012). Gal-3 is synthesised in the cytoplasm and is a monomer at low concentrations. Gal-3 is preferentially bound to glycoproteins such as CD44. Eventually forming a pentamer on the surface of cell membrane with CD44 and GSL to orchestrate membrane bending and clathrin-dependent endocytosis (Stanley, 2014). Function of Gal-3 appears to be context dependent. In cytoplasm, it promotes cell proliferation (Song et al, 2009), apoptosis inhibition (Mazurek et al, 2012), and cell cycle promotion (Wang et al, 2009). On the surface of cell membrane, it promotes apoptosis in cytotoxic T-cells (Xue et al, 2017). Gal-3 is pro-angiogenic and facilitates adhesion/metastases (Wesley et al, 2013). In the nucleus, it facilitates transcription regulation and mRNA splicing. Finally, circulating Gal-3 facilitates cancer cell dissemination, adhesion, and anti-apoptotic functions.
Gal-3 overexpression has been documented in pancreatic cancer, breast cancer, hepatocellular carcinoma, prostate cancer, head and neck cancer, among others (Moon et al, 2001; Song et al, 2012; Jiang et al, 2014; Salajegheh et al, 2014), but its role in GAC has been addressed in a limited manner. Okada et al (2006) reported that lower expression of Gal-3 was associated with poor survival. Dong et al, (2008) reported that Gal-3 mRNA levels were higher in GAC patients with higher stage, with poor differentiation, and with node positivity. Prognostic implications were not mentioned. In our report, which has novel information compared to prior reports, for the first time we document the significance of Gal-3 overexpression, which differs in geographically unique populations, with particular relevance to North American GAC patients. We also provide supporting preclinical data to delineate its mechanisms of action. Finally, we provide evidence that high expression Gal-3 is a potential therapeutic target by novel inhibitors.
Materials and methods
Patient population
GAC specimens from the North American (University of Texas M. D. Anderson Cancer Center in Houston [UTMDACC], Texas, USA) and Japanese patients (St. Lukes Hospital in Tokyo, Japan) were obtained under an agreement approved by Ethics Committees of both institutions. UTMDACC Investigation Review Board has approved this analysis. Clinical parameters of all patients were collected in a database and personal information was de-identified.
Cells and reagents
The human GC cell lines Snu1 and GT5 were obtained from Ju-Seog Lee, UTMDACC and were previously described (Soldes et al, 1999; Raju et al, 2006; Wang et al, 2012). All cell lines are genetically confirmed in the cell line core facility every 6 months (UTMDACC policy). Cells were cultured in DMEM supplemented with 10% foetal bovine serum (FBS) and antibiotics (100 μg ml−A streptomycin and 100 IU ml−U of penicillin). Anti-Gal-3 antibody was obtained as described previously (Song et al, 2005, 2009, 2012). Anti-RalA monoclonal antibody, anti-phospho-S6, and anti-YAP1 antibodies were from Cell Signaling Technology, Inc. (Beverly, MA, USA). Anti-c-MYC antibody was from Santa Cruz Biotechnology, Inc. (Santa Cruz, CA, USA).
Protein isolation and immunoblotting analysis
Protein isolation and western blot analyses were performed as previously described (Song et al, 2009).
Generation of stable Gal-3 overexpressing GC cell lines
To produce lentivirus for Gal-3 overexpression, pLVTHM-Gal-3 and control vector were co-transfected with the packaging plasmid (MD2G) and envelope plasmid (PAX2) required for viral production into 293FT cells by using lipofectamine 2000 reagent (Invitrogen, Carlsbad, CA, USA). A medium containing lentivirus with Gal-3 overexpression (named Gal-3) and control vector (named V) were used to transduce Snu1 and GT-5 GC cell lines with none or low Gal-3 expression. Cell sorting was conducted in a fluorescence-activated cell sorting ARIA flow cytometer (BD Biosciences, San Jose, CA, USA). Gal-3 expression of stable cell lines was further confirmed by Western blotting.
Cell proliferation assay
Cell viability was measured using the CellTiter Aqueous One Solution Cell Proliferation Assay kit as previously described (Song et al, 2009).
Co-immunoprecipitation
Co-immunoprecipitation in control and Gal-3 overexpressed cells from both Snu1 and GT-5 cells were performed using RalBP1 antibody pulldown and immonoblotting with YAP1 antibody as described (Song et al, 2009).
RalA activity assay
Equal amounts of cell lysate protein from control cells or Gal-3 stably expressed cells from Snu1 and GT-5 were incubated for 45 min at 4 °C, with agarose beads coated with RalBP1 agarose for total Ral A activity (Upstate Biotechnology, Inc., Lake Placid, NY, USA). Beads were then washed and bound protein was eluted with 2 × Laemmli sample buffer that had been preheated to 95 °C and analysed by immunoblotting for RalA activity using anti-RalA monoclonal antibody.
Matrigel invasion and migration assays
The invasive capability of cells was determined by using Matrigel-coated invasion chambers with 0.8 μm pore size (BD Biosciences). A single-cell suspension containing 1 × 105 cells was added to the inner chamber. After 16 h of incubation at 37 °C in 5% CO2, cells on the upper surface of the inner chamber were removed with cotton swabs. Invaded cells that adhered on the lower surface of the membrane were fixed, stained with Diff-Quik (Dade Behring), and counted.
Transient transfection and luciferase reporter assays
The 5 × -UAS-luciferase reporter and Gal4-TEAD4 constructs (reflected YAP1/Tead transcriptional activity), described previously (Zhao et al, 2009), were obtained from Dr. Johnson of MD Anderson. The c-MYC luciferase reporter described previously (He et al, 1998; Song et al, 2013) was obtained from Addgene (Cambridge, MA, USA). Transient co-transfection GC cells (V vs Gal-3) with 5 × -UAS-luciferase reporter and Gal4-TEAD4 or c-MYC luciferase reporter plasmid with or without YAP1 cDNA as well as a Renilla vector (pCH110) were performed as described previously (Song et al, 2013). Luciferase reporter activity was measured after 48 h of transfection. For all experiments, values shown represent the mean and SD of at least triplicate assays (*P<0.05; **P<0.01).
Real-time PCR
To quantify the changes in Gal-3, c-MYC, YAP1, CTGF mRNA levels, real-time RT-PCR was performed on the ABI Prism 7900 (Applied Biosystems, Foster City, CA, USA) using primers as followings: of genes for Gal-3 (Mm00802901_m1), and the cyclophilin A (4326316E; Applied Biosystems) as described previously (Song et al, 2009). The 7900-Sequence Detection System 2.2 software (Applied Biosystems) automatically determined the fold-change for Gal-3 in each sample by using the δδCt method with 95% confidence.
Reverse-phase protein arrays and gene set enriched analysis
Reverse-phase protein arrays (RPPA) analysis was performed in cell lysate from GAC cell lines (Snu1 and GT-5) with Gal-3 overexpression (named Gal-3) and control vector (named V) in RPPA core UTMDACC facility. Samples were serially diluted two-fold for five dilutions and probed with 275 antibodies and arrayed on nitrocellulose-coated slides. Relative protein levels were normalised for protein loading and determined by interpolation of each dilution curve from the standard curve as previously described. Gene set enriched analysis (GSEA) was conducted by a specialised bioinformatician (BL) as previously described (Wang et al, 2012).
Immunohistochemistry
Immunohistochemical staining for Gal-3 was performed on microarray tissue slides consisting of specimens from 212 GAC patients who had surgery at UTMDACC or at St. Lukes Hospital in Japan. After antigen retrieval and endogenous peroxidase blockage, the sections were incubated with antibodies against Gal-3 (1 : 100 dilution) at 4 °C overnight, then incubated with secondary antibody at room temperature for 60 min. Standard avidin–biotin immunohistochemical analyses were performed according to the manufacturer’s recommendations (Vector Laboratories, Burlingame, CA, USA). The staining results were evaluated by pathologist (CQ) based on the percentage of staining in tumour cells (0, no staining; 1, ⩾10%; 2, 10%–50% and 3, >50%) and the staining intensity (0—negative, 1—weak, 2—moderate and 3—strong). GACs were categorised into Gal-3-low (combined scores <3.5) and Gal-3-high (combined score β⩾3.5).
Statistical analysis
Two biostatisticians (AC and LX) independently analysed the results of Gal-3 immunohistochemistry with regards to their clinical relevance. The Fisher’s exact test and Wilcoxon rank sum test/Kruskal-Wallis test were used to evaluate the association of Gal-3 with categorical and continuous covariates, respectively. The Kaplan–Meier method was used to estimate probability of survival. The log rank test and Cox model were used to determine the association between markers and survival outcomes. Other assays are presented in graphs as mean±s.d. and represent the results of at least three experiments. Significance of differences between the groups was judged using a two-tailed Student t-test. Results were considered statistically significant if the P value was <0.05.
Results
Gal-3 is highly upregulated in human GACs
Previously, we reported that Gal-3 promotes cell growth and invasion in colon and pancreas by promoting Wnt/β-catenin (Song et al, 2009) and activation of Ras (Song et al, 2012). To explore its role in GAC, we first examine Gal-3 expression level in patient samples and cell lines. Patient characteristics for the entire TMA cohort are shown in Supplementary Table 1. There was predominance of men, poorly differentiated GAC, and Japanese patients. Gal-3 expression was high in the majority of GACs (including intestinal and diffused phenotypes), while there was no or low expression of Gal-3 in normal gastric tissues (Figure 1A). Fifty one of 62 (82%) poorly differentiated GACs had high levels (Gal-3-high), while only 18% well-moderate GACs had Gal-3-high. Gal-3 expression was higher in diffuse type (63%) than intestinal type (37%; P<0.022). Interestingly, we found that the expression of Gal-3 is significantly higher in the North American cohort (72%) than that in the Japanese cohort (49.7%) (Supplementary Figure 1). Gal-3 expression was present in most of GAC cell lines (Supplementary Figure 1A).
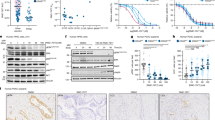
Gal-3 is highly up-regulated in human GAC tissues and associated with poor survival in North American GAC patients. (A) Human gastric tissue microarray slides consisting of 215 Gastric adenocarcinomas and non-neoplastic gastric tissue samples were immunohistochemically stained using a monoclonal Gal-3 antibody as described in Materials and Methods. Gal-3 expression was increased in both diffused and intestinal GAC tissues compared to normal stomach. Representative images show IHC staining for Gal-3 in tissues of normal, intestinal and diffused type GAC tissues. (B) Cumulative Survival Probability was analysed in All GAC patients by specialised statisticians (A.C) using Kaplan–Meier methods; Gal-3 expression high (>3.5) or low (<3.5) as a cut off as indicated in meterials and Methods (P=0.025). (C) Cumulative Relapse-Free Survival was analysed using Kaplan–Meier methods in All GAC patients; Gal-3 expression high (>3.5) or low (<3.5) as a cut off as indicated in materials and Methods. High Gal-3 expression is associated with shorter survival (P=0.029). (D) Cumulative Survival Probability was analysed in American GAC patients by specialised statisticians (A.C) using Kaplan-Meier methods (P=0.001). (E) Cumulative Survival Probability was analysed in Japanese GAC patients by specialised statisticians (A.C) using Kaplan–Meier methods (P=0.335).
Univariate and multivariate survival analyses
We noted that Gal-3 levels were elevated in poorly differentiated GACs and in diffuse type of GAC (Supplementary Table1). In univariate Cox Regression analysis (Table 1), poorly differentiated GAC (P=0.007), higher T stage (P=<0.0001), nodes positive tumours (P=<0.0001), M1 stage (P=<0.0001) and overexpression of Gal-3 (P=0.026) were associated with higher risk of death in all patients (Table 1). The Kaplan–Meier survival analysis demonstrated that OS was significantly shorter for patients with higher Gal-3 than patients with lower Gal-3 level (P=0.025, Figure 1B). Disease-free survival also decreased in patients with high Gal-3 expression than that with low Gal-3 expression (P=0.029, Figure 1C). More intriguingly, when we separately analyse the OS between US patients and Japanese patients, we found that Gal-3 overexpression was highly associated with higher risk of death in North American patients (P=0.001; Figure 1D) but not in Japanese patients (P=0.335; Figure 1E). In the multivariate Cox regression analysis, Gal-3 (P=0.001) and pN+ (P=<0.001) were independently associated with shorter OS in North American patients (Table 2) but not in Japanese patients.
Gal-3 upregulates c-MYC and increases GAC cell proliferation, invasion and migration
To define whether Gal-3 regulation of c-MYC facilitates malignant phenotype, we stably transfected Gal-3 cDNA into Snu1 and GT-5 GAC cells with no or low Gal-3 expression among cell lines (Supplementary Figure 2A) using Gal-3 cDNA lentivirus transduction and selection using GFP FACS sorting. RPPA was performed on these cells and their Gal-3 stable clones. Figure 2A shows that many oncogenic transcriptional factors were increased by Gal-3 stable transfection. c-MYC was the most induced of all oncogenes (Red arrow), GSEA showed that c-MYC regulated genes, cytoskeleton related, and mTOR pathway genes were upregulated (Figure 2B). We noted that transfected Gal-3 into Snu1 and GT-5 dramatically increased c-MYC protein (western blot; Figure 2C) and its mRNA levels by the quantitative–PCR assay (Figure 2D). By using MTS proliferation assay and Matrigel invasion assay, we document that transfected Snu1 and GT-5 cells had higher capacity for proliferation (Figure 2E), invasion (Figure 2F), and migration (Figure 2G).
Gal-3 upregulates c-MYC and increases GAC cell proliferation, invasion and migration. (A) Heat map shows the gene expression change induced by Gal-3 in GT-5 cells. (B) Gene set enriched analysis (GSEA) conducted by a specialised bioinformatist (BL) demonstrated that c-MYC and many other oncogenic genes are up-regulated by Gal-3. (C) Overexpression Gal-3 in Snu1 and GT-5 cells with no or lower Gal-3 expressed cells by lentivirus infection of Gal3 cDNA (Gal3) or vector control (V) was confirmed by immunoblots. Strong induction of c-MYC expression was detected in Gal-3 overexpression GAC cells by immunobloting as described in Materials and Methods. (D) Real-time quantitative PCR for Gal-3 and c-MYC RNA expression using Gal-3 and c-MYC specific primers after total RNA extraction from GAC cells with without Gal-3 stably transfection, **P<0.01. (E) Effects of Gal-3 on cell proliferation. Proliferation of Snu1 and GT5 cells in which over-expressed by Gal-3 cDNA was determined by using the CellTiter Aqueous One Solution Cell Proliferation Assay kit as described in Materials and Methods, **P<0.01. (F) Effect of Gal-3 on GAC cell invasion. Representative fields (top panel) and quantification (low panel) are shown in Snu1 and GT-5 GAC cells (1 × 105). Snu1 and GT-5 cells with Gal-3 overexpression (Gal3) and vector transfected control cells (V) were seeded onto Matrigel-coated invasion chambers; 24 h later, invaded cells that adhered on the lower surface of the membrane were fixed, stained with Diff Quick set, and count as described in Materials and Methods. Representative fields (top panel) and quantification (low panel) are **P<0.01. (G) Effect of Gal-3 on GAC cell migration. GAC cells (1 × 105) in which Gal-3 overexpression (Gal-3) and vector transfected control cells (V) were seeded onto six-well plates and migration assay was performed as described in Materials and Methods. Representative fields of migration of GAC cell with or without Gal-3 over-expression are shown.
Gal-3 correlated with RalA expression in GACs and increases RalA activity
It has been reported that Gal-3 is associated with activated K-Ras in COS cells and in pancreatic cancer cells (Elad-Sfadia et al, 2004; Song et al, 2012, 2013). We, therefore, hypothesised that Gal-3 may activate RalA, a major effector, downstream of Ras to enable proliferation and invasion. RalA can bind RalBP1 and induce migration, invasion, tumour progression, metastasis, and gene expression. We manipulated Gal-3 expression and observed RalA activity. Overexpression of Gal-3 in Snu1 and GT-5 cells (Figure 2C) increased RalA GTP activity after RalBP1 pull-down since only activated RalA is able to bind RalBP1 (Figure 3A, top panel). To determine the relationship between Gal-3 and RalA, we first searched mRNA expression of both Gal-3 and RalA in TCGA GAC database. As shown in Figure 3B, expression of Gal-3 is correlated with RalA mRNA expression in 289 GACs. In another 69 GAC patients shown in Supplementary Figure 2B, expression of Gal-3 is associated with expression of RalA. We note that 10% RalA and 11% of RalBP1 alterations are present in TCGA GAC database (www.cbioportal.org; Supplementary Figure 2C). GAC patients with alterations of RalA have a shorter OS than those without alterations (Supplementary Figure 2D) indicating its high relevance in GAC.
Gal-3 Increases RalA activity and Increases YAP1/TEAD and c-MYC transcriptional activity. (A) Active RalA GTP activity was determined in Snu1 and GT-5 Gal-3 cells and control cells (V). Equal amounts of protein from Snu1 and GT-5 control cells (V) or Gal3 overexpressed cells (Gal-3) were incubated with agarose beads coated with RalBP1-RBD, and active RalA-GTP and YAP1 was detected in the complex of RalA and RalBP1 as described in Materials and Methods. YAP1 strongly bind RalA/RalBP1 only in Gal-3 stably transfected GAC cells (Gal3) which is concomitant to increased RalA activity. (B) Correlation expression of Gal-3 and RalA in GAC patients from TCGA database (http://www.cbioportal.org/). (C) Real-time quantitative PCR for Gal-3 and YAP1 and CTGF RNA expression in Snu1 GAC cells with (Gal-3) or Vector (V) stably transfection were performed as described in Materials and methods. (D) YAP1/TEAD, c-MYC and CTGF luciferase promoter activities were determined by co-transfection of Gal4-Tead and 5XUAS-luciferase; c-MYC and CTGF luciferase promoter plasmids respectively. Luciferase reporter activity was measured after 48 h of transfection. For all experiments, values shown represent the mean and s.d. of at least triplicate assays (**P<0.01). (E) YAP1/TEAD transcriptional activity was determined by co-transfection of Gal4-Tead and 5XUAS-luciferase and YAP1 cDNA in Snu1 GAC cells with or without Gal-3 stably transfection; (F). c-MYC luciferase promoter activity was determined by co-transfection c-MYC luciferase promoter construct and YAP1 cDNA in Snu1 GAC cells with or without Gal-3 stably transfection; Luciferase reporter activity was measured after 48 h of transfection. For all experiments, values shown represent the mean and s.d. of at least triplicate assays (*P<0.05; **P<0.01).
Gal-3 enhances YAP1 binding to RalA/RalBP1 complex and increases YAP1/TEAD and c-MYC transcriptional activity
Activated Ral A (GTP-bound state) activates downstream effectors including RalBP1, Filamin, and others to induce proliferation, invasion, and metastasis. Using pull-down assay with RalBP1 antibody only in Gal-3 stably overexpressed Snu1 and GT-5 cells, we, serendipitously, found that YAP1 binds to the RalBP1 and Ral A complex concomitantly with increased RalA activity (Figure 3A). In addition, YAP1 mRNA and its target CTGF were increased by stably Gal-3 expression in GAC cells (Figure 3C). Gal-3 also increased YAP1/TEAD, c-MYC and CTGF transcriptional activity demonstrated by transient transfection of their promoter luciferase, respectively (Figure 3D). Interestingly, Gal-3 significantly enhances YAP1/TEAD transcriptional activity (Figure 3E) and c-MYC transcriptional activity (Figure 3F). This would indicate that Gal-3 and YAP1 collaborate to increase YAP1’s transcriptional activity and its target genes (CTGF and c-MYC).
BET and YAP1 inhibitors blocked GAC cell proliferation, invasion and migration induced by Gal-3
JQ1, a c-MYC transcription inhibitor through inhibition of BRD4, was recently shown to inhibit YAP1 (Zhang et al, 2017) and a novel YAP1 inhibitor CA3, identified through a chemical library screen, can reduce interaction between YAP1 and RalA/RalBP1. As shown in Supplementary Figure 3 stable Gal-3 expression in both Snu1 and GT-5 cells dramatically increased YAP1 binds to RalBP/RalA using RalBP1 pulldown assay, while JQ1 or CA3 partially reduced this binding in both Snu1 and GT5 cells. We assessed if JQ1 and CA3 reduced aggressive phenotype induced by Gal-3. As shown in Figure 4A, both JQ1 and CA3 preferentially reduced Gal-3 stably overexpressed Snu1 and GT-5 proliferation in a dose-dependent manner. Importantly, CA3 in combination with JQ1 significantly inhibited Snu1 and GT-5 proliferation, particularly in cells overexpressing Gal-3 (Figure 4B). In addition, Gal-3-induced cell invasion in both Snu1 and GT-5 cells (Figure 4C), was blocked by YAP1 inhibitor CA3 or BET inhibitor JQ1 (Figure 4C) as well as migration (Figure 4D).
BET and YAP1 inhibitors blocked GC cell proliferation, invasion and migration induced by Gal-3. (A) Cell growth of Snu1 and GT5 cells in which over-expressed by Gal-3 cDNA or Vector (V) treated with JQ1 (A) or CA3 (B) as indicated dosage was determined by using the CellTiter Aqueous One Solution Cell Proliferation Assay kit as described in Materials and Methods, **P<0.01; ***P<0.001. (B) Cell growth of Snu1 and GT5 cells in which over-expressed by Gal-3 cDNA or Vector (V) treated with JQ1 (1, 5 μM) or CA3 (0.5 μM) alone or the combination of CA3 with JQ1 as indicated dosage was determined using MTS. (C). Invasion assay was performed in Snu1 and GT5 with Gal-3 stably transfection and treated with CA3 and JQ1 as described in Materials and Methods; representative fields (top panel) and quantification (low panel) are shown. (D). Migration assay was performed in Snu1 and GT5 with Gal-3 stably transfection and treated with CA3 and JQ1 as described in Materials and Methods; Representative fields of migration of GAC cell with or without Gal-3 over-expression are shown.
Discussion
Patients with metastatic GAC and their families are always devastated to learn the current treatment options and their limited potential. Only 3 drugs (docetaxel, trastuzumab, and ramucirumab) have been approved in the last 35 years to treat such patients. Molecular understanding of GAC has lagged behind other common tumour types creating a major impediment in our understanding of the underpinnings of GAC and how to make advances. TCGA has provided first genotype-based subgroups but no immediate therapeutics. Novel targets need to be discovered to help the patients. In this report, we demonstrated that Gal-3 overexpression is particularly associated with diffuse type GAC histology in North Americans. Gal-3 expression is significantly higher in North American patients than in Japanese patients and is associated with shorter survival. This dramatic difference in Gal-3 expression pattern may be one of the explanations for varied rates of poor differentiation or diffuse histology as well as OS in patients in Asia vs North America. Our preclinical data show that Gal-3 upregulates c-MYC overexpression and enables aggressive phenotype through increasing RalA activity and enhance RalA/RalBP1 binding to YAP1. YAP1/BET bromodomain inhibitors effectively counteract the aggressiveness mediated by Gal-3. Thus, targeting the Gal3/RalA/YAP/c-MYC axis could be a novel therapeutic strategy for GAC patients, particularly with diffuse type histology.
One of the novel findings is to clarify the significance of the Gal-3 expression in GACs. There are conflicting results of clinical relevance in the literature. Several independent reports observed that Gal-3 expression significantly higher in poorly differentiated GAC and in metastatic lymph nodes (Miyazaki et al, 2002; Dong et al, 2008; Kim et al, 2014). While Okada et al, (2006) reported that lower expression Gal-3 was associated with poor survival. We noted that Gal-3 is independently prognostic for shorter OS and more common in North American patients with diffuse histology. This alone may explain some of the published results. This is the first report to our knowledge showing geographical differences in Gal-3 expression and its therapeutic implications. This may also reflect the difference of etiology and pathogenesis of GAC in different regions. In North America, Gal-3 may be activated by bile reflux (which happens in more obese patients) and play more oncogenic role than that patients in other regions. Larger and in-depth study are warranted in the future.
Gal-3 has been linked to aggressive phenotype and drug resistance in breast, prostate, gastric and pancreatic cancers through variety of mechanisms (Kim et al, 2010; Song et al, 2012; Wang et al, 2013; Zhang et al, 2014). Our preclinical experiments provide novel observations that Gal-3 upregulates c-MYC, increases RalA activity, and promotes RalA binding to YAP1. Gal-3 augments the YAP1/RalA/RalBP complex to mediate its oncogenic function. We found that the BET bromodomain inhibitor, JQ1, can block c-MYC and YAP1 expression in cell lines. These results are consistent with a prior report by Zhang H et al (2017) who reported that JQ1 increased P21 by suppression of YAP1 and thereby reduced chondrosarcoma cell growth. We demonstrated that a novel YAP-1 inhibitor (CA3) suppresses YAP1-mediated transcription, YAP1 expression, and partially blocks YAP1/RalA interaction while reducing aggressive phenotype. In addition, the Gal3/RalA/RalBP/YAP/c-MYC axis is relevant to human GAC from TCGA database. Together 54% of GACs have alterations and alterations are associated with shorter OS. RalA, RalBP, YAP1, and MYC are important oncogenes and play an important role in mediating cell growth and invasive phenotype (Song et al, 2014; Hsieh et al, 2015; Moroishi et al, 2015; Shirakawa and Horiuchi, 2015; Song et al, 2015). Thus, the Gal3/RalA/YAP1/c-MYC axis is an emerging target.
Collectively, our study reveals that Gal-3 is an independent prognosticator in North American GAC patients, particularly with diffused type histology. We demonstrated that Gal-3 upregulates c-MYC through increasing RalA activity and its binding to YAP1 which contribute to aggressive phenotype; novel YAP1 inhibitor and BET inhibitor effectively block Gal-3-mediated phenotype. Therefore, our study provides new insights into the molecular basis for the oncogenic role of Gal-3 in GAC cells and uncovers the Gal3/RalA/YAP1/c-MYC axis as a novel therapeutic target.
Change history
09 January 2018
This paper was modified 12 months after initial publication to switch to Creative Commons licence terms, as noted at publication
References
Ajani JA (2007) Recent developments in cytotoxic therapy for advanced gastric or gastroesophageal carcinoma: The phase III trials. Gastrointest Cancer Res 1 (2 Suppl): S16–S21.
Ajani JA, Takiuchi H (1999) Recent developments in oral chemotherapy options for gastric carcinoma. Drugs 58 (Suppl 3): 85–90.
Dong WG, Yu QF, Xu Y, Fan LF (2008) Li-cadherin is inversely correlated with galectin-3 expression in gastric cancer. Digest Dis Sci 53 (7): 1811–1817.
Elad-Sfadia G, Haklai R, Balan E, Kloog Y (2004) Galectin-3 augments K-Ras activation and triggers a Ras signal that attenuates ERK but not phosphoinositide 3-kinase activity. J Biol Chem 279 (33): 34922–34930.
He TC, Sparks AB, Rago C, Hermeking H, Zawel L, da Costa LT, Morin PJ, Vogelstein B, Kinzler KW (1998) Identification of c-MYC as a target of the APC pathway. Science 281 (5382): 1509–1512.
Hsieh AL, Walton ZE, Altman BJ, Stine ZE, Dang CV (2015) MYC and metabolism on the path to cancer. Semin Cell Dev Biol 43: 11–21.
Jemal A, Bray F, Center MM, Ferlay J, Ward E, Forman D (2011) Global cancer statistics. CA Cancer J Clin 61 (2): 69–90.
Jiang SS, Weng DS, Wang QJ, Pan K, Zhang YJ, Li YQ, Li JJ, Zhao JJ, He J, Lv L, Pan QZ, Xia JC (2014) Galectin-3 is associated with a poor prognosis in primary hepatocellular carcinoma. J Transl Med 12: 273.
Jiang Y, Ajani JA (2010) Multidisciplinary management of gastric cancer. Curr Opin Gastroenterol 26 (6): 640–646.
Kim SJ, Choi IJ, Cheong TC, Lee SJ, Lotan R, Park SH, Chun KH (2010) Galectin-3 increases gastric cancer cell motility by up-regulating fascin-1 expression. Gastroenterology 138 (3): 1035–45 e1-2.
Kim SJ, Lee HW, Gu Kang H, La SH, Choi IJ, Ro JY, Bresalier RS, Song J, Chun KH (2014) Ablation of galectin-3 induces p27(KIP1)-dependent premature senescence without oncogenic stress. Cell Death Differ 21 (11): 1769–1779.
Mazurek N, Byrd JC, Sun Y, Hafley M, Ramirez K, Burks J, Bresalier RS (2012) Cell-surface galectin-3 confers resistance to TRAIL by impeding trafficking of death receptors in metastatic colon adenocarcinoma cells. Cell Death Differ 19 (3): 523–533.
Miyazaki J, Hokari R, Kato S, Tsuzuki Y, Kawaguchi A, Nagao S, Itoh K, Miura S (2002) Increased expression of galectin-3 in primary gastric cancer and the metastatic lymph nodes. Oncol Rep 9 (6): 1307–1312.
Moon BK, Lee YJ, Battle P, Jessup JM, Raz A, Kim HR (2001) Galectin-3 protects human breast carcinoma cells against nitric oxide-induced apoptosis: implication of galectin-3 function during metastasis. Am J Pathol 159 (3): 1055–1060.
Moroishi T, Hansen CG, Guan KL (2015) The emerging roles of YAP and TAZ in cancer. Nat Rev 15 (2): 73–79.
Newlaczyl AU, Yu LG (2011) Galectin-3—a jack-of-all-trades in cancer. Cancer Lett 313 (2): 123–128.
Okada K, Shimura T, Suehiro T, Mochiki E, Kuwano H (2006) Reduced galectin-3 expression is an indicator of unfavorable prognosis in gastric cancer. Anticancer Res 26 (2B): 1369–1376.
Radosavljevic G, Volarevic V, Jovanovic I, Milovanovic M, Pejnovic N, Arsenijevic N, Hsu DK, Lukic ML (2012) The roles of Galectin-3 in autoimmunity and tumor progression. Immunol Res 52 (1-2): 100–110.
Raju U, Ariga H, Koto M, Lu X, Pickett J, Valdecanas D, Mason KA, Milas L (2006) Improvement of esophageal adenocarcinoma cell and xenograft responses to radiation by targeting cyclin-dependent kinases. Radiother Oncol 80 (2): 185–191.
Salajegheh A, Dolan-Evans E, Sullivan E, Irani S, Rahman MA, Vosgha H, Gopalan V, Smith RA, Lam AK (2014) The expression profiles of the galectin gene family in primary and metastatic papillary thyroid carcinoma with particular emphasis on galectin-1 and galectin-3 expression. Exp Mol Pathol 96 (2): 212–218.
Shirakawa R, Horiuchi H (2015) Ral GTPases: crucial mediators of exocytosis and tumourigenesis. J Biochem 157 (5): 285–299.
Siegel RL, Miller KD, Jemal A (2017) Cancer statistics, 2017. CA Cancer J Clin 67 (1): 7–30.
Soldes OS, Kuick RD, Thompson Ii IA, Hughes SJ, Orringer MB, Iannettoni MD, Hanash SM, Beer DG (1999) Differential expression of Hsp27 in normal oesophagus, Barrett/'s metaplasia and oesophageal adenocarcinomas. Br J Cancer 79 (3-4): 595–603.
Song S, Ajani JA, Honjo S, Maru DM, Chen Q, Scott AW, Heallen TR, Xiao L, Hofstetter WL, Weston B, Lee JH, Wadhwa R, Sudo K, Stroehlein JR, Martin JF, Hung MC, Johnson RL (2014) Hippo coactivator YAP1 upregulates SOX9 and endows stem-like properties to esophageal cancer cells. Cancer Res 74 (15): 4170–4182.
Song S, Byrd JC, Mazurek N, Liu K, Koo JS, Bresalier RS (2005) Galectin-3 modulates MUC2 mucin expression in human colon cancer cells at the level of transcription via AP-1 activation. Gastroenterology 129 (5): 1581–1591.
Song S, Honjo S, Jin J, Chang SS, Scott AW, Chen Q, Kalhor N, Correa AM, Hofstetter WL, Albarracin CT, Wu TT, Johnson RL, Hung MC, Ajani JA (2015) The hippo coactivator YAP1 mediates EGFR overexpression and confers chemoresistance in esophageal cancer. Clin Cancer Res 21 (11): 2580–2590.
Song S, Ji B, Ramachandran V, Wang H, Hafley M, Logsdon C, Bresalier RS (2012) Overexpressed galectin-3 in pancreatic cancer induces cell proliferation and invasion by binding Ras and activating Ras signaling. PLoS ONE 7 (8): e42699.
Song S, Maru DM, Ajani JA, Chan CH, Honjo S, Lin HK, Correa A, Hofstetter WL, Davila M, Stroehlein J, Mishra L (2013) Loss of TGF-beta adaptor beta2SP activates notch signaling and SOX9 expression in esophageal adenocarcinoma. Cancer Res 73 (7): 2159–2169.
Song S, Mazurek N, Liu C, Sun Y, Ding QQ, Liu K, Hung MC, Bresalier RS (2009) Galectin-3 mediates nuclear beta-catenin accumulation and Wnt signaling in human colon cancer cells by regulation of glycogen synthase kinase-3beta activity. Cancer Res 69 (4): 1343–1349.
Stanley P (2014) Galectins CLIC cargo inside. Nat Cell Biol 16 (6): 506–507.
Wadhwa R, Elimova E, Shiozaki H, Sudo K, Blum MA, Estrella JS, Chen Q, Song S, Ajani JA (2014) Anti-angiogenic agent ramucirumab: meaningful or marginal? Expert Rev Anticancer Ther 14 (4): 367–379.
Wadhwa R, Taketa T, Sudo K, Blum-Murphy M, Ajani JA (2013) Ramucirumab: a novel antiangiogenic agent. Future Oncol 9 (6): 789–795.
Wang Y, Balan V, Gao X, Reddy PG, Kho D, Tait L, Raz A (2013) The significance of galectin-3 as a new basal cell marker in prostate cancer. Cell Death Dis 4: e753.
Wang Y, Ding Q, Yen CJ, Xia W, Izzo JG, Lang JY, Li CW, Hsu JL, Miller SA, Wang X, Lee DF, Hsu JM, Huo L, Labaff AM, Liu D, Huang TH, Lai CC, Tsai FJ, Chang WC, Chen CH, Wu TT, Buttar NS, Wang KK, Wu Y, Wang H, Ajani J, Hung MC (2012) The crosstalk of mTOR/S6K1 and Hedgehog pathways. Cancer Cell 21 (3): 374–387.
Wang Y, Nangia-Makker P, Tait L, Balan V, Hogan V, Pienta KJ, Raz A (2009) Regulation of prostate cancer progression by galectin-3. Am J Pathol 174 (4): 1515–1523.
Wesley UV, Vemuganti R, Ayvaci ER, Dempsey RJ (2013) Galectin-3 enhances angiogenic and migratory potential of microglial cells via modulation of integrin linked kinase signaling. Brain Res 1496: 1–9.
Xue H, Liu L, Zhao Z, Zhang Z, Guan Y, Cheng H, Zhou Y, Tai G (2017) The N-terminal tail coordinates with carbohydrate recognition domain to mediate galectin-3 induced apoptosis in T cells. Oncotarget 8: 49824–49838.
Zhang H, Luo M, Liang X, Wang D, Gu X, Duan C, Gu H, Chen G, Zhao X, Zhao Z, Liu C (2014) Galectin-3 as a marker and potential therapeutic target in breast cancer. PLoS One 9 (9): e103482.
Zhang HT, Gui T, Sang Y, Yang J, Li YH, Liang GH, Li T, He QY, Zha ZG (2017) The BET Bromodomain Inhibitor JQ1 Suppresses Chondrosarcoma Cell Growth via Regulation of YAP/p21/c-Myc Signaling. J Cell Biochem 118 (8): 2182–2192.
Zhao B, Kim J, Ye X, Lai ZC, Guan KL (2009) Both TEAD-binding and WW domains are required for the growth stimulation and oncogenic transformation activity of yes-associated protein. Cancer Res 69 (3): 1089–1098.
Acknowledgements
We thank Ms. Xiaolian Chun, an experienced biostatistician in the Department of Bioinformatics of MDACC to analyse our IHC staining (Gal-3) independently. Generous grants from the Caporella, Dallas, Sultan, Park, Smith, Frazier, Oaks, Vanstekelenberg, Planjery, and Cantu Families. From the Schecter Private Foundation, Rivercreek Foundation, Kevin Fund, Myer Fund, Dio Fund, Milrod Fund, and Multidisciplinary Grants from the University of Texas M. D. Anderson Cancer Center, Houston, USA. Supported in part by the National Cancer Institute and Department of Defense awards CA138671, CA172741, CA15033 and CA129926 CA160445 (JAA) and CA160433 (Song S).
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Additional information
This work is published under the standard license to publish agreement. After 12 months the work will become freely available and the license terms will switch to a Creative Commons Attribution-NonCommercial-Share Alike 4.0 Unported License.
Supplementary Information accompanies this paper on British Journal of Cancer website
Rights and permissions
From twelve months after its original publication, this work is licensed under the Creative Commons Attribution-NonCommercial-Share Alike 4.0 Unported License. To view a copy of this license, visit http://creativecommons.org/licenses/by-nc-sa/4.0/
About this article
Cite this article
Ajani, J., Estrella, J., Chen, Q. et al. Galectin-3 expression is prognostic in diffuse type gastric adenocarcinoma, confers aggressive phenotype, and can be targeted by YAP1/BET inhibitors. Br J Cancer 118, 52–61 (2018). https://doi.org/10.1038/bjc.2017.388
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/bjc.2017.388
Keywords
This article is cited by
-
YAP-galectin-3 signaling mediates endothelial dysfunction in angiotensin II-induced hypertension in mice
Cellular and Molecular Life Sciences (2023)
-
Signaling pathways and therapeutic interventions in gastric cancer
Signal Transduction and Targeted Therapy (2022)
-
The immune microenvironment in gastric adenocarcinoma
Nature Reviews Gastroenterology & Hepatology (2022)
-
Activation of AMPK inhibits Galectin-3-induced pulmonary artery smooth muscle cells proliferation by upregulating hippo signaling effector YAP
Molecular and Cellular Biochemistry (2021)