Abstract
Β-lactamases-producing Escherichia coli are a widely distributed source of antimicrobial resistance (AMR), for animals and humans. Little is known about the sensitivity profile and genetic characteristics of E. coli strains isolated from domestic cats. We report a cross-sectional study that evaluated E. coli strains isolated from domestic cats in Panama. For this study the following antibiotics were analyzed: ampicillin, amoxicillin-clavulanate cefepime, cefotaxime, cefoxitin, ceftazidime, aztreonam, imipenem, gentamicin, kanamycin, streptomycin, tetracycline, ciprofloxacin, nalidixic acid, trimethoprim-sulfamethoxazole, and chloramphenicol. The data obtained were classified as resistant, intermediate, or sensitive. MDR strains were established when the strain presented resistance to at least one antibiotic from three or more antimicrobial classes. Forty-eight E. coli isolates were obtained, of which 80% presented resistance to at least one of the antibiotics analyzed, while only 20% were sensitive to all (p = 0.0001). The most common resistance was to gentamicin (58%). Twenty-nine percent were identified as multidrug-resistant isolates and 4% with extended spectrum beta-lactamase phenotype. The genes blaTEM (39%), blaMOX(16%), blaACC (16%) and blaEBC (8%) were detected. Plasmid-mediated resistance qnrB (25%) and qnrA (13%) are reported. The most frequent sequence types (STs) being ST399 and we reported 5 new STs. Our results suggest that in intestinal strains of E. coli isolated from domestic cats there is a high frequency of AMR.
Similar content being viewed by others
Introduction
Escherichia coli is among the main bacteria responsible for infections in both humans and animals. It is one of the key pathogens contributing to the burden of antimicrobial resistance (AMR) and resistance-associated deaths1. While some pathogenic strains of E. coli can cause intestinal infections, the majority of strains can potentially cause extra-intestinal infections, mainly of the urinary tract2. Based on distinctive genotypes, the main phylogenetic groups of E. coli have been established (A, B1, B2, and D). The pathogenic extra-intestinal strains belong mainly to Group B2 and, to a lesser extent, to Group D, while the strains of Groups A and B1 are usually intestinal pathogens or commensals3.
Escherichia coli infections are widely treated with β-lactam antibiotics and fluoroquinolones in both human and veterinary medicine. A study conducted in Latin America reported the antibiotics most used in this region for the treatment of infections in pets corresponded to β-lactams, prescribed in 65.3% and quinolones in 36.2% of the cases treated4. However, resistance to these groups of antibiotics has increased worldwide, which is why the World Health Organization (WHO) has devised a list of priority pathogens to combat AMR, where multidrug-resistant (MDR) E. coli made the list as a critical priority 1 pathogen5.
Among E. coli strains, the main mechanism of AMR to β-lactams is the production of β-lactamase enzymes, among which are the extended-spectrum β-lactamase (ESBL) enzymes and the AmpC type encoded by plasmids (pAmpC). ESBL/AmpC–producing E. coli constitutes a widely distributed source of AMR in both humans and animals6. On the other hand, within the mechanisms of AMR to quinolones, chromosomal amino acid substitutions have been mainly described within the quinolone resistance determining regions (QRDR) of the genes gyrA and parC, which are targets of quinolones, in addition to the acquisition of plasmid-mediated quinolone resistance (PMQR) genes, among which the qnr genes (qnrA, qnrB, qnrC, qnrD, and qnrS) have been described7.
Transmission of E. coli with ESBL/AmpC has become a One Health issue, as these strains can be transmitted between humans, animals, and the environment. Close contact between companion animals and humans increases the risk of transmission of ESBL/AmpC-carrying bacteria through horizontal transfer and clonal spread8,9. Worldwide, around 56% of people have at least one pet, with domestic cats (Felis catus) among the most popular in the world. For example, in a survey conducted in the United States (US), 45.3 million households reported having at least one cat10, reflecting the magnitude of its presence in domestic cats.
Healthy cats are recognized as a reservoir of E. coli strains carrying ESBL/AmpC, with a global prevalence of 5.04%, predominantly identifying the CTX-M type for ESBL and CMY for pAmpC11. Studies carried out in Latin America (i.e., in Argentina and Brazil) on E. coli isolates obtained from cats have detected ESBL of the CTX-M-15 and CTX-M-2 types and for the plasmid type pAmpC, the CMY-2 has been reported12,13.
In both pets and humans, quinolone resistance has been most frequently reported in β-lactam–resistant Enterobacteriaceae. Studies have demonstrated the association between genes encoding ESBL and gene variants encoding PMQR, such as qnrA, qnrB, and qnrS genes14,15.
In companion animals, around 171 different sequence types (STs) of E. coli have been reported, with ST38 and ST131 found in all continents11. In Latin America, ST90, ST457, ST155, and ST131 have been predominantly identified in cat samples14,15.
Studies carried out in Latin America, specifically in South America, show a high percentage of resistance to multiple antibiotics in strains of E. coli isolated from cats14,15. However, as far as we know, there are no reports published for Central America. The purpose of this study is, then, to characterize E. coli strains isolated from fecal samples of domestic cats in the central region of Panama, with the objective of investigating the AMR phenotype by identifying sensitivity profiles, characterizing the ESBL/AmpC–producing strains and their molecular typing using the multilocus sequence typing (MLST) scheme and phylogenetic group analysis.
Results
Escherichia coli strains isolated from fecal samples of domestic cats
A total of 48 E. coli strains were isolated from domestic cats in the central region of Panama, of which 75% (36/48) were obtained from females and 25% (12/48) from males, with a mean (SD) age of 13.2 (14.5) months. The cat breeds were all undefined and the type of feeding was distributed in 56% (27/48) for kibbles, 40% (19/48) varied and 4% (2/48) a mixture of varied and kibbles. Ten percent (5/48) had a history of previous antibiotic use, including: penicillin (2/5), metronidazole/spiramycin (1/5), amoxicillin (1/5), and enrofloxacin (1/5).
Sensitivity profile of E. coli strains
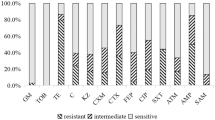
Of the E. coli strains analyzed, 80% (38/48) showed resistance to at least one of the antibiotics and 20% (10/48) were sensitive to all antibiotics analyzed (P < 0.001). Figure 1 presents the sensitivity to the antibiotics analyzed with the E. coli isolates. We note greater resistance to gentamicin (58%), kanamycin (25%), and tetracycline (21%) and no resistance to amoxicillin-clavulanate and imipenem. No significant differences were evident when comparing the proportion of resistance by age, sex, breed, type of diet, and history of previous antibiotic use.
Sensitivity profile of E. coli strains isolated from domestic cats in central Panama. AMC, amoxicillin–clavulanate; AMP, ampicillin; ATM, aztreonam; CAZ, ceftazidime; CIP, ciprofloxacin; CPL, chloramphenicol; CTX, cefotaxime; FEP, cefepime; FOX, cefoxitin; GEN, gentamicin; IPM, imipenem; KAN, kanamycin; NAL, nalidixic acid; STS, streptomycin; SXT, trimethoprim–sulfamethoxazole; TET, tetracycline.
Proportion of antimicrobial resistance in the identified phylogenetic groups
Table 1 shows the phylogeny classification of the isolated E. coli strains, with 65% commensals from phylogenetic groups A or B1 and 33% pathogenic from phylogenetic group B2; no strains from phylogenetic Group D were identified. Regarding the distribution of isolates with antibiotic resistance by phylogroup, Table 2 shows that gentamicin was the antibiotic presenting the highest proportion of resistance in the three phylogenetic groups: for Group A, 33% of the strains presented resistance to three or more antibiotics, for Groups B1 and B2, respectively, 30% and 38% presented resistance to three or more antibiotics, with no significant difference between phylogenetic groups (P = 0.182).
Phenotypes and genotypes
Molecular typing using Pasteur's MLST technique identified 18 STs (Table 3). We observed ST399 (17%), ST1242 (17%), ST1243 (9%), and 1% for each of the remaining STs. Five 5 new STs are reported: ST1242, ST1243, ST1244, ST1245, and ST1247.
The characterization of MDR strains was detected in 29% (14/48), among which 36% (5/14) presented resistance to three, 36% (5/14) to four, and 28% (4/14) to five or more types of antibiotics. We identified that 27% (13/48) of the strains were resistant to at least one of the β-lactams analyzed and 4% (2/48) presented the ESBL phenotype. Regarding the genotypes of the strains showing β-lactam resistance, the blaTEM gene was identified in 39% (5/13), while blaCTX-M and blaSHV were not detected. Within pAmpC genes, blaMOX was identified in 16% (2/13), blaACC in 16% (2/13), and blaEBC in 8% (1/13).
Table 4 shows the PMQR and QRDR for the strains showing quinolone and fluoroquinolone resistance, with 38% (3/8) showing changes in gyrA and ST355 (GLS02) with changes in parC. We observed the following changes: Ser83Leu (38%, 3/8), Asp87Asn (13%, 1/8), and Ser80Ile (13%, 1/8). Among the strains with quinolone resistance, we identified qnrA genes in 13% (1/8) and qnrB in 25% (2/8).
Discussion
Escherichia coli with ESBL has become a One Health problem, since it can be transmitted among humans, animals, and the environment and represents one of the main pathogens contributing to the AMR burden and resistance-associated mortality1. The present study shows bacterial phenotypes in combination with genetic characteristics of E. coli strains isolated from healthy domestic cats from the central region of Panama. We observed that 80% of the E. coli strains presented resistance to at least one of the antibiotics analyzed, among which we identified the ESBL phenotype in 4%. Worldwide, it has been estimated that the prevalence of ESBL in cats is 5.04% and in dogs it is 6.87%11. This prevalence may vary by region, for example, in America it has been reported in cats 8.15% and in dogs 6.79%11. Therefore, the ESBL prevalence identified in our study concurs with region’s reported prevalence rates. Globally, no significant differences have been observed between the prevalence of ESBL between dogs and cats. Similarly in Panama, a previous study conducted on E. coli strains isolated from healthy dogs identified an 8% ESBL phenotype16. Worldwide, the most identified ESBL genotypes are blaCTX-M, blaSHV, and blaTEM, which are uniformly distributed between dogs and cats. However, the prevalence rates of these phenotypes may vary between regions11. In our study, the blaTEM , blaCTX-M and blaSHV were not identified among the ESBL strains tested. These findings are not in agreement with data from studies conducted in Latin America where ESBL of the CTX-M-2 and CTX-M-15 genotypes have been identified in E. coli strains isolated from cats12,13.
Different groups of pAmpC enzymes have been described, including: CMY, ACC, DHA, CIT, EBC, FOX, and MOX, which have been detected in both humans and animals17. The CMY-2 type is the pAmpC most identified in domestic animals. However, in our study, pAmpC of the MOX, ACC, and EBC were the main groups identified8. Previous studies carried out on E. coli strains isolated from cats have identified pAmpC of the EBC and ACC groups17, however the report of MOX group enzymes in companion animals is unusual—there are reports in production animals, such as chickens and goats18,19. In our study, two strains (GLS01 and GLS17) co-expressed the blaACC and blaMOX genes, which shows an exchange of E. coli strains carrying important interspecies resistance genes, since the MOX group has been described mainly in production animals. This represents an important source of MDR dissemination, given that encoding genes are carried in mobile elements and confer resistance to broad-spectrum cephalosporins.
Of the strains analyzed in this study, 29% were MDR within all phylogenetic groups (A, B1, and B2) with no significant differences between groups. This finding is of particular concern, due to the potential for opportunistic infections, zoonotic transmission, and transmission of antibiotic-resistant determinants from commensal isolates to potential pathogenic bacteria20. MDR E. coli strains have been previously described in studies carried out in healthy domestic animals11,13,14,15,16,17 as well as from environmental strains21. This aspect has varied in diverse countries where regulatory standards based on non-prescription antibiotics used in veterinary medicine have been regulated due to the emergence of MDR strains22. On the other hand, aspects such as diet, use of antibiotics without volumetric determination, commonly used antibiotics (e.g., doxycycline and enrofloxacin) all contribute to the emergence of resistant strains23. MDR E. coli poses a challenge to global health systems due to the treatment of infectious diseases and increased therapeutic complications24.
Around 171 different STs have been described in cats and dogs, with ST38 and ST131 identified on all continents, followed by ST68, ST405, and ST61711. In Latin America, the prevalence of STs obtained by the MLST technique from E. coli strains isolated in cats varies between countries. For example, in Brazil and Argentina, ST90, ST457, and ST131 have been identified as the most predominant12,13. In our study, ST399 was the most prevalent and interestingly, this same ST was mostly identified in a study of E. coli strains isolated from healthy dogs in central Panama16, which could be due to the close coexistence of dogs and cats in the same households. Likewise, ST399 has been described in isolates from dogs and cats in Oceania and Asia11. In addition, ST532, previously described in Europe, and ST367 and ST132—described in the Americas, Asia, and Europe11 were identified. The above evidences the existence of an important distribution of circulating genetic diversity.
The ST88 identified in this study has been previously reported in clinical isolates from hospitalized patients in our country25 and also in isolates from healthy dogs16, which allows us to infer that there may be exchange of E. coli strains carrying genes of resistance between owners, pets, and the environment26. ST132 and ST83 presented the expression of PMQR genes, with ST83 presenting co-expression of resistance genes qnrB and qnrS. As stated, it is paramount that pet owners and their relatives may be at risk of acquiring bacteria carrying antibiotic resistance genes and plasmid-mediated genes. A study showed that the transport of community-acquired ESBL/pAmpC was attributed in 7.9% to companion animals, constituting the most common nonhuman sources6.
The WHO establishes among its critically important antibiotics the quinolones and β-lactams (including 3rd, 4th, and 5th generation cephalosporins and amino penicillins with and without beta-lactamase inhibitors) in addition to aminoglycosides and others27. Many of these are also used in veterinary medicine. Our data showed resistance to several of the aforementioned critically important antibiotics (Fig. 1), which draws attention to the potential difficulties in managing infections, as well as the importance of knowing the composition and distribution of antibiotic resistance genotype as an important step to delineate the impact of MDR E. coli infections in our population.
The limitations of this study have to do with the small number of strains analyzed due to the high cost of inputs in Panama. Limitations aside and to the best of our knowledge, this is the first study in Panama and Central America on patterns of antimicrobial sensitivity and resistance, as well as molecular epidemiology data of E. coli strains isolated from fecal samples of domestic cats showing MDR is important in commensal strains of E. coli in healthy cats widely distributed among other carrier species, some of them of clinical interest, as well as the new STs described for E. coli.
Methods
Strain isolates
A cross-sectional study was conducted in August 2022, where fecal samples were obtained from domestic cats from the central provinces of Herrera and Los Santos in the Republic of Panama, with the informed consent of their owners. Samples were obtained by rectal swabbing of domestic cats in Cary-Blair media (COPAN Diagnostics Inc.; Murrieta, CA) for E. coli isolates. For each sample collected, a technical sheet was completed with the following variables, if present: age, sex, breed, diet, and previous use of antibiotics. One sample was obtained per individual. E. coli isolates were obtained using Chromocult® agar (Merck Millipore; Darmstadt, Germany) and on eosin-methylene blue (EMB) agar (Merck Millipore; Darmstadt, Germany) for confirmation. The study protocol was reviewed and approved by the Animal Bioethics Subcommittee at the University of Panama’s Committee of Ethics of Research and Animal Welfare (No. CEIBA-UP-027-2022, dated 11 July 2022), all methods were performed in accordance with the relevant guidelines and regulations.
Antibiotic sensitivity testing
Antibiotic sensitivity was determined by the disk diffusion method on Muller Hinton Agar (Merck Millipore; Darmstadt, Germany)28. Sensitivity profiles were interpreted according to the Clinical and Laboratory Standards Institute29. The following antibiotics were analyzed: ampicillin (AMP 10 μg), amoxicillin-clavulanate (AMC 20/10 μg), cefepime (FEP 30 μg), cefotaxime (CTX 30 μg), cefoxitin (FOX 30 μg), ceftazidime (CAZ 30 μg ), aztreonam (ATM 30 µg), imipenem (IPM 10 µg), gentamicin (GEN 10 µg), kanamycin (KAN 30 µg), streptomycin (STS 300 µg), tetracycline (TET 30 µg), ciprofloxacin (CIP 5 µg) , nalidixic acid (NAL 30 µg), trimethoprim-sulfamethoxazole (SXT 1.25/23.75 µg), and chloramphenicol (CPL 30 µg). The data obtained were classified as resistant, intermediate, or sensitive. MDR strains were established when the strain presented resistance to at least one antibiotic from three or more antimicrobial classes30.
Molecular typing
Molecular typing using the MLST technique was conducted on the strains showing resistance to more than one antibiotic and four strains with resistance to one antibiotic were chosen at random (GLS20, GLS39, GLS47, and GLS10) in addition to a strain sensitive to all antibiotics (GLS13). DNA extraction from the isolates was carried out using the boiling method. The extracted DNA was stored at − 20 °C to perform molecular tests. The Pasteur Institute MLST technique was used to study eight genes: dinB, icdA, polB, pabB, putP, trpA, trpB, and uidA31. Samples were amplified from chromosomal DNA, analyzed by polymerase chain reaction (PCR) in a MiniAmp Plus Thermal Cycler (Applied Biosystems; Waltham, MA) and visualized in electrophoresis with 1.5% agarose gel (Thermo Fisher Scientific; Waltham, MA). Sequencing of PCR products was performed using Macrogen services (Macrogen Inc.; Seoul, Korea). The sequences of the eight genes were analyzed in Geneious Prime v.2020 2.4. (Dotmatics; Boston, MA) and were run on the Institut Pasteur MLST Bacterial Isolate Genome Sequence Database (BIGSDB) website for particular allele profiles and STs (https://bigsdb.pasteur.fr/ecoli/ecoli.html). A search was conducted for the STs identified in our study and the corresponding STs using Achtman’s MLST Method (https://pubmlst.org/bigsdb?db=pubmlst_escherichia_seqdef).
Amplification of resistance genes
The isolates based on the resistance phenotype to β-lactams and quinolones were analyzed by PCR to determine the presence of blaCTX-M, blaTEM, blaSHV, blaAMPC genes, using specific PCR primers, as previously described and summarized in Table 516,31,32,33. The fragments obtained from the PCR product were analyzed by electrophoresis in a 1.5% agarose gel and visualized under ultraviolet light. The presence of qnrA, qnrB, and qnrS was determined for PMQR and QRDR with gyrA and parC34,35. The results obtained from the sequencing of gyrA and parC were analyzed in Geneious Prime v.2020 2.4, BLASTN nucleotide search program of the National Center for Biotechnology Information (NCBI) (https://blast.ncbi.nlm.nih.gov/Blast.cgi) and compared with partial sequences of E. coli K12 (X57174.1, M58408.1).
Phylogenetic analysis
The distribution of phylogenetic groups of commensal and pathogenic strains was carried out based on methodologies described by Clermont’s group3. The fragments obtained were analyzed by electrophoresis in 2% agarose gel and visualized under ultraviolet light.
Data analyses
Data were recorded in MS Excel (The Microsoft Corporation; Redmond, WA, US). Data analysis was performed in Stata v. 11.0 (StataCorp, LLC; College Station, TX). We applied descriptive statistics and estimated their respective 95% confidence intervals (CIs). We used Fisher’s exact test to compare proportions. An alpha at 0.05 was set for a significant statistic to compare antibiotic resistance frequencies by age, sex, and previous antibiotic group use.
Institutional review board statement
The study protocol was reviewed and approved by the Animal Bioethics Committee at the University of Panama: Comité de Ética de la Investigación y el Bienestar de los Animales de la Universidad de Panamá CEIBAUP (No. CEIBA-UP-027-2022, dated 11 July 2022).
Informed consent statement
Informed consent was obtained from all owners of cats participating in the study.
Use of generative artificial intelligence (AI)
The authors declare that no generative AI tools or services were used in the preparation of this manuscript.
Data availability
All data generated or analysed during this study are included in this published article. The study is reported in accordance with ARRIVE guidelines.
References
Antimicrobial Resistance Collaborators. Global burden of bacterial antimicrobial resistance in 2019: A systematic analysis. Lancet 399, 629–655 (2022).
Cundon, C. C., Ameal, A., Maubecín, E. & Bentancor, A. Characterization of extraintestinal pathogenic Escherichia coli strains isolated from household dogs and cats in Buenos Aires, Argentina. Rev. Argent. Microbiol. 50, 290–294 (2018).
Clermont, O., Bonacorsi, S. & Bingen, E. Rapid and simple determination of the Escherichia coli phylogenetic group. Appl. Environ. Microbiol. 66, 4555–4558 (2000).
Galarce, N. et al. Antimicrobial use in companion animals: Assessing veterinarians’ prescription patterns through the first national survey in Chile. Animals. 11, 348 (2021).
World Health Organization. Global priority list of antibiotic-resistant bacteria to guide research, discovery, and development of new antibiotics. https://www.who.int/news/item/27-02-2017-who-publishes-list-of-bacteria-for-which-new-antibiotics-are-urgently-needed (Accessed 30 Oct 2023).
Mughini-Gras, L. et al. Attributable sources of community-acquired carriage of Escherichia coli containing β-lactam antibiotic resistance genes: A population-based modelling study [published correction appears in Lancet Planet Health 2019, 3, e408]. Lancet Planet Health 3, e357–e369 (2019).
Chong, Y., Shimoda, S. & Shimono, N. Current epidemiology, genetic evolution and clinical impact of extended-spectrum β-lactamase-producing Escherichia coli and Klebsiella pneumoniae. Infect. Genet. Evol. 61, 185–188 (2018).
Hong, J. S. et al. Molecular characterization of fecal extended-spectrum β-lactamase- and AmpC β-lactamase-producing Escherichia coli from healthy companion animals and cohabiting humans in South Korea. Front. Microbiol. 11, 674 (2020).
Jin, M. et al. Evidence for the transmission of antimicrobial resistant bacteria between humans and companion animals: A scoping review. One Health. 17, 100593 (2023).
American Pet Products Association. 2021–2022 National pet Owner's Survey. https://www.americanpetproducts.org/press_industrytrends.asp (Accessed 30 Oct 2023).
Salgado-Caxito, M., Benavides, J. A., Adell, A. D., Paes, A. C. & Moreno-Switt, A. I. Global prevalence and molecular characterization of extended-spectrum β-lactamase producing-Escherichia coli in dogs and cats—A scoping review and meta-analysis. One Health. 12, 100236 (2021).
Rumi, M. V. et al. Co-occurrence of clinically relevant β-lactamases and MCR-1 encoding genes in Escherichia coli from companion animals in Argentina. Vet. Microbiol. 230, 228–234 (2019).
Melo, L. C. et al. Prevalence and molecular features of ESBL/pAmpC-producing Enterobacteriaceae in healthy and diseased companion animals in Brazil. Vet. Microbiol. 221, 59–66 (2018).
Tolun, V. et al. Relationship between ciprofloxacin resistance and extended-spectrum beta-lactamase production in Escherichia coli and Klebsiella pneumoniae strains. Clin. Microbiol. Infect. 10, 72–75 (2004).
Corkill, J. E., Anson, J. J. & Hart, C. A. High prevalence of the plasmid-mediated quinolone resistance determinant qnrA in multidrugresistant Enterobacteriaceae from blood cultures in Liverpool, UK. J. Antimicrob. Chemother. 56, 1115–1117 (2005).
Núñez-Samudio, V., Pimentel-Peralta, G., De La Cruz, A. & Landires, I. Genetic diversity and new sequence types of Escherichia coli coharboring β-lactamases and PMQR genes isolated from domestic dogs in central Panama. Genes. 14, 73 (2022).
Seo, K. W. Development of a method for the fast detection of extended-spectrum β-lactamase- and plasmid-mediated Ampc β-lactamase-producing Escherichia coli and Klebsiella pneumoniae from dogs and cats in the USA. Animals. 13, 649 (2023).
Abd El-Aziz, N. K. & Gharib, A. A. Coexistence of plasmid-mediated quinolone resistance determinants and AmpC-Beta-Lactamases in Escherichia coli strains in Egypt. Cell Mol. Biol. 61, 29–35 (2015).
Osman, K. M. et al. Poultry hatcheries as potential reservoirs for antimicrobial-resistant Escherichia coli: A risk to public health and food safety. Sci. Rep. 8, 5859 (2018).
Schmidt, V. M. et al. Antimicrobial resistance risk factors and characterisation of faecal E. coli isolated from healthy Labrador retrievers in the United Kingdom. Prev. Vet. Med. 119, 31–40 (2015).
Carattoli, A. Animal reservoirs for extended spectrum beta-lactamase producers. Clin. Microbiol. Infect. 14, 117–123 (2008).
Ramadan, H. et al. Circulation of emerging NDM-5-producing Escherichia coli among humans and dogs in Egypt. Zoonoses Public Health. 67, 324–329 (2020).
Dolejska, M. et al. Plasmids carrying blaCTX-M-1 and qnr genes in Escherichia coli isolates from an equine clinic and a horseback riding centre. J. Antimicrob. Chemother. 66, 757–764 (2011).
Villegas, M. V., Blanco, M. G., Sifuentes-Osornio, J. & Rossi, F. Increasing prevalence of extended-spectrum-beta-lactamase among Gram-negative bacilli in Latin America—2008 update from the Study for Monitoring Antimicrobial Resistance Trends (SMART). Braz. J. Infect. Dis. 15, 34–39 (2011).
Núñez-Samudio, V. et al. Molecular epidemiology of Escherichia coli clinical isolates from central Panama. Antibiotics. 10, 899 (2021).
Palmeira, J. D., Haenni, M., Metayer, V., Madec, J.-Y. & Ferreira, H. M. N. Epidemic spread of IncI1/pST113 plasmid carrying the extended-spectrum beta-lactamase (ESBL) blaCTX-M-8 gene in Escherichia coli of Brazilian cattle. Vet. Microbiol. 243, 108629 (2020).
World Health Organization. Critically important antimicrobials for human medicine: Categorization for the development of risk management strategies to contain antimicrobial resistance due to non-human antimicrobial use. Report of the Second WHO Expert Meeting; Copenhagen, Denmark: 29–31 May 2007. https://apps.who.int/iris/bitstream/handle/10665/43765/9789241595742_eng.pdf (Accessed 30 Oct 2023).
Bauer, A. W., Kirby, W. M., Sherris, J. C. & Turck, M. Antibiotic susceptibility testing by a standardized single disk method. Am. J. Clin. Pathol. 45, 493–496 (1966).
Clinical and Laboratory Standards Institute. Performance Standards for Antimicrobial Susceptibility Testing (30th ed). CLSI supplement M100. (Clinical and Laboratory Standards Institute, 2020) https://clsi.org/standards/products/microbiology/documents/m100/ (Accessed 30 Oct 2023).
Rafailidis, P. I. & Kofteridis, D. Proposed amendments regarding the definitions of multidrug-resistant and extensively drug-resistant bacteria. Expert Rev. Anti Infect. Ther. 20(2), 139–146 (2022).
Jaureguy, F. et al. Phylogenetic and genomic diversity of human bacteremic Escherichia coli strains. BMC Genomics. 9, 560 (2008).
Dallenne, C., Da Costa, A., Decré, D., Favier, C. & Arlet, G. Development of a set of multiplex PCR assays for the detection of genes encoding important β-lactamases in Enterobacteriaceae. J. Antimicrob. Chemother. 65, 490–495 (2010).
Gundran, R. S. et al. Prevalence and distribution of blaCTX-M, blaSHV, blaTEM genes in extended-spectrum β-Lactamase-producing E. coli isolates from broiler farms in the Philippines. BMC Vet. Res. 15(1), 1–8 (2019).
Gonggrijp, M. A. et al. Prevalence and risk factors for extended-spectrum β-lactamase-and AmpC-producing Escherichia coli in dairy farms. J. Dairy Sci. 99(11), 9001–9013 (2016).
Dierikx, C. M. et al. Occurrence and characteristics of extended-spectrum-β-lactamase-and AmpC-producing clinical isolates derived from companion animals and horses. J. Antimicrob. Chemother. 67, 1368–1374 (2012).
Liao, C. H., Hsueh, P. R., Jacoby, G. A. & Hooper, D. C. Risk factors and clinical characteristics of patients with qnr-positive Klebsiella pneumoniae bacteraemia. J. Antimicrob. Chemother. 68, 2907–2914 (2013).
Acknowledgements
The authors would like to acknowledge Humberto López Castillo, MD, PhD, CPH, CMI, for his review of the manuscript. I.L. and V.N.-S. are members of the Sistema Nacional de Investigación (SNI), which is supported by Panama’s Secretaría Nacional de Ciencia, Tecnología e Innovación (SENACYT).
Author information
Authors and Affiliations
Contributions
Conceptualization, V.N.-S.; G.P.-P. and I.L.; methodology, V.N.-S.; G.P.-P.; A.D.L.C. and I.L.; software, V.N.-S. and I.L.; validation, V.N.-S.; G.P.-P.; A.D.L.C. and I.L.; formal analysis, V.N.-S.; G.P.-P.; A.D.L.C. and I.L.; investigation, V.N.-S.; G.P.-P.; A.D.L.C. and I.L.; resources, V.N.-S. and I.L.; data curation, V.N.-S.; G.P.-P. and I.L.; writing—original draft preparation, V.N.-S.; G.P.-P. and I.L.; writing—review and editing, V.N.-S.; G.P.-P. and I.L.; visualization, V.N.-S.; G.P.-P. and I.L.; supervision, V.N.-S. and IL; project administration, V.N.-S. and I.L.; funding acquisition, V.N.-S. and I.L. All authors have read and agreed to the published version of the manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Núñez-Samudio, V., Pimentel-Peralta, G., De La Cruz, A. et al. Multidrug-resistant phenotypes of genetically diverse Escherichia coli isolates from healthy domestic cats. Sci Rep 14, 11260 (2024). https://doi.org/10.1038/s41598-024-62037-8
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-024-62037-8
Keywords
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.