Abstract
The golden-striped salamander, Chioglossa lusitanica, is a streamside species distributed in the northwest of the Iberian Peninsula. We gathered cytochrome b mitochondrial DNA sequence data from samples across the species range and used nested clade distance analysis to assess evolutionary processes explaining the geographical distribution of mtDNA diversity in Chioglossa lusitanica. A significant association was observed between genetic structure and geography. The geographical patterns were explained by past fragmentation and restricted gene flow. Inferences were concordant with previous hypotheses (from allozyme data) involving an old fragmentation event and secondary contact near the Mondego River in central Portugal. A range expansion to the north from a southern refuge was supported by descriptive interpretations of mtDNA data and by complementary analyses suggesting population expansions, but not by formal nested clade analysis. The power of nested clade analysis to detect the genetic signature of range expansion deserves further testing.
Similar content being viewed by others
Introduction
Intraspecific patterns of genetic diversity are determined by both historical and contemporary processes. Conventional approaches to describing population structure and estimating gene flow interpret spatial patterns of allelic distribution as a function of migration and genetic drift (Wright, 1931, see Neigel, 1997 for a review). Single summary statistics (such as F statistics) fail to discriminate between recurrent gene flow and historical processes such as fragmentation and range expansion. However, given that each process will leave a distinct signature, it is possible, at least in theory, to disentangle the contributions of history and recurrent gene flow to the observed pattern of haplotype composition. Templeton et al (1995) proposed a new class of phylogeographic statistics that takes advantage of information contained in molecular genetic data by testing the geographical associations at different hierarchical levels of an estimated gene tree. Predictions from coalescent theory are then used to interpret the results of this ‘nested clade analysis’ (see Templeton, 1998 for a review) and to make spatio-temporal inferences about the evolutionary processes (fragmentation, range expansion or restricted gene flow) likely to have contributed to the observed patterns.
Geographic patterns of genetic diversity in salamanders may reflect historical rather than contemporary processes because of the low mobility and strict ecological requirements of these organisms (Slatkin, 1981; Larson et al, 1984). Indeed, salamanders of the genus Ambystoma were first used to exemplify nested clade analysis because they provide an adequate model, showing the joint effects of fragmentation, range expansion and isolation by distance (Templeton et al, 1995). The golden-striped salamander, Chioglossa lusitanica Bocage 1864, is a peculiar streamside species with a distribution restricted to north-western Iberia. These salamanders exhibit a suite of morpho-physiological traits that determine a strong dependence on brook vicinities and other moist habitats which is likely to result in directional dispersal along watercourses (Arntzen, 1981, 1994). We have recently analysed allozyme and mitochondrial DNA (mtDNA) variation in C. lusitanica and discovered two distinct evolutionary lineages, with the spatial pattern of genetic variation reflecting historical fragmentation and range expansion (Alexandrino et al, 2000).
We here extend our work through sequencing of the mitochondrial cytochrome b gene to 700 bp in 124 individuals from across the species range. We apply nested clade analysis to assess the relative contribution of historical vs contemporary evolutionary processes to the geographical patterns of mtDNA variation. In addition, we test the hypothesis that populations within the area of range expansion show the genetic characteristics that are typical of demographic expansion. In this way we test, refine and expand upon hypotheses put forward under a more descriptive framework (Alexandrino et al, 2000).
Materials and methods
Data collection
Salamander tail tips were collected from 17 localities across north-western Iberia covering the species range (Figure 1). DNA extraction, PCR amplification of cytochrome b and sequencing protocols are as in Alexandrino et al (2000). An 840 bp fragment of the cytochrome b mtDNA gene was amplified by PCR using the primers MVZ15 (Moritz et al, 1992; Tan and Wake, 1995) and cytb702 (5′-GGCAAATAGGAAGTATCATTCTG-3′, Moritz et al, 1992, modified). Both primers were used for direct sequencing of the PCR products. Sequences of 700 bp, corresponding to 16 309 (5′ end) and 17 008 (3′ end) of the Xenopus laevis mtDNA (Roe et al, 1985) were obtained for 124 individuals (two to 11 individuals per population). The sequences were aligned using BioEdit, version 1.8 (Hall, 1999) with no indels. Nucleotide diversity (π), its standard error and sequence divergence (d) between haplotypes were calculated using DnaSP version 3.14 (Rozas and Rozas, 1999). The nucleotide sequence data have been deposited in GenBank under accession numbers AF329285-AF329314.
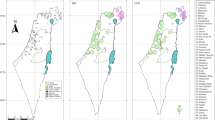
The distribution of Chioglossa lusitanica in the northwestern Iberian Peninsula (shaded areas, from Arntzen, 1999) and the study localities: 1, Muradal, Foz de Giraldo; 2, Lousã, C. Pêra; 3, Lousã, Vilarinho; 4, Açor, Margaraça; 5, Várzeas; 6, Buçaco; 7, Saide; 8, Covelo; 9, Tarouca; 10, Montemuro; 11, Valongo; 12, Bom Jesus; 13, Barroso; 14, Gerês; 15, Pontevedra; 16, Fonsagrada; 17, Salas. Dotted polygons represent six population groups identified by analysis of molecular variance (details see text): (1,4) Açor-Muradal; (2,3) Lousã; (5,6) Várzeas-Buçaco; (6,7) Buçaco-Saide; (9,10) Montemuro; and (8,11–17) Covelo and populations north of the Douro River.
Nested clade analysis
Haplotype network estimation:
The intraspecific haplotype network was reconstructed using the algorithm described by Templeton et al (1992). The method was designed to estimate phylogenies at low levels of divergence and begins by calculating the overall limits of parsimony to reconstruct intraspecific relationships of haplotypes using a statistic from neutral coalescent theory (Hudson, 1989). The parameter θ (Watterson, 1975) was estimated for our sample with DnaSP version 3.14 (Rozas and Rozas, 1999) and used to solve H in equation 10 of Hudson (1989). Once the use of parsimony was validated, the haplotype network was estimated by finding haplotype connections with probabilities above the 0.95 limit. TCS version 1.0 (Clement et al, 2000) implements the algorithm in Templeton et al (1992). It was used to estimate those probabilities and for network construction.
Nested design:
We used the rules given in Templeton et al (1987) and Templeton and Sing (1993) to define an evolutionary clade hierarchy upon the intraspecific haplotype network. The method starts from the tips of the network by nesting haplotypes (‘zero step clades’) separated by one mutational change within ‘one-step clades’, and proceeds step by step to identify higher level clades until the final level of nesting includes the entire network.
Testing for geographical association:
The nested design was used to investigate geographical association of clades and nested clades in two ways. First, clades with genetic and/or geographical variation were tested against their geographical locations through a permutational contingency analysis (Templeton and Sing, 1993; Templeton et al, 1995). A more objective analysis was conducted using information on geographical distances as described in Templeton et al (1995): clade distance (Dc) for the geographical spread of a clade and nested clade distance (Dn) for the distance of a clade from the geographical centre of the nested clade. These distances were then contrasted between interior clades and tip clades by calculating within each nested level the difference between the average interior distance and the average tip distance (I-TDc and I-TDn). This interior vs tip contrast of clades corresponds to an old vs young contrast and, to a lesser extent, common vs rare, under expectations from neutral coalescent theory (Crandall and Templeton, 1993). We used these predictions to help determine interior vs tip status by calculating outgroup weights as described in Castelloe and Templeton (1994). Observed Dc and Dn values were then compared to a distribution of Dc and Dn generated by random permutations of clades against sampling locations with GeoDis version 2.0 (Posada et al, 2000). One thousand permutations were performed to obtain statistical inferences at α = 5%. The null hypotheses were (i) no geographical association of clades and (ii) clade dispersal distances are not significantly different from random. Inferences about the population processes underlying the observed clade dispersal patterns followed the Templeton et al (1995) inference key.
Testing for neutral evolution and population expansion
The neutrality of cytochrome b evolution was analysed at the level of the haplotype network. Following Templeton (1996), the null hypothesis of homogeneity of four mutational categories (silent vs replacement and tip vs interior substitutions) was tested with Fisher's exact test on two-by-two contingency tables. We also performed Fu's (1997) Fs test with the Arlequin 2.001 software (Schneider et al, 2000) by comparing Fs against a distribution generated from 10 000 random samples under the hypothesis of selective neutrality and population equilibrium. Fs tends to be negative under an excess of recent mutations and a significant negative value will be taken as evidence of population growth and/or selection. Finally, we obtained maximum-likelihood estimates of ancestral population parameters using the Metropolis–Hastings sampling procedure as implemented in the program FLUCTUATE (Kuhner et al, 1998). This coalescent-based approach produces a likelihood surface and maximum likelihood estimates for θ (2 × Ne − the female effective population size × μ − the per site per generation mutation rate) and for the exponential growth rate (g). Because θ and g are biased upwards we did not use them to calculate Ne. Instead we determined if population size change (growth or decline) or population stasis could be inferred from the genealogy, by looking at the 95% confidence interval of g (g ± s.d.) taken from the likelihood curve, which was assumed to be normally distributed (Kuhner et al, 1998). The latter two analyses were conducted for higher level clades as identified by nested clade analysis and for groups of populations such that between-group population structure was maximized, following results obtained by analysis of molecular variance (Excoffier et al, 1992) with Arlequin 2.001. Accordingly, groups of populations considered were (1,4), (2,3), (5,6), (6,7), (9,10) and (8,11–17).
Results
MtDNA variation
Cytochrome b sequencing of 124 individuals of Chioglossa lusitanica yielded 34 variable positions describing 30 distinct haplotypes. Sequence divergence ranged from 0.14% to 2.14% with an overall nucleotide diversity of 0.99%. Seven nucleotide substitutions were observed at the first codon position, five at the second position, and 22 at the third position, underlying nine aminoacid replacements at eight positions (Table 1, Figure 2).
Network for cytochrome b haplotypes observed in 17 populations of Chioglossa lusitanica. The network was estimated under the 95% statistical limits of parsimony using the algorithm in Templeton et al (1992). Circle size represents haplotype frequency and dots represent hypothetical haplotypes. Solid retangles represent replacement substitutions.
Twenty-two haplotypes were found at single localities and eight (h2, h5, h8, h15, h16, h21, h23 and h27) were observed in two or more localities (Table 1). Haplotype h16 was the most frequently observed haplotype (n = 47) and was found in eight populations north of the Mondego River. The otherwise most frequent haplotype (h8, n = 13) was found in populations south of the Mondego. All but two of 12 localities with more than five individuals sampled were polymorphic with two to five haplotypes differing by one to four mutational changes (π ranging from 0.0003 ± 0.0004 to 0.0026 ± 0.0018). The exceptions were locality 17 which was monomorphic for haplotype h16 and locality 6 with one haplotype differing from the other three by 10–12 mutational changes (π = 0.0093 ± 0.0055).
Nested clade analysis
Haplotype network and nested design:
The parsimony criterion appeared to be justified to reconstruct the haplotype relationships of C. lusitanica. With θ = 0.012 (n = 30) H was estimated at 0.036, indicating that a difference observed at any given site between two randomly drawn haplotypes was unlikely to have arisen from more than one mutational step. The maximum number of mutational steps between haplotypes allowing parsimonious connections with a probability P > 0.95 was 11 steps. Haplotype network estimation using parsimony within this limit resulted in a single network (Figure 2). The nesting design is shown in Figure 3.
Maximum parsimony network and corresponding nested design for cytochrome b haplotypes observed in 17 populations of Chioglossa lusitanica. Note that the numbers are given without the ‘h’ prefix. Hypothetical haplotypes are represented by ‘o’. Thin-lined polygons enclosed 1-step clades and are designated by ‘1-n’; thick-lined polygons enclosed 2-step clades ‘2-n’); the thick line separates 3-step clades (3-1 and 3-2).
Nested contingency analysis:
Nested contingency analysis revealed significant association of clades and sampling locations at all clade levels (Table 2). Two 1-step clades (1–3 and 1–4) and two 2-step clades (2–1 and 2–4) showed significant levels of geographical association, while the null hypothesis of no geographical association could be rejected at all higher-level categories (3-step clades and the entire cladogram). The geographical distance analysis showed significant differences for clade (Dc) and nested clade (Dn) distances (Figure 4). The inferences made from the key in Templeton et al (1995) and the geographical distribution of clades (Figure 5) are given in Table 3. Restricted gene flow with isolation-by-distance was inferred for three nested clades (1–3, 2–1 and 3–2) whereas past fragmentation was the most likely explanation for the patterns observed at nested levels 1–4, 2–4, 3–1 and for the entire network.
Nested clade distance analysis of cytochrome b haplotypes observed in Chioglossa lusitanica. Brackets reflect the nesting structure (see Figure 3). Dc and Dn are clade and nested clade distances, respectively (for details see Templeton et al, 1995). Interior vs tip contrasts for Dc and Dn are indicated with ‘I–T’ in the corresponding clade, with interior clades given in italics bold type. For clades 1–8 and 2–1 the tip interior/status is inferred from coalescent theory (Castelloe and Templeton, 1994), and the results of ‘I–T’ are given in italics. Superscript S and superscript L indicate that distance measures are significantly smaller and larger, respectively, than expected under random distribution of haplotypes.
Schematic spatial representation of nested clade hierarchical design for the cytochrome b haplotype network of Chioglossa lusitanica. Numbers identify clades within a certain clade-level (see Figure 3). Note that geographical ranges are the same for clades 1–3 and 2–1, 1–4 and 2–2 and 1–6 and 2–3. For topographical names see Figure 1.
Neutral evolution and population expansion
The haplotype network accomodates 11 silent and eight replacement substitutions at the tips vs 19 silent and one replacement substitutions at the interior. Accordingly, the null hypothesis of neutral cytochrome b evolution was rejected (Fisher's exact test, P < 0.01). Fu's Fs test showed significant deviations for Fs from values expected under population stasis and/or neutrality at the level of clades 3–1 and 3–2 and for population groups (1,4), (9,10) and (8,11–17). The maximum likelihood estimate of g indicated significant increases in population size for each of these five groups (Table 4).
Discussion
Population structure vs population history
Nested clade distance analysis of mtDNA sequence variation revealed a high level of phylogeographic structure in the salamander Chioglossa lusitanica. The rejection of the null hypothesis of random distribution of haplotypes was common at the higher-level nestings, reflecting deep lineage divergence and statistical power (Templeton, 1998). Both population structure and population history shaped the observed geographical patterns of mtDNA variation in C. lusitanica. Past fragmentation and restricted gene flow were the processes inferred to explain the geographical associations of haplotypes (Table 3). The oldest event inferred for C. lusitanica was past fragmentation with the diverged clades, 3–1 and 3–2, distributed south and north of the Buçaco Mountains respectively (near the Mondego; Figure 5). Within these two regions, populations appear to have been subject to past fragmentation and restricted gene flow. In the south, the observations indicate fragmentation over the mountains of Muradal-Açor-Lousã vs Lousã-Buçaco (clades nested in 1–4 and 3–1) and restricted gene flow within this area (clades nested in 1–3 and 2–1). In the north, restricted gene flow is likely to have been widespread from Buçaco to northwestern Spain (clades nested in 3–2). Within the northern range, fragmentation appears to have affected the southern parts between Buçaco and the Douro River (clades nested in 2–4).
Vicariance and secondary contact
The deep mtDNA genetic divergence observed between two assemblages of C. lusitanica was concordant with allozyme genetic diversity (Alexandrino et al, 2000). The inferred fragmentation and the depth of divergence (d = 1.5%, for clades 3–1 clade 3–2) support the hypothesis of a late Pliocene/early Pleistocene vicariance across the Mondego River (Alexandrino et al, 2000). The two mtDNA lineages were only found together at Buçaco (population 6). This is more likely due to secondary contact than to insufficient time for coalescence to occur. Allozyme variation in populations from between Lousã (population 3) and Buçaco also suggested the direct contact between the southern and the northern group of populations (Alexandrino et al, 2000). This is concordant with restricted gene flow inferred for this relatively small area (clades 1–3 and 2–1) from nested clade analysis. A low level of gene flow in areas adjacent to the Mondego indicates a barrier to gene flow, composed by the river and an associated zone of generally unfavourable habitat (Alexandrino et al, 2000; Teixeira et al, 2001), strengthening the hypothesis of a local south–north separation under adverse climatic conditions during the Pleistocene (Alexandrino et al, 2000). After climatic amelioration contact between populations was probably re-established at a low level of gene flow and without extensive introgression. Recurrent events of fragmentation/restricted gene flow during the Pleistocene in a zone of decreased salamander density may have contributed to the further divergence of populations.
The fragmentation events inferred for the areas south and north of the Mondego, indicate that additional barriers have shaped the present phylogeographic structure of C. lusitanica. Two populations immediately south of the Douro River, in particular, showed high genetic distinctiveness (populations 9 and 10, clade 1–10, see Figure 5), which parallels the observed high frequency of private alleles at certain allozyme loci (Alexandrino et al, 2000). This supports the hypothesis that during the Pleistocene the Montemuro area harboured several isolated refugial populations of the northern lineage (Alexandrino et al, 2000).
Population and range expansion
The significant excess of recent mutations observed in the mtDNA polymorphism of C. lusitanica and the maximum likelihood estimates of population growth rate indicated an overall population expansion for the species (Table 4). Similar results were obtained at the local level, for population groups (1,4) from Açor-Muradal, (9,10) from Montemuro and (8,11–17) from north of the river Douro. For the other population groups, representing Lousã and Buçaco-Várzeas, the hypothesis of population expansion was not supported These areas, adjacent to the Mondego river, where population sizes appear to have been more or less stable, are therewith good candidates to have been refugia for C. lusitanica. The Montemuro mountains were previously identified as a potential refugial area (Alexandrino et al, 2000). The local populations, however, do also show the genetic characteristics of a recently expanded population.
High levels of genetic divergence in the south contrast with low levels of genetic divergence in the northern part of the C. lusitanica range. The relative genetic uniformity in populations north of the Douro, with a haplotype interior to the haplotype network predominating (Figure 2, Table 1), suggests a relatively recent origin, through colonization by a small number of founders. A similar but geographically more limited range expansion may have occurred in the areas to the southeast of Lousã as suggested by a low genetic diversity at nuclear loci (Alexandrino et al, 2000). Interestingly, the inferences of range expansion could not be not made from nested clade analysis. This may be due to lack of statistical power in the detection of geographical associations of rare haplotypes at clades with low levels of divergence (Templeton, 1998).
An important assumption underlying our inferences on population demographic expansions is that the signals obtained from mtDNA polymorphisms are not due to selection. We can however not firmly reject the possibilities for selective sweeps, hitchhiking and background selection. Indeed, the significant excess of replacement substitutions at the (young) tips of the haplotype network suggests that background selection does occur and that replacement substitutions are deleterious. Nevertheless, we argue that our results are best explained by population expansions and not by selection because: (i) we found concordant patterns of allele diversity at several unlinked nuclear enzyme loci (Alexandrino et al, 2000) and selective pressure is unlikely to have the same effect across unlinked loci; (ii) selection through the environment would produce similar patterns of polymorphism in neighbouring populations and this is not what we found (eg, population groups (1,4) and (2,3), see Table 4 and Figure 1]; and (iii) other, well-documented cases of mildly deleterious evolution of mtDNA involve expanding populations (mostly humans and their commensals) suggesting that selection may be relaxed (Zouros and Rand, 2000).
Conclusions
The historical scenario of Pleistocene vicariance for C. lusitanica put forward by Alexandrino et al (2000) was confirmed by nested clade analysis of a substantially enlarged mtDNA data set. Hitherto unrecognized phylogeographic structure suggests that habitat fragmentation may have been quite common in the southern range of this salamander. The pattern of mtDNA sequence polymosphism combined with the distribution of diversity and the presence of rare alleles at protein loci (Alexandrino et al, 2000) were used to identify specific areas as past refugia of C. lusitanica. Range expansion inferred from allozyme studies was supported by the descriptive interpretations of mtDNA data and by statistical tests revealing several cases of population expansion, but not from formal nested clade analysis. Showing range expansion with nested clade analysis may be problematic because (i) not all genetic characteristics expected to arise from range expansion are incorporated in the inference key, and (ii) lack of statistical power in recently colonized areas (as described above). A situation similar to that of C. lusitanica was found in Drosophila buzzatii – but not in the nested clade analysis for another 12 species with strong a priori evidence of expansion events (Templeton, 1998). In both species founder effects caused the near fixation of ancestral haplotypes in areas of range expansion, defying the methodology of nested clade analysis (Cann et al, 1987; Templeton, 1998). The inference key for nested clade analysis deserves further testing through the modelling of simulated data sets and through the analysis of actual data from organisms representing a range of ecological and population dynamic characteristics.
The observed concordance of allozyme and mtDNA variation across two diverged population assemblages in C. lusitanica brings up again the question of their taxonomic status. We described morphological differentiation for the two groups here recognized (Alexandrino et al, submitted). Any taxonomic revision should not only rely on patterns of variation concordant across the population assemblages, but also assess the level of gene flow in the putative secondary contact zone. This requires a fine scale genetic study of this area, for example through the use of highly variable nuclear markers such as microsatellites. Such data along with information on ecological cohesiveness (sensu Crandall et al, 2000) would also provide guidance on how these two groups of C. lusitanica should be managed for conservation.
References
Alexandrino, J, Froufe, E, Arntzen, JW, Ferrand, N (2000). Genetic subdivision, glacial refugia and postglacial recolonizationin the golden-striped salamander, Chioglossa lusitanica (Amphibia: Urodela). Molec Ecol, 9: 771–781.
Arntzen, JW (1981). Ecological observations on Chioglossa lusitanica (Caudata, Salamandridae). Amphibia-Reptilia, 1: 187–203.
Arntzen, JW (1994). Speedy salamanders: sedentariness and migration of Chioglossa lusitanica. Revista Española Herpetol, 8: 81–86.
Arntzen, JW (1999). Chioglossa lusitanica Bocage, 1864 – Goldstreifensalamander. In: Grossenbacher K, Thiesmeier B (eds) Handbuch der Amphibien und Reptilien Europas, AULA-Verlag: Wiesbaden pp 301–321.
Cann, RL, Stoneking, M, Wilson, AC (1987). Mitochondrial DNA and human evolution. Nature, 325: 31–36.
Castelloe, J, Templeton, AR (1994). Root probabilities for intraspecific gene trees under neutral coalescent theory. Molec Phylogenet Evol, 3: 102–113.
Clement, M, Posada, D, Crandall, K (2000). TCS: a computer program to estimate gene genealogies. Molec Ecol, 9: 1657–1660.
Crandall, KA, Templeton, AR (1993). Empirical tests of some predictions from coalescent theory with applications to intraspecific phylogeny reconstruction. Genetics, 134: 959–969.
Crandall, KA, Bininda-Edmonds, ORP, Mace, GM, Wayne, RK (2000). Considering evolutionary processes in conservation biology. Trends Ecol Evol, 15: 290–295.
Excoffier, L, Smouse, PE, Quattro, JM (1992). Analysis of molecular variance inferred from metric distances among DNA haplotypes: application to human mitochondrial DNA restriction data. Genetics, 131: 479–491.
Fu, YX (1997). Statistical neutrality of mutations against population growth, hitchhiking and background selection. Genetics, 147: 915–925.
Hall, TA (1999). BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucl Acids Symp Ser, 41: 95–98.
Hudson, RR (1989). How often are polymorphic restriction sites due to single mutation? Theoret Pop Biol, 36: 23–33.
Kuhner, MK, Yamato, J, Felsenstein, J (1998). Maximum likelihood estimation of population growth rates based on the coalescent. Genetics, 149: 429–434.
Larson, A, Wake, DB, Yanev, KP (1984). Measuring gene flow among populations having high levels of genetic fragmentation. Genetics, 106: 293–308.
Moritz, C, Schneider, CJ, Wake, DB (1992). Evolutionary relationships within the Ensatina escholtzii complex confirm the ring species interpretation. Systematic Biol, 41: 273–291.
Neigel, JE (1997). A comparison of alternative strategies for estimating gene flow from genetic markers. Ann Rev Ecol Systemat, 28: 105–128.
Posada, D, Crandall, KA, Templeton, AR (2000). GeoDis: a program for the cladistic nested analysis of the geographical distribution of genetic haplotypes. Molec Ecol, 9: 487–488.
Roe, B, Ma, D, Wilson, R, Wong, J (1985). The complete nucleotide sequence of the Xenopus laevis mitochondrial genome. J Biolog Chem, 260: 9759–9774.
Rozas, J, Rozas, R (1999). DnaSP, Version 3: an integrated program for molecular population genetics and molecular evolution analysis. Bioinformatics, 15: 174–175.
Schneider, S, Roessle, D, Excoffier, L (2000). Arlequin: a software for population genetics data analysis. Version 2.001. Genetics and Biometry Laboratory, Department of Anthropology, University of Geneva.
Slatkin, M (1981). Estimating levels of gene flow in natural populations. Genetics, 99: 323–335.
Tan, AM, Wake, DB (1995). MtDNA phylogeography of the California newt, Taricha torosa (Caudata, Salamandridae). Molec Phylogenet Evol, 4: 383–394.
Teixeira, J, Ferrand, N, Arntzen, JW (2001). Biogeography of the golden-striped salamander, Chioglossa lusitanica: a field survey and spatial modelling approach. Ecography, 24: 618–623.
Templeton, AR (1996). Contingency tests of neutrality using intra/interspecific gene trees: the rejection of neutrality for the evolution of the mitochondrial cytochrome oxidase II gene in hominoid primates. Genetics, 144: 1263–1270.
Templeton, AR (1998). Nested clade analysis of phylogeographic data: Testing hypotheses about gene flow and population history. Molec Ecol, 7: 381–397.
Templeton, AR, Boerwinkle, E, Sing, CF (1987). A cladistic analysis of phenotypic associations with haplotypes inferred from restriction endonuclease mapping. I. Basic theory and an analysis of alcohol dehydrogenase activity in Drosophila. Genetics, 117: 343–351.
Templeton, AR, Crandall, KA, Sing, CF (1992). A cladistic analysis of phenotypic associations with haplotypes inferred from restriction endonuclease mapping. III. Cladogram estimation. Genetics, 132: 619–633.
Templeton, AR, Routman, E, Phillips, CA (1995). Separating population structure from population history: a cladistic analysis of the geographical distribution of mitochondrial DNA haplotypes in the tiger salamander, Ambystoma tigrinum. Genetics, 140: 767–782.
Templeton, AR, Sing, CF (1993). A cladistic analysis of phenotypic associations with haplotypes inferred from restriction endonuclease mapping. IV. Nested analyses with cladogram uncertainty and recombination. Genetics. 134: 659–669.
Watterson, GA (1975). On the number of segregating sites in genetical models without recombination. Theoret Pop Biol, 7: 256–276.
Wright, S (1931). Evolution in Mendelian populations. Genetics, 16: 97–159.
Zouros, E, Rand, DM (2000). Population genetics and evolution of animal mitochondrial DNA. In: Singh RS, Krimbas CB (eds) Evolutionary Genetics, Cambridge University Press: Cambridge, UK pp 430–455.
Acknowledgements
This work was supported by the ‘Instituto da Conservação da Natureza’ (ICN), the ‘LIFE’ programme of the European Community and the ‘Fundação para a Ciencia e a Tecnologia’ (PRAXIS XXI/BD/5917/95 PhD grant to JA and PRAXIS/P/BIA/11174/1998). Licensing to collect was provided by ICN for Portugal and by the Galician and Asturian ‘Junta de Medio Ambiente’ for Spain. We thank D Tarkhnishvili, M García-París, P Alexandrino, M Branco, R Godinho, C Oliveira, F Sequeira and J Teixeira for support and discussions. We thank D Posada for his help with TCS 1.0 and AR Templeton, E Lessa, J Brookfield and an anonymous reviewer for comments and suggestions on an earlier version of the manuscript.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Alexandrino, J., Arntzen, J. & Ferrand, N. Nested clade analysis and the genetic evidence for population expansion in the phylogeography of the golden-striped salamander, Chioglossa lusitanica (Amphibia: Urodela). Heredity 88, 66–74 (2002). https://doi.org/10.1038/sj.hdy.6800010
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/sj.hdy.6800010
Keywords
This article is cited by
-
What can genetics tell us about the history of a human-mediated introduction of the golden-striped salamander south of its native range?
European Journal of Wildlife Research (2022)
-
The Asian plethodontid salamander preserves historical genetic imprints of recent northern expansion
Scientific Reports (2021)
-
Is Malaysia’s banded langur, Presbytis femoralis femoralis, actually Presbytis neglectus neglectus? Taxonomic revision with new insights on the radiation history of the Presbytis species group in Southeast Asia
Primates (2019)
-
A review of the palaeoclimatic inference potential of Iberian Quaternary fossil batrachians
Palaeobiodiversity and Palaeoenvironments (2016)
-
Phylogeography and demographic history of Lacerta lepida in the Iberian Peninsula: multiple refugia, range expansions and secondary contact zones
BMC Evolutionary Biology (2011)