Abstract
Aim:
To investigate the potential effects of chronic ethanol intake on protein-tyrosine phosphatase-1B (PTP1B) and the insulin receptor signaling pathway in rat skeletal muscle.
Methods:
Rats received ethanol treatment at a daily dose of 0 (control), 0.5 (group L), 2.5 (group M) or 5 g·kg−1 (group H) via gastric gavage for 22 weeks. In vivo insulin sensitivity was measured using a hyperinsulinemic-euglycemic clamp. Expression of PTP1B in skeletal muscles was examined at both the mRNA (real-time PCR) and protein (Western blot) levels. PTP1B activity was assayed with a p-nitrophenol phosphate (PNPP) hydrolysis method. Changes of insulin signaling in skeletal muscle were analyzed with Western blotting.
Results:
The activity and expression of PTP1B were dose-dependently elevated 1.6 and 2.0 fold in the skeletal muscle by ethanol, resepctively, at the doses of 2.5 and 5 g·kg−1·d−1. Total IRβ and IRS-1, as well as their phosphorylated forms, were decreased by ethanol at the two higher doses. Moreover, chronic ethanol consumption resulted in a significant inhibition of the association between IRS-1 and the p85 subunit of phosphatidylinositol 3-kinase, inhibition of Akt phosphorylation and reduced levels of mitogen-activated protein kinase phosphorylation.
Conclusion:
Chronic ethanol intake at 2.5 and 5 g·kg−1·d−1 sufficient doses can down-regulate the expression of IRβ, P-IRβ, and IRS-1, as well as the phosphorylated forms of IRS-1 and Akt, in rat skeletal muscle, possibly through increased PTP1B activity.
Similar content being viewed by others
Introduction
Alcohol consumption is associated with insulin resistance1, 2. Long-term exposure to excessive alcohol may lead to glucose intolerance3, 4. Insulin produces a diverse array of metabolic actions by binding to a heterotetrameric receptor protein consisting of two α and two β subunits5. Phosphorylated insulin receptor (IR) binds to and activates IR substrate (IRS), which in turn leads to translocation of glucose transporters to the cell surface via a complicated cascade of signaling events that include phosphatidylinositol 3-kinase (PI3K) and Akt/protein kinase B (PKB)6, 7.
The insulin signaling pathway is tightly regulated by the balance of phosphorylation and dephosphorylation at several key molecules. Not surprisingly, protein-tyrosine phosphatases (PTPases) play a critical role in the regulation of carbohydrate metabolism8, 9. PTP1B is a member of the PTPase family10, 11. PTP1B can inhibit the insulin-signaling pathway by dephosphorylating IR and IRS. PTP1B has been reported to be elevated in diabetes and in insulin-resistant states12. Transgenic overexpression of PTP1B decreased glucose uptake in muscle13. Ablation of PTP1B, in contrast, improves glucose uptake14. Mice lacking PTP1B are hyper-responsive to insulin and resistant to diet-induced obesity15.
Previous studies have demonstrated that ethanol may impair mitochondrial function and inhibit insulin-stimulated survival of cultured neuronal cells16. Monte et al17, for example, found that increased PTP1B contributes to impaired insulin sensitivity in brain tissue. Skeletal muscle is the largest organ that utilizes glucose in an insulin-dependent manner and therefore represents a key site for the pathogenesis of diabetes, regardless of the specific events that trigger glucose intolerance. The present study was designed to examine whether insulin intolerance induced by chronic ethanol exposure is correlated with PTP1B expression/activity in skeletal muscle in a rat model of chronic ethanol exposure. Molecular targets of PTP1B (eg, IR and IRS-1) and downstream molecules of the insulin signaling pathway (eg, PI3K and Akt) were also examined.
Materials and methods
Reagents and antibodies
Horseradish peroxidase (HRP)-labeled anti-rabbit and anti-mouse immunoglobulin G (IgG) antibodies were purchased from Upstate Biotechnology (Lake Placid, NY, USA). Total and phosphorylated (P) IRβ, IRS-1, Akt, and mitogen-activated protein kinase (MAPK) as well as PTP1B antibodies were purchased from Santa Cruz Biotechnology (Santa Cruz, CA, USA). All other reagents were purchased from Sigma (St Louis, MO).
Animal care and feeding
Sixty adult male Wistar rats (180–200 g, provided by the Animal Research Center of Shandong University) were housed individually at 25 °C and 50% humidity under a 12:12 h light/dark cycle. The rats had unlimited access to standard rat chow and tap water.
Chronic ethanol feeding model
Rats were acclimated for one week prior to receiving daily ethanol (C, control, 0 g·kg−1·d−1; H, 5 g·kg−1·d−1; M, 2.5 g·kg−1·d−1; L, 0.5 g·kg−1·d−1 via gastric gavage; n=15 in each group) for 22 weeks. Distilled water or edible (50% v/v; Ji-nan Baotu Spring Distillery, Shandong, China) was delivered once daily at 8:00–9:00 AM. Food and water consumption as well as body weight were monitored weekly. At the end of the 22-week treatment period, food was withdrawn 12 h, and blood samples were taken for biochemical analysis. Insulin sensitivity was determined using a hyperinsulinemic-euglycemic clamp. At the completion of the clamp, rats were euthanized with sodium pentobarbital (100 mg/kg ip; Abbott Laboratories). The gastrocnemius muscle was quickly removed, washed in cold phosphate-buffered saline (PBS, pH 7.4) and stored in liquid nitrogen for subsequent in vitro analysis. Experimental procedures were approved by the Shandong University Institutional Animal Care and Use Committee.
Hyperinsulinemic-euglycemic clamp
Insulin sensitivity was determined using a hyperinsulinemic-euglycemic clamp18. Rats were anesthetized with pentobarbital sodium administered intraperitoneally after fasting for 8 h. Insulin (Nordisk, Denmark) was infused at 8 milliunits·kg−1·min−1 for 2 h via the left jugular and left femoral vein. Blood glucose level was measured every 5 min via a catheter in the left jugular vein. Twenty percent glucose solution was infused via the right femoral vein. The blood glucose concentration was maintained at 5.2±0.2 mmol/L by adjusting the rate of glucose infusion. The glucose infusion rate in the second hour of the experiment was considered to reflect insulin sensitivity.
Biochemical analysis
Plasma ethanol concentrations were measured using a commercial kit (Sigma, St Louis, MO). Blood glucose levels were determined by a glucose oxidase method after 12-h fasting. Serum insulin levels were measured by radioimmunoassay (Northern Bioengineering Institute, China). Serum concentrations of aspartate aminotransferase, alanine aminotransferase, triglyceride and cholesterol were measured using an automated biochemistry analyzer (Hitachi, Japan).
Real-time PCR
Total RNA was extracted from the gastrocnemius muscle using the standard Trizol RNA isolation method. Reverse transcription of 2 μg of RNA was carried out using a TaKaRa RT-PCR kit. The qualities of RNA and cDNA were checked using a DU640 nucleic acid analyzer (Beckman, USA). The specific primer sequences used in the real-time PCR are: PTP1B forward: CGAGGGTGCAAAGTTCATCAT, reverse: GGTCTTCATGGGAAAGCTCCTT; GAPDH forward: TGGTGGACCTCATGGCCTAC, reverse: CAGCAACTGAGGGCCTCTCT. One hundred nanograms (ng) of cDNA was used as the template in a 25 μL reaction volume and was amplified by real-time PCR assay using a QuantiTect SYBR Green kit (Qiagen, USA) and the ABI 7500 Prism real-time PCR instrument and software (Prism 7500; ABI, USA). GAPDH was used as the internal control. The relative quantification of gene expression was analyzed by the 2−ΔΔCt method19, 20, and the results were expressed as the level of change with respect to control values.
Western blotting
Aliquots (80 μg protein) of gastrocnemius muscle homogenates were subjected to 7.5% SDS-PAGE and then transferred electrophoretically onto nitrocellulose membranes (Millipore, Billerica, MA). The membrane was blocked with 5% nonfat milk in 10 mmol/L Tris containing 150 mmol/L NaCl and 0.02% Tween-20 for 1 h at room temperature. The nitrocellulose blots were incubated overnight at 4 °C with a primary antibody (Santa Cruz Biotechnology; Santa Cruz, CA). Incubation with secondary antibody, horseradish peroxidase (HRP)-labeled anti-rabbit or anti-mouse IgG antibody (Upstate Biotechnology; Lake Placid, NY) lasted for 1 h. Target proteins were quantified using an enhanced chemiluminescence detection system (Amersham), followed by autoradiography with preflashed Kodak XAR films (Manaus, Amazonas-Brazil) using an Alphaimager 2200 system. All experiments included β-actin as an internal control.
PTP1B activity assay
PTP1B activity was assayed by a p-nitrophenol phosphate (PNPP) hydrolysis method. Briefly, gastrocnemius muscles were removed and homogenized in a buffer containing (mmol/L): Tris 20 (pH 7.6), EDTA 5, PMSF 2, EGTA 1 and NaCl 130, with aprotinin 0.1 mg/mL−1 and 1% Triton X-100. The lysates were centrifuged at 15 000×g for 25 min at 4 °C. PTP1B was immunoprecipitated with anti-PTP1B antibody (Upstate Biotechnology). The immunoprecipitates were incubated in a phosphatase reaction buffer (20 mmol/L HEPES, pH 7.4, 150 mmol/L NaCl, 5 mmol/L dithiothreitol, 1 mmol/L PNPP) for 20 min at 37 °C. The reactions were stopped with 0.2 mol/L NaOH. The absorbance was measured at 410 nm. The assay was run in triplicate.
Statistical analysis
All experiments were repeated at least five times. Values are reported as means ± standard deviations (SD). Data were analyzed by using SPSS 13.0 software (SPSS, Chicago, IL, USA). Statistical significance was assessed by ANOVA and unpaired Student's t-tests. Differences were considered statistically significant when P<0.05.
Results
Characterization of chronically ethanol-fed rats
Chronic ethanol treatment decreased body weight and increased serum alanine and aspartate aminotransferase levels, as well as cholesterol and triglyceride levels (Table 1).
Hyperinsulinemic-euglycemic clamps
To investigate whether rats receiving ethanol were insulin resistant, we measured the peripheral insulin sensitivity with a hyperinsulinemic-euglycemic clamp. In this technique, plasma glucose is maintained at a steady state within the euglycemic range while plasma insulin levels are elevated to a desired plateau. The higher the glucose infusion rate needed to maintain euglycemia, the greater the insulin sensitivity. The hyperinsulinemic-euglycemic clamp experiments revealed impaired insulin sensitivity (Table 1). The rates of glucose infusion needed to maintain glucose levels at 5.2±0.2 mmol/L were 11.51±1.32 mg·kg−1·min−1 in the vehicle control rats and were decreased by 36% (P<0.05 vs control) and 51% (P<0.01 vs control) in rats receiving 2.5 and 5 g·kg−1·d−1 ethanol, respectively. However, the lower ethanol dose of 0.5 g·kg−1·d−1 did not affect glucose infusion rate significantly.
Ethanol up-regulates the expression of PTP1B
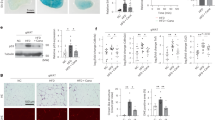
To investigate whether PTP1B expression changes during the decreased glucose uptake in rat skeletal muscle after chronic ethanol feeding, we measured PTP1B mRNA and protein levels using real-time PCR (Figure 1A) and Western blotting (Figure 1B). Our findings indicate that ethanol increased PTP1B protein and mRNA levels in skeletal muscles in a dose-dependent manner (P<0.05 and <0.01 for rats receiving 2.5 and 5 g·kg−1·d−1 ethanol, respectively, vs vehicle controls). The lower ethanol dose of 0.5 g·kg−1·d−1 did not affect the expression of PTP1B significantly.
Effects of ethanol on PTP1B in rat skeletal muscle. (A) PTP1B mRNA levels were determined using RT-PCR. (B) PTP1B protein levels were determined by Western blot analysis. (C) PTP1B activities were assayed using PTP1B assay kit. Compared to the control group, the PTP1B levels in groups M and H increased significantly (bP<0.05, cP<0.01 vs group C). n=15. Values are given as means±SD. C, control; H, 5 g·kg−1·d−1 ethanol treatment; M, 2.5 g·kg−1·d−1 ethanol treatment; and L, 0.5 g·kg−1·d−1 ethanol treatment.
PTP1B activity in muscles of control and ethanol-fed rats
In this study, PTP1B activities were assessed using a PTP1B assay kit (Figure 1C). Our results indicated that PTP1B activities were significantly elevated in both rats receiving 5 (2-fold, P<0.01) and 2.5 g·kg−1·d−1 (1.6-fold, P<0.05) ethanol, compared to PTP1B activities in the vehicle control rats. However, the lower dose of 0.5 g·kg−1·d−1 did not change PTP1B activity in the skeletal muscles significantly.
Ethanol down-regulates IRβ and IRS-1 protein expression
We examined a potential dose-response effect of ethanol on IRβ and IRS-1 content by Western blotting. Our results showed that IRβ and IRS-1 protein levels were significantly decreased (Figure 2, 3) in rats receiving ethanol at 2.5 and 5 g·kg−1·d−1 but not 0.5 g·kg−1·d−1. The phosphorylated forms of IRβ and IRS-1 (P-IRβ and P-IRS-1) were also decreased by chronic ethanol in a similar manner. Furthermore, ethanol exposure caused a dose-dependent decline in the P-IRβ/IRβ protein ratios (P<0.01, 0.05 and 0.05 for the dose of 5, 2.5, and 0.5 g·kg−1·d−1, respectively) and a decline in the P-IRS-1/IRS-1 protein ratios with 5 (P<0.01) and 2.5 g·kg−1·d−1 doses (P<0.05, Figure 2, 3).
Expression of IRβ and P-IRβ protein levels in rat skeletal muscle. Levels of IRβ and P-IRβ protein were determined by Western blot analysis. Compared to the control group, the IRβ and P-IRβ proteins decreased significantly in groups M and H, while P-IRβ proteins decreased significantly in all ethanol-treated groups (bP<0.05, cP<0.01 vs group C). n=15. Values are given as means±SD.
Effects of ethanol on PI3K
The association between IRS-1 and the p85 subunit of PI3K in skeletal muscle was also decreased (P<0.05 and P<0.01 for 2.5 and 5 g·kg−1·d−1, respectively) by chronic ethanol treatment (Figure 4), which was consistent with changes in phosphorylation of IRS-1. In contrast, p85 protein levels were similar in control and ethanol-treated groups (Figure 4).
Expression of PI3K protein levels in rat skeletal muscle. Levels of p85 subunit of PI3K and p85-associated IRS-1 protein were determined by Western blot analysis. Compared to the control group, the p85-associated IRS-1 proteins in groups M and H decreased significantly (bP<0.05, cP<0.01 vs group C). In contrast, p85 protein levels were similar in control and ethanol-treated groups. n=15. Values are given as means±SD.
Effects of ethanol on Akt
We found that phosphorylation of Akt was inhibited by chronic ethanol exposure, as shown in Figure 5. This regulation was significant in rats receiving ethanol at 5 (P<0.01) and 2.5 g·kg−1·d−1 (P<0.05), but not significantly different at 0.5 g·kg−1·d−1 (P>0.05). However, there were no differences in total Akt protein levels among the groups (Figure 5).
Expression of Akt and P-Akt protein levels in rat skeletal muscle determined by Western blot analysis. Compared to the control group, the P-Akt proteins in groups M and H decreased significantly (bP<0.05, cP<0.01 vs group C). There were no differences in Akt protein levels among the groups. n=15. Values are given as means±SD.
Effects of ethanol on MAPK
We next examined the protein expression of MAPK by western blotting. Chronic ethanol consumption resulted in significantly reduced levels of P-MAPK (P<0.05 and P<0.01 for 2.5 and 5 g·kg−1·d−1, respectively). The lower dose of 0.5 g·kg−1·d−1 did not affect the expression of P-MAPK (Figure 6). In contrast, the levels of MAPK proteins were similar in control and ethanol-treated groups (Figure 6).
Expression of MAPK and P-MAPK protein levels in rat skeletal muscle were determined by Western blot analysis. Compared to the control group, the P-MAPK proteins in groups M and H decreased significantly (bP<0.05, cP<0.01 vs group C). There were no differences in MAPK protein levels among the groups. n=15. Values are given as means±SD.
Discussion
The Lieber-Decarli ethanol diet (where the ratio of calories supplied by ethanol is assigned as a fixed value) is more widely used in animal studies. However, this method also has a shortcoming that cannot be ignored—if the quantity of food consumed daily by the animals varies, there is variability in the daily quantity of ethanol consumed. In the present study, we were most concerned with standardizing the quantity of ethanol fed daily and, therefore, administered a 50% ethanol solution to each animal according to its body weight. Previous studies from our laboratory21 and from another group22 revealed compromised insulin-stimulated glucose uptake upon chronic ethanol exposure. Using hyperinsulinemic-euglycemic clamps, the current study confirmed decreased insulin sensitivity in rats receiving chronic ethanol at relatively high doses of 2.5 to 5 g·kg−1·d−1, but not at the lower dose of 0.5 g·kg−1·d−1. Consistent with the reported increase in PTB1B in diabetic patients, we found a significantly up-regulated expression of PTP1B in rats chronically exposed to high doses of ethanol, suggesting that increased PTP1B expression/activity plays an important role in the development of glucose intolerance induced by ethanol. The most compelling evidence supporting a role for PTP1B in the literature is increased insulin sensitivity and resistance to diet-induced obesity in PTP1B knockout mice15. Insulin receptors in these mice are highly phosphorylated and present in large amounts in the cell membrane of skeletal muscle and liver15.
Onishi et al provided preliminary evidence suggesting that ethanol exposure impairs PI3K22, a key step in the signaling cascade that mediates insulin action. Results from the current study demonstrated reduced P-IRS-1, p85-IRS-1 association, P-Akt and P-MAPK in skeletal muscle from rats chronically exposed to high doses of ethanol, indicating that the detrimental effects of ethanol on insulin sensitivity are mediated by the PI3K and MAPK pathways. Impaired signaling through IRS-2 is another candidate for mediating the IRS-dependent effects as phosphorylation of IRS-2 could activate PI3 kinase in the skeletal muscle23. Potential involvement of IRS-2 in ethanol-induced glucose intolerance is currently under investigation in our laboratory.
Our results are consistent with studies showing that excessive consumption of alcohol can lead to glucose intolerance4, 5. These results are contrary to previous in vitro studies showing that regular, moderate alcohol drinkers were more insulin-sensitive than abstainers24. In this study, we found that low ethanol intake cannot increase insulin sensitivity. We believe that the reason may be attributed to the different alcohol intervention time, different testing methods and tissue specificity. Thus, some compensatory mechanism(s) may exist to regulate the efficiency of signaling effectors. Further investigation is necessary to explore these possibilities.
For decades it has been argued whether alcohol exerts its toxic effects directly on skeletal muscle or indirectly through its metabolites, and the regulatory role of PTP1B activity in this process is not clear. In the past decade, several studies have documented the fact that cardiomyocyte loss is a critical factor in the development and progression of ventricular dysfunction and failure, and apoptosis of myocardial cells has been associated with the progression of cardiomyopathy25. Moreover, animal experiments indicated that chronic prenatal ethanol exposure increases neuronal cell apoptosis in the hippocampus of a term fetus, which appears to occur via an intrinsic, mitochondrial-directed mechanism initiated by leakage of pro-apoptotic cytochrome c from mitochondria into the cytoplasm26. Interestingly, both acetaldehyde and ethanol have been shown to accelerate apoptotic cell death in various cells27. Therefore, apoptosis may be one of the possible mechanisms through which ethanol down-regulates gene expression. A previous study of PTP1B expression in different tissues suggested that small molecules in the body, such as NO or H2O2, may regulate PTP1B activity28. Sreejayan et al demonstrated that NO could enhance PTP1B activity in smooth muscles and down-regulate insulin signal transduction29. Mahadev et al showed that H2O2 could inhibit PTP1B activity in hepatoma and fat cells30, suggesting that redox status could affect PTP1B activity. Whether the observed action of ethanol on insulin sensitivity is attributed to its reducing properties remains to be investigated.
The results of our experiments linked chronic consumption of ethanol to aberrantly increased expression and enzymatic activity of PTP1B, which has a pivotal role in regulating PI3K-activated insulin signaling, and this effect is accompanied by down-regulation of P-IRβ, P-IRS-1, P-Akt, and P-MAPK protein expression. Our results suggest a role for PTP1B specifically in mediating the IRS-1/PI3K/Akt pathway of insulin resistance induced by excessive ethanol consumption in rat skeletal muscle. However, the mechanism by which ethanol feeding increases PTP1B expression is unclear and needs further investigation. It remains to be seen if modulating PTP1B levels via gene transfer can ameliorate the symptoms of insulin resistance in animal models of this disease. If so, this may prove to be a potential therapeutic avenue for human patients.
Author contribution
Jia-jun ZHAO and Ling GAO designed the research; Li GAO, Xu ZHANG and Ming-feng CAO performed the research; Fu-rong WANG, Nan-nan SUN and Jie ZHANG analyzed the data; Li GAO, Xiu-juan ZHANG and Ling GAO wrote the paper.
References
Fowman DT . The effect of ethanol and its metabolites on carbohydrate, protein and lipid metabolism. Ann Clin Lab Sci 1988; 18: 181–9.
Rimm EB, Chan J, Stampfer MJ, Colditz GA, Willett WC . Prospective study of cigarette smoking, alcohol use, and the risk of diabetes in men. BMJ 1995; 310: 555–9.
Dornhorst A, Ouyang A . Effect of alcohol on glucose tolerance. Lancet 1971; 2(7731): 957–9.
Shelmet JJ, Reichard GA, Skutches CL, Hoeldtke RD, Owen OE, Boden G, et al. Ethanol causes acute inhibition of carbohydrate, fat, and protein oxidation and insulin resistance. J Clin Invest 1988; 81: 1137–45.
Cttensmeyer FP, Beniac DR, Luo RZ . Mechanism of transmembrane signaling: insulin binding and the insulin receptor. Biochemistry 2000; 39: 12103–12.
Shpakov AO, Pertseva MN . Structural and functional characterization of insulin receptor substrate proteins and the molecular mechanisms of their interaction with insulin superfamily tyrosine kinase receptors and effector proteins. Membr Cell Biol 2000; 13: 455–84.
Lam K, Carpenter CL, Ruderman NB, Friel JC, Kelly KL . The phosphatidylinositol 3-kinase serine kinase phosphorylates IRS-1: stimulation by insulin and inhibition by wortmannin. J Biol Chem 1994; 269: 20648–52.
Koren S, Fantus IG . Inhibition of the protein tyrosine phosphatase PTP1B: potential therapy for obesity, insulin resistance and type-2 diabetes mellitus. Best Pract Res Clin Endocrinol Metab 2007; 21: 621–40.
Zhang Y, Li Y, Guo YW, Jiang HL, Shen X . A sesquiterpene quinone, dysidine, from the sponge Dysidea villosa, activates the insulin pathway through inhibition of PTPases. Acta Pharmacol Sin 2009; 30: 333–45.
Tonks NK, Diltz CD, Fischer EH . Characterization of the major protein-tyrosine phosphatase of human placenta. J Biol Chem 1988; 263: 6722–37.
Charbonneau H, Tonks NK, Kumar S, Diltz CD, Harrylock M, Cool DE, et al. Human placenta protein-tyrosine phosphatase: amino-acid sequence and relationship to a family of receptor-like proteins. Proc Natl Acad Sci USA 1989; 86: 5252–6.
Lalli CA, Pauli JR, Prada PO, Cintra DE, Ropelle ER, Velloso LA, et al. Statin modulates insulin signaling and insulin resistance in liver and muscle of rats fed a high-fat diet. Metabolism 2008; 57; 57–65.
Zabolotny JM, Haj FG, Kim YB, Kim HJ, Shulman GI, Kim JK, et al. Transgentic overexpression of protein-tyrosine phosphatase 1B in muscle causes insulin resistance, but overexpression with leukocyte antigen related phosphatase does not additively impair insulin action. J Biol Chem 2004; 279: 24844–51.
Delibegovic M, Bence KK, Mody N, Hong EG, Ko HJ, Kim JK, et al. Improved glucose homeostasis in mice with muscle-specific deletion of protein-tyrosine phosphatase 1B. Mol Cell Biol 2007; 27: 7727–34.
Xu J, Li L, Qian Z, Hong J . Reduction of PTPlB by RNAi upregulates the activity of insulin controlled fatty acid synthase promoter. Biochem Biophys Res Commun 2005; 329: 538–43.
de la Monte SM, Wands JR . Chronic gestational exposure to ethanol impairs insulin-stimulated survival and mitochondrial function in cerebellar neurons. Cell Mol Life Sci 2002; 59: 882–93.
de la Monte SM, Xu J, Wands J . Ethanol inhibits insulin expression and actions in the developing brain. Cell Mol Life Sci 2005, 62: 1131–45.
Margaret EG, Melissa JM, Gary WC, Kim B, Nicole B, Dennis L, et al. Free fatty acid-induced insulin resistance is associated with activation of protein kinase C u and alterations in the insulin signaling cascade. Diabetes 1999; 48: 1270–4.
Schmittgen TD, Zakrajsek BA, Mills AG, Gorn V, Singer MJ, Reed MW . Quantitative reverse transcription-polymerase chain reaction to study mRNA decay: comparison of endpoint and real-time methods. Anal Biochem 2000; 285: 194–204.
Livak KJ, Schmittgen TD . Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCt method. Methods 2001; 25: 402–8.
Wan Q, Liu Y, Guan Q, Gao L, Lee KO, Zhao JJ . Ethanol feeding impairs insulin-stimulated glucose uptake in isolated rat skeletal muscle: role of Gs alpha and cAMP. Alcohol Clin Exp Res 2005; 29: 1450–6.
Onishi M, Honda T, Oqihara T, Sakoda H, Anai M, Fujishiro M . Ethanol feeding induces insulin resistance with enhanced PI3-kinase activation. Biochem Biophys Res Commun 2003; 303: 788–94.
Welham MJ, Bone H, Levings M, Learmonth L, Wang LM, Leslie KB, et al. Insulin receptor substrate-2 is the major 170-kDa protein phosphorylated on tyrosine in response to cytokines in murine lymphohemopoietic cells. J Biol Chem 1997; 272: 1377–81.
Furuya DT, Binsack R, Machado UF . Low ethanol consumption increases insulin sensitivity in Wistar rats. Braz J Med Biol Res 2003; 36: 125–30.
Narula J, Haider N, Virmani R, DiSalvo TG, Kolodgie FD, Hajjar RJ . Apoptosis in myocytes in end-stage heart failure. N Engl J Med 1996; 335: 1182–9.
Green CR, Kobus SM, Ji Y, Bennett BM, Reynolds JN, Brien JF . Chronic prenatal ethanol exposure increases apoptosis in the hippocampus of the term fetal guinea pig. Neurotoxicol Teratol 2006; 28: 296–7.
Baroni GS, Marucci L, Benedetti A, Mancini R, Jezequel AM, Orlandi F . Chronic ethanol feeding increases apoptosis and cell proliferation in rat liver. J Hepatol 1994; 20: 508–13.
Wu X, Zhu L, Zilbering A, Mahadev K, Motoshima H, Yao J . Hyperglycemia potentiates H2O2 production in adipocytes and enhances insulin signal transduction: potential role for oxidative inhibition of thiol-sensitive protein tyrosine phosphatases. Antioxid Redox Signal 2005; 7: 526–37.
Sreejayan N, Lin Y, Hassid A . NO attenuate insulin signaling and motility in aortic smooth muscle cells via tyrosine phosphatase1B—mediated mechanism. Arterioscler Thromb Vasc Biol 2002; 22: 1086–92.
Mahadev K, Zilbering A, Zhu L, Goldstein BJ . Insulin-stimulated hydrogen peroxide reversibly inhibits protein-tyrosine phosphatase Ib in vivo and enhances the early insulin action cascade. J Biol Chem 2001; 276: 21938–42.
Acknowledgements
This work was supported by a grant from the National Natural Science Foundation of China (Grant No 30940038) and Shandong Province (Grant No Q2006C15). The Science Center of Shandong Provincial Hospital provided technical support for this study.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Gao, L., Zhang, X., Wang, Fr. et al. Chronic ethanol consumption up-regulates protein-tyrosine phosphatase-1B (PTP1B) expression in rat skeletal muscle. Acta Pharmacol Sin 31, 1576–1582 (2010). https://doi.org/10.1038/aps.2010.161
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/aps.2010.161
Keywords
This article is cited by
-
Skeletal Muscle Atrophy in Men with Chronic Alcoholic Myopathy (impairment to signal pathways)
Neuroscience and Behavioral Physiology (2019)
-
Behavioral, Metabolic, and Immune Consequences of Chronic Alcohol or Cannabinoids on HIV/AIDs: Studies in the Non-Human Primate SIV Model
Journal of Neuroimmune Pharmacology (2015)