Abstract
Glioblastoma (GB) is a devastating tumor of the central nervous system characterized by a poor prognosis. One of the best-established predictive biomarker in IDH-wildtype GB is O6-methylguanine-DNA methyltransferase (MGMT) methylation (mMGMT), which is associated with improved treatment response and survival. However, current efforts to monitor GB patients through mMGMT detection have proven unsuccessful. Small extracellular vesicles (sEVs) hold potential as a key element that could revolutionize clinical practice by offering new possibilities for liquid biopsy. This study aimed to determine the utility of sEV-based liquid biopsy as a predictive biomarker and disease monitoring tool in patients with IDH-wildtype GB. Our findings show consistent results with tissue-based analysis, achieving a remarkable sensitivity of 85.7% for detecting mMGMT in liquid biopsy, the highest reported to date. Moreover, we suggested that liquid biopsy assessment of sEV-DNA could be a powerful tool for monitoring disease progression in IDH-wildtype GB patients. This study highlights the critical significance of overcoming molecular underdetection, which can lead to missed treatment opportunities and misdiagnoses, possibly resulting in ineffective therapies. The outcomes of our research significantly contribute to the field of sEV-DNA-based liquid biopsy, providing valuable insights into tumor tissue heterogeneity and establishing it as a promising tool for detecting GB biomarkers. These results have substantial implications for advancing predictive and therapeutic approaches in the context of GB and warrant further exploration and validation in clinical settings.
Similar content being viewed by others
Introduction
Treatment of central nervous system (CNS) tumors is a significant unmet medical need. According to the WHO 2016 Classification of gliomas, IDH wild-type glioblastoma (GB) is the most common CNS tumor and has an inherently poor prognosis1. The WHO CNS tumor classification of 2016 was updated in 20212. Conventional treatment of GB consists of maximum surgical tumor resection, followed by radiotherapy with concomitant temozolomide (TMZ) and adjuvant TMZ according to the STUPP protocol3. GB is the most aggressive glioma, and despite advances in surgical techniques and antitumoural treatments, patients have a median overall survival (OS) of 15 months4.
Currently, one of the best-established predictive biomarker in IDH-wildtype GB is O6-methylguanine-DNA methyltransferase (MGMT) gene promoter methylation, which is associated with TMZ response and better survival4,5,6. The 2-year survival rate is 12% in patients harboring unmethylated MGMT tumors compared to 49% in methylated MGMT (mMGMT) tumors4. The cytotoxic actions of TMZ have been associated with its ability to form DNA adducts that cause tumor cell death. The repair process in tumor cells is carried out by the MGMT protein, so an unmethylated MGMT gene promoter, leading to high protein expression, is associated with resistance to TMZ treatment. However, the mMGMT gene promoter is associated with the silencing of MGMT expression and thus the inability of the cell to maintain the repair process5,7.
The tumor MGMT methylation status is routinely obtained from surgical resection or biopsy in clinical practice. However, due to tumor location or tumor necrosis, it is sometimes difficult to obtain enough tissue for biomarker testing8. Tumors are known to release markers into the peripheral blood in various ways; thus, liquid biopsy is emerging as an alternative tool that could overcome the boundaries associated with tumor sampling9,10. Circulating tumor DNA (ctDNA), small extracellular vesicles (sEVs) and circulating tumor cells are being used to detect and monitor cancer biomarkers10,11 but are heavily influenced by the type, stage, and location of the tumor12. However, detecting mMGMT in blood remains a challenge for patients with glioma because the amount of ctDNA is reduced, making it necessary to identify new approaches that increase the sensitivity of liquid biopsy in these patients12,13,14,15. A sEV-based liquid biopsy approach may provide new insights in this regard, as IDH mutations have been detected in the sEV DNA of patients with GB16. Therefore, this study aimed to determine the utility of sEV-based liquid biopsy to analyze mMGMT status as a predictive biomarker and disease monitoring tool in patients with glioblastoma.
Methods
Study design and patients
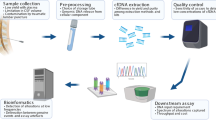
We conducted a prospective exploratory study at Hospital La Paz (Madrid, Spain) between January 2017 and March 2021. 50 patients were included in the study. Eligibility criteria included patients with histopathological confirmation of IDH wild-type GB according to WHO 2016 Classification, age ≥ 18 years, signed informed consent and an Eastern Cooperative Oncology Group (ECOG) performance status of 0 to 317 and first-line STUPP treatment candidates. Once patients had progressed on the STUPP protocol, further treatment decisions were left to the choice of the prescribing physician. In addition, we included eight healthy donors in the study from volunteers at our center. Eligibility criteria for healthy donors were absence of oncological disease at the time of sample collection, ECOG 0, age ≥ 18 years and signed informed consent. The exclusion criteria for both groups included not meeting the inclusion criteria or failing to sign the informed consent form. The primary endpoint was to assess the sensitivity and specificity of sEV-DNA mMGMT with respect to tumor methylation results. The association between sEV-DNA mMGMT and OS was also evaluated, as well as its usefulness in patient follow-up according to MRI images. The MRI response was interpreted based on the modified criteria for radiographic response assessment recommendations proposed by Ellingson et al.18. Surgical complete resection was defined based on absence of tumor contrast enhancement on early postoperative MRI (< 72 h from surgery). This study was approved by the institutional Ethics Committee (PI-2887) according to the Helsinki Declaration.
Specimen characteristics
Blood samples were obtained in EDTAK2 BD Vacutainer tubes (Becton Dickinson, USA) after surgery and before starting the concomitant treatment with STUPP protocol. Namely, between 3 and 9 weeks after surgery, depending on each patient's recovery time. Plasma was obtained by a first centrifugation at 2500g for 10 min at 4 °C. The supernatant was transferred to a new tube followed by a second step of centrifugation at 35,000g for 20 min at 4 °C. Plasma samples were stored in 1.5 ml aliquots at − 80 °C until further processing.
Assay methods
sEV isolation
Ultracentrifugation (UC) is the most common method used for sEV isolation19. Between 1–1.5 ml of plasma was thawed and filtered through a 0.2 µm polyether sulfone membrane (Fisher Scientific, Massachusetts, USA) to eliminate larger particles. Samples were diluted in filtrated phosphate buffered saline (PBS) up to a volume of 7 ml to reduce viscosity and increase the yield of sEV isolation20. Then, the PBS-diluted plasma was filled into 13.5 ml thick-walled polycarbonate reusable tubes specific for UC processing (Beckman Coulter, USA). Samples were ultracentrifuged at 100,000×g for 70 min at 4 °C using a fixed-angle rotor (Rotor 70.1 Ti, K factor of 36) in an Optima L-100 XP ultracentrifuge (Beckman Coulter, USA). The pellet was washed with 7 ml of filtrated PBS and ultracentrifuged again under the same conditions. The pellet containing the sEVs was suspended in 150 μl of filtered PBS.
sEV characterization
Two independent methods, nanoparticle tracking analysis (NTA) and transmission electron microscopy (TEM), were used on a subset of 18 samples to characterize sEVs as recommended19.
Nanosight LM10 (Malvern, UK) employs NTA technology that enables precise analysis of the size distribution and concentration of all types of suspended nanoparticles. From 10 μl of the sEV suspension obtained after UC, a 1:100 dilution was carried out with filtered PBS to achieve the concentration range recommended by the manufacturer (108–109 particles/ml). This dilution was injected into the device, and two 60-s videos for each sample were generated and analyzed by NTA 3.0 software, providing particle size and distribution data.
The sEV morphology was confirmed in a few samples by TEM (n = 4). Fifty microliters of sEV suspension obtained after UC was fixed with 4% paraformaldehyde (PFA) (pH 7.2). Five microliters of this suspension was placed on parafilm and a formvar/carbon grid. The grid was washed out with PBS and then fixed with 1% glutaraldehyde. Once the sEVs were fixed on the grid, they were stained with uranil-oxalate, embedded in methylcellulose-uranil and visualized with TEM (JEOL JEM 1010, Japan) at 100 kV and 100,000×. Digital Micrograph software was used for image visualization and processing.
Nucleic acid isolation and MGMT methylation analysis
DNA from formalin-fixed and paraffin-embedded (FFPE) tissue samples was processed in routine clinical practice. Samples were fixed in 10% formalin and subsequently embedded in paraffin within the pathology department. Hematoxylin and eosin staining was performed on each sample to confirm the presence of tumor nuclei exceeding 20%. The FFPE samples were stored at room temperature until DNA isolation. FFPE tissue samples were deparaffinised using xylene. DNA extraction from tissue samples followed established protocols using phenol–chloroform and chloroform, culminating in suspension in 20 μl of Tris–EDTA buffer solution (TE 1×), as previously described7. sEV-DNA was isolated by organic extraction (phenol:chloroform) according to standard protocols and finally resuspended in TE 1×21.
For mMGMT analysis, sEV-DNA was modified using sodium bisulfite according to standard protocols22. Next, nested PCR was performed. The reaction mixture contained 6 μl of bisulfite-modified DNA, 3 μl of buffer (Biotools Buffer 10×), 4 μl of dNTPs (10 mM), 1 μl of MgCl2 (50 nM), 2 μl of primers (100 ng/µL) and 0.75 μl of enzyme (Biotools DNA polymerase 1 U/µl). The nested PCR conditions were 94 °C for 5 min, followed by 40 cycles at 94 °C for 1 min, 54 °C for 1 min, 72 °C for 1 min and ending with 1 cycle at 72 °C for 8 min. The product of the reaction was a 166 bp amplicon. The sequences (5′–3′) used were as follows: GGATATGTTGGGATAGTT (forward primer) and CCTACAAAACCACTCRAAACT (reverse primer). Six microliters of the product resulting from this PCR was used as a template for quantitative methylation-specific PCR (qMSP). The analysis of qMSP in tumors was performed according to the previously published formula7, while the analysis of qMSP in sEV-DNA was performed according to the formula log(−∆Ct). Supplementary Fig. 2 shows a summary of all primers used.
Statistical analysis
mMGMT was recorded as a continuous variable, but in oncology, it is common to convert it into a categorical form by using cutoff points23. The primary endpoint was to assess the sensitivity and specificity of sEV-DNA mMGMT in relation to tumor methylation results. To categorize the mMGMT results obtained in tumor tissue, we used the cutoff point established and validated in our previous study7. For sEV-DNA samples, the quantification of methylated tumor DNA is challenging in blood samples due to its low abundance, typically estimated to be less than 1%. To overcome this limitation, we employed the log(−∆Ct) formula, where any positive ∆Ct value was considered a methylated result to minimize false negatives reported in liquid biopsy24. This allows us to detect methylated DNA when the FAM probe initiates amplification, even if it occurs significantly later than the amplification of the VIC probe (representing unmethylated DNA) as expected. This approach enables the accurate quantification of tumor DNA methylation levels in blood samples with higher sensitivity and precision.
Concordance between dichotomous variables was studied using sensitivity, specificity, positive predictive value (PPV), negative predictive value (NPV) and the percentage of observed agreements (concordance percentage) with their respective 95% confidence intervals (95% CI). The kappa index has also been analyzed with its respective significance, and it is considered a more robust method because it takes into account coincidences that occur by chance25. Kappa index result was interpreted as follows: values ≤ 0 as indicating no agreement and 0.01–0.20 as none to slight, 0.21–0.40 as fair, 0.41–0.60 as moderate, 0.61–0.80 as substantial, and 0.81–1.00 as almost perfect agreement26. The association between mMGMT and clinicopathological status (qualitative variables) was analyzed by the chi-squared test (surgical resection extension, ECOG) or Fisher's exact test (sex). For the comparison between qualitative mMGMT (qualitative) and age (quantitative data), Student's t test was used for independent data. The normality of these variables was assessed using the Kolmogorov–Smirnov test. The OS analysis was performed using Kaplan‒Meier tests. A p value less than 0.05 was considered statistically significant. Given that the extension of resection has been previously reported to have an impact on OS27,28,29,30, a subgroup analysis of OS was performed among those patients who underwent incomplete resection. The statistical analysis was carried out using R (version 4.0.2) and survival, KMsurv and survminer packages.
Ethics approval and consent to participate
This study was conducted under the approval of the ethics committee of the La Paz University Hospital with the ethics number PI-2887. The study was performed according to the Helsinki Declaration. Informed consent was signed by all patients.
Results
Patient characteristics
From January 2017 to March 2021, 50 patients were included in the study, with a median follow-up time of 13 months. Three patients had an insufficient tumor cell percentage in the sample (< 20%), and one patient was diagnosed at another hospital, so the tumor was not available to perform MGMT studies. The blood sample collection flow chart and the clinical and demographic characteristics of the patients are shown in Fig. 1 and Table 1, respectively.
Extracellular vesicle characterization
NTA analysis was performed on those samples where there was sufficient plasma to perform both determinations, MGMT methylation and NTA assays. We also confirmed the presence of sEVs by EM. The use of both methods confirmed that most of the EVs observed were less than 200 nm in size and had the characteristic cup shape of sEVs (Fig. 2).
Extracellular vesicle characterization. (A) Visualisation of circulating sEVs obtained from the plasma of GB patients using NTA. Examples of extracellular vesicle size distribution profiles in four out the fourteen samples obtained by NTA. NTA confirms the presence of extracellular vesicles below 150 nm. ID36 and ID37 are used to identify patients according to Table 2. (B) Visualisation of circulating sEVs obtained from the plasma of GB patients using transmission electron microscopy (EM). Images were taken at a magnification of 120,000. EM images showed small vesicles of approximately 120–130 nm in diameter, clustered in some cases. The arrow indicates a larger vesicle (> 200 nm).
Evaluation of mMGMT in liquid biopsy using sEV-DNA is consistent with the biomarker analysis in tumor tissue DNA
As expected, mMGMT methylation was not detected in the sEV DNA of any of the eight healthy donors (Supplementary Fig. 1).
The tissue sample was available for 46 out of the 50 patients for whom blood was available. MGMT promoter methylation was detected in 19 (41.3%) of these patients using DNA of tumor tissue origin and in 14 (28%) patients using sEV-DNA. There were seven cases (14%) in which the presence of methylation was detected in the tumor DNA but not in the matching sEV DNA samples. In contrast, there were two patients in whom mMGMT was detected in sEV-DNA but not in tumor tissue-DNA (Table 2).
Of the first seven cases, six corresponded to patients who had undergone macroscopically complete resection as described intraoperatively by the neurosurgeon. Complete resection was confirmed in two of these patients (ID22 and ID44) by early postoperative magnetic resonance imaging (MRI). Although the targeted surgical lesion was completely resected for patient ID21, this is a case of multifocal GB, where a known residual lesion was confirmed by postoperative MRI. The extent of resection could not be confirmed by early postoperative MRI for patient ID45, given that images were not assessable due to the patient’s movements when the images were taken. Last, although intraoperative complete resection was described for patient ID46, tumor remnants were observed in the early postoperative MRI. Postsurgical MRI was not available for the final patient.
The concordance percentage obtained between sEV-DNA and tumor tissue-DNA was 80.4% (95% CI: 66.1–90.6). The sensitivity to detect methylation in sEV-DNA according to the methylation observed in tumor tissue-DNA was 63.2% (95% CI: 38.4–83.7), while the specificity was 92.6% (95% CI: 74.7–99.1). The PPV and NPV were 85.7% (95% CI: 60.2–95.9) and 78.1% (95% CI: 62.2–86.7), respectively (Table 2). The kappa index showed moderate concordance (k = 0.580) between tumor tissue-DNA and sEV-DNA (p < 0.001). If we only considered the subgroup of patients who underwent incomplete resection or biopsy, we achieved a concordance of 90.0% (95% CI: 68.3–98.8), a sensitivity to detect methylation in sEV-DNA of 87.5% (95% CI: 47.3–99.7) and a specificity of 91.7% (95% CI: 61.5–99.8), with a PPV of 87.5% (95% CI: 51.3–97.9) and an NPV of 91.7% (95% CI: 63.6–98.6). The kappa index was 0.792 (substantial concordance) between tumor tissue-DNA in the subgroup of patients who underwent incomplete resection or biopsy and sEV-DNA (p < 0.001).
Determination of mMGMT status in sEV-DNA as a survival predictor biomarker
In our patient cohort, mMGMT tumor status discriminated patients with respect to OS without reaching statistical significance (p = 0.06). After a median follow-up of 24 months, the median OS in patients with MGMT-unmethylated tumors was 18 months (95% CI: 9.1–26.9), while the median OS was not reached in patients with MGMT-methylated tumors after 24 months of follow-up (Fig. 3A). When overall survival was analyzed with respect to the MGMT methylation status in the DNA of the sEVs, regardless of the extent of resection, no significant differences were observed (p = 0.735) (Fig. 3B).
Overall Survival curves in our patient cohort. (A) Kaplan–Meier estimates of overall survival according to MGMT methylation in tumor sample. (B) Kaplan–Meier estimates of overall survival according to MGMT methylation in sEVs-DNA. (C) Kaplan–Meier estimates of overall survival according to MGMT methylation in sEVs-DNA in the subgroup of patients without complete resection. n.a. not available.
This result led us to evaluate the median OS in the group of patients who did not undergo complete resection, in which a poorer OS was achieved in patients with MGMT-unmethylated versus MGMT-methylated sEVs-DNA (10.9 months; 95% CI: 5.3–16.5 versus not reached (p value not assessable due to lack of events) (Fig. 3C).
mMGMT in sEV-DNA as a patient monitoring tool
To study the implications of measuring mMGMT status in blood along the course of the disease, we collected 20 baseline and follow-up samples from eight patients (Fig. 4A).
Serial monitoring sEVs-DNA methylation status. (A) sEVs-DNA methylation status of all samples from the 8 monitored patients. (B) Changes in MGMT methylation ∆Ct levels detected in sEVs-DNA-based liquid biopsy in patients ID1. (C) Changes in MGMT methylation ∆Ct levels detected in sEVs-DNA-based liquid biopsy in patients ID2.
Patients ID1, ID2, ID28, ID32 and ID34 had an MGMT methylated tumor. Patient ID1 was diagnosed with an unresectable mMGMT tumor. sEV-DNA at baseline showed mMGMT, which decreased over time during treatment and was undetectable in the final follow-up samples. The mMGMT profile in sEV-DNA was observed with a MRI image compatible with a partial response in the first follow up and complete response after treatment. Consequently, the mMGMT levels of patient ID1 showed consistency with the imaging data, suggesting that methylation levels may underpin the results obtained through MRI (Fig. 4B). Patient ID28 had a similar course to patient ID1. However, in this case, it was not possible to obtain further follow-up samples to confirm a favorable evolution, with a PFS of 32 months.
Patients ID2 and ID32 also carried an MGMT methylated tumor sample. mMGMT was also detected in sEV-DNA in all blood samples obtained during the follow-up. Both patients progressed without significantly decreasing methylation levels in their blood. Specifically, patient ID2 was diagnosed with MGMT methylated GB and underwent tumor resection. Postoperative MRI revealed remaining tumor tissue and baseline sEV-DNA showed mMGMT. Although there was no evidence of disease progression on the first follow-up MRI scan, the monitoring sample revealed that mMGMT ∆Ct was increased. After 3 months, MRI showed disease progression, suggesting that mMGMT ∆Ct elevation could predict tumor recurrence. The patient started a new treatment which slightly decreased the ∆Ct values, but eventually the patient progressed and died (Fig. 4C). Patient ID32 had a similar situation to the previous patient. Therefore, the dynamics of mMGMT in sEV-DNA were closely associated with the MRI results and patient prognoses, providing a valuable tool for the follow-up of patients with mMGMT.
Patient ID34 underwent complete resection of a methylated MGMT tumor. The methylation remained at the baseline sEV-DNA sample, but was no longer observed in the remaining follow-up. MRI performed one month before sample collection showed no progression; however, the patient progressed and died two months after the sample was obtained; therefore, it was not possible to obtain a second follow-up at that stage to corroborate the mMGMT status.
Patients ID12 and ID41 had an absence of mMGMT in the tumor and, as expected, in all sEV-DNA samples obtained. In the case of patient ID40, who underwent complete resection of an unmethylated MGMT GB tumor, sEV-DNA methylation in the baseline sample was detected. In the first follow-up sample, the patient no longer showed mMGMT and remained stable until 9 months.
Discussion
Cancer management involves challenges associated with diagnosis, prognosis, and prediction of treatment response. mMGMT is a well-known predictive biomarker in patients with GB4,7,31, and its assessment is an important factor in choosing the best treatment strategy or selecting patients for clinical trials. Its reliable assessment in liquid biopsy would therefore be a robust tool of great value in the clinical follow-up of GB patients24.
Liquid biopsy results are highly dependent on tumor type, stage and location and can achieve sensitivities of over 90% in metastatic patients32. However, in contrast to other solid tumors, the procedure of conducting liquid biopsies relying on ctDNA in glioma patients presents notable challenges due to the persistently low levels of detectable biomarkers in their bloodstream. Achieving the highest sensitivities typically involves assays utilizing cerebrospinal fluid (CSF); however, these methods necessitate invasive sampling33,34. Initially, these challenges were attributed to the presence of the blood–brain barrier (BBB). Nonetheless, recent studies have shed light on disruptions in the BBB in glioma patients, complicating our understanding of this limitation35. On the other hand, García-Romero et al. have provided evidence that sEVs can effectively cross the intact BBB and be detected in peripheral blood. This discovery overcomes the previous limitation and introduces a new way to perform liquid biopsy in glioma patients, potentially improving diagnostic accuracy and clinical management strategies16.
The field of EVs has not been without its challenges, requiring the development and refinement of methodologies for EV isolation and characterization. This diversity of methodologies poses significant hurdles to standardization efforts36. As emphasized by the International Society of Extracellular Vesicles, it is imperative to use at least two different and complementary techniques for sample characterization, such as electron microscopy, nanoparticle tracking analysis or protein marker detection. In our study, we chose the first two methods19. The comprehensive characterization obtained using these techniques confirms the successful isolation of EVs. While we acknowledge the potential presence of platelets or lipoproteins, we maintain that this inherent limitation does not compromise the integrity of our results. Our focus on mMGMT, which is exclusively associated with oncological processes, remains unaffected by these considerations.
To date, studies to determine mMGMT status in blood-based liquid biopsy using ctDNA in patients with gliomas obtained a sensitivity ranging from 11 to 76.6%10,13,14,15,37,38,39,40,41, achieving the best results when the blood sample was obtained prior to surgical resection. For example, the methylation status of MGMT and other genes was investigated in a cohort of 28 glioblastoma patients treated with 1,3-bis(2-chloroethyl)-1-nitrosourea or temozolomide plus cisplatin, and compared to tissue, the sensitivity for detecting MGMT methylation in serum was 62.5%, with a specificity of 92.3%41. Other study showed that the methylation status of the MGMT promoter in serum had a sensitivity of 66.7% and a specificity of 100.0%40. Similarly, Lavon et al. reported a sensitivity of 59% and a specificity of 100% for MGMT methylation38. Notably, the study by Gong et al. showed the best performance with a sensitivity of 76.61% and a specificity of 98.28%15. Utilizing an alternative approach focused on analyzing RNA content within EVs, Mut, Melike et al. developed a LASSO-penalized binomial regression model. This model, employing 17 out of 569 differentially expressed genes as predictors in EVs, achieved a predictive accuracy of 91% sensitivity and 73% specificity in determining the MGMT methylation status42. Additionally, it is noteworthy to mention that no studies evaluating the methylation of MGMT using DNA in EVs have been identified to date. Therefore, our study provides an unprecedented increase in the percentage of marker agreement between tissue and plasma, reaching 90% concordance with a sensitivity of 87.5% and a specificity of 92% in subtotal resection cases. Furthermore, given the primary objective of a predictive biomarker is forecast treatment response, our results suggest that MGMT methylation in tumor approaches statistical significance in predicting survival although our limited sample size. Additionally, patients undergoing subtotal resection may also benefit from the mMGMT biomarker in EVs in terms of survival, albeit without reaching statistical significance. While acknowledging the limitations of the limited sample size, our data suggest that mMGMT in EVs may have potential significance as a predictive biomarker in glioblastoma. Therefore, further investigation is needed to support our observations and facilitate the integration of the mMGMT biomarker into clinical contexts.
We observed that lower concordance in MGMT methylation between tumor and sEV was associated with patients who underwent complete resection. This suggests that detecting MGMT methylation in sEV is more challenging in cases where the tumor has been previously removed. Thus, considering our results in tumors with incomplete resection and the results obtained in monitored patients, our study showed promising results, achieving the highest sensitivity reported to date for sEV. Therefore, we speculate that if preoperative blood samples had been available in our study, the sensitivity achieved in sEV DNA analysis would likely have been further increased. This speculation is supported by the established sensitivity of preoperative samples demonstrated by previous liquid biopsy research15,39. The potential increase in sensitivity underscores the critical role of timing of sample collection and highlights the importance of optimizing sample collection protocols to increase the potential utility of liquid biopsy approaches.
To our knowledge, only one study has previously used sEV-DNA-based liquid biopsy to investigate IDH status in patients with GB16. Alternatively, two studies have compared sEV-DNA-based liquid biopsy with cDNA-based liquid biopsy in paired samples. The study by Allenson et al. detected a higher rate of KRAS mutations in sEV-DNA compared with ctDNA with sensitivities of 66.7%, 80% and 85% of patients with localized, locally advanced and metastatic pancreatic cancer, respectively, whereas the results obtained in ctDNA were 45.5%, 30.8% and 57.9%, respectively43. Bernard et al. achieved similar results, obtaining higher sensitivity using sEV-DNA than ctDNA44. Furthermore, serum analysis has revealed that EGFRvIII can be detected non-invasively in EV, with an overall clinical sensitivity of 81.58% and specificity of 79.31%45. sEV also contain abundant miRNAs and proteins associated with proliferation, angiogenesis, cell migration, immune response and histone modification, which could be used as biomarkers in this disease46,47. Nevertheless, each of these studies employed varying sample types (serum/plasma), DNA isolation methodologies, and techniques for mMGMT determination. Consequently, it is paramount to exercise caution when interpreting and comparing absolute values, recognizing the inherent limitations associated with these methodological disparities.
Reaching 100% sensitivity in liquid biopsy tests is difficult due to the low concentration of tumor markers in the pool of molecules in the blood. However, this same condition makes the lack of specificity an uncommon event. In our study, we found two patients in whom MGMT promoter was methylated in sEV-DNA but was not detected in tumor tissue DNA (ID14 and ID40). Since false positives in liquid biopsy are rare, it leads us to suspect that it might have been a false negative in the tumor tissue determination mainly due to tumor heterogeneity, one of the main limitations of tissue sampling, given that it is difficult for a single sample to represent the entire lesion8,32,48.
The role that liquid biopsy can play in the monitoring of patients with GB is not well established to date, given that the results published on this disease have been very limited. In a subgroup of 12 patients, Bagley et al. reported that ctDNA levels obtained prior to initial tumor resection in adult patients with newly diagnosed GB increased with progression and remained stable in patients who did not progress49, data that were corroborated by Fontanilles et al.50. Muralidharan et al. monitored TERT mutations in ctDNA in five patients, concluding that their frequency reflects the clinical course of the disease with levels that decrease after surgery and increase with tumor progression51. In our study, we recruited twenty samples from eight patients with baseline and at least one follow-up sample. Five of these patients had mMGMT GB. We found that patients with mMGMT GB harboring detectable mMGMT at baseline and during follow-up in blood samples were patients who showed tumor progression. However, patients with mMGMT in sEV-DNA in the baseline sample that decreased in the follow-up samples were patients with radiographic evidence of tumor response. Therefore, changes in ∆Ct levels in sEV-DNA provide relevant predictive information to manage and monitor the outcome of patients with GB.
Methylation in sEV DNA in cancer patients has only been detected in the study by Zavridou et al., who detected methylation in the GSTP1 and RASSF1A genes52. To our knowledge, our study shows for the first time that detection of mMGMT in sEV-DNA-based liquid biopsy in patients with GB is feasible and greatly improves on the previously published sensitivity calculated using circulating blood DNA. These results are supported by a recent study conducted on GB cell culture-derived sEVs in a genome-wide methylation profiling approach. In this study, Maire et al. were able to correctly categorize tumors according to the Heidelberg classification, including the MGMT promoter methylation status53. Thus, the in vitro study conducted by Maire et al. together with our in vivo data provide the basis for the diagnostic characterization of genome-wide methylation in sEV-DNA, which would allow the molecular classification and disease monitoring of CNS tumors in patients in whom tumor samples are not available.
Conclusions
We report for the first time the detection of mMGMT in sEV-DNA with a sensitivity and specificity of 87.5% and 90%, respectively. The presence of MGMT in sEV-DNA appeared to show an association with patient OS and facilitated patient monitoring; however, statistical power limitations prevent a definitive determination. Therefore, the results obtained here represent an important contribution to the field of extracellular vesicle-based liquid biopsy, suggesting that it reflects the heterogeneity of tumor tissue and represents a promising tool for biomarker detection. However, further larger studies are required to confirm our findings.
Data availability
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.
Abbreviations
- CNS:
-
Central nervous system
- WHO:
-
World Health Organization
- IDH:
-
Isocitrate dehydrogenase
- GB:
-
Glioblastoma
- TMZ:
-
Temozolomide
- OS:
-
Overall survival
- MGMT:
-
O6-methylguanine-DNA methyltransferase
- mMGMT:
-
Methylated MGMT
- ctDNA:
-
Circulating tumor DNA
- sEVs:
-
Extracellular vesicles
- BBB:
-
Blood–brain barrier
- sEV-DNA:
-
DNA extracted from sEVs
- ECOG:
-
Eastern Cooperative Oncology Group
- UC:
-
Ultracentrifugation
- PBS:
-
Phosphate buffered saline
- NTA:
-
Nanoparticle tracking analysis
- TEM:
-
Transmission electron microscopy
- TE 1×:
-
Tris-EDTA buffer solution
- PPV:
-
Positive predictive value
- NPV:
-
Negative predictive value
- 95% CI:
-
95% Confidence intervals
- DP:
-
Disease progression
- MRI:
-
Magnetic resonance imaging
References
Louis, D. N. et al. The 2016 World Health Organization Classification of Tumors of the Central Nervous System: A summary. Acta Neuropathol. 131(6), 803–820 (2016).
Louis, D. N. et al. The 2021 WHO Classification of Tumors of the Central Nervous System: A summary. Neuro Oncol. 23(8), 1231–1251 (2021).
Stupp, R. et al. Radiotherapy plus concomitant and adjuvant temozolomide for glioblastoma. N. Engl. J. Med. 352(10), 987–996 (2005).
Stupp, R., Brada, M., van den Bent, M. J., Tonn, J. C. & Pentheroudakis, G. High-grade glioma: ESMO Clinical Practice Guidelines for diagnosis, treatment and follow-up. Ann. Oncol. 25(Suppl 3), iii93–iii101 (2014).
Esteller, M. et al. Inactivation of the DNA-repair gene MGMT and the clinical response of gliomas to alkylating agents. N. Engl. J. Med. 343(19), 1350–1354 (2000).
Wick, W. et al. Prognostic or predictive value of MGMT promoter methylation in gliomas depends on IDH1 mutation. Neurology 81(17), 1515–1522 (2013).
Rosas-Alonso, R. et al. Clinical validation of a novel quantitative assay for the detection of MGMT methylation in glioblastoma patients. Clin. Epigenetics 13(1), 52 (2021).
Parker, N. R. et al. Intratumoral heterogeneity identified at the epigenetic, genetic and transcriptional level in glioblastoma. Sci. Rep. 6, 22477 (2016).
Crowley, E., Di Nicolantonio, F., Loupakis, F. & Bardelli, A. Liquid biopsy: Monitoring cancer-genetics in the blood. Nat. Rev. Clin. Oncol. 10(8), 472–484 (2013).
Soffietti, R. et al. Liquid biopsy in gliomas: A RANO review and proposals for clinical applications. Neuro Oncol. 24(6), 855–871 (2022).
Siravegna, G., Marsoni, S., Siena, S. & Bardelli, A. Integrating liquid biopsies into the management of cancer. Nat. Rev. Clin. Oncol. 14(9), 531–548 (2017).
Bettegowda, C. et al. Detection of circulating tumor DNA in early- and late-stage human malignancies. Sci. Transl. Med. 6(224), 224ra24 (2014).
Estival, A. et al. Pyrosequencing versus methylation-specific PCR for assessment of MGMT methylation in tumor and blood samples of glioblastoma patients. Sci. Rep. 9(1), 11125 (2019).
Wang, Z. et al. MGMT promoter methylation in serum and cerebrospinal fluid as a tumor-specific biomarker of glioma. Biomed. Rep. 3(4), 543–548 (2015).
Gong, M. et al. Alu hypomethylation and MGMT hypermethylation in serum as biomarkers of glioma. Oncotarget 8(44), 76797–76806 (2017).
Garcia-Romero, N. et al. DNA sequences within glioma-derived extracellular vesicles can cross the intact blood-brain barrier and be detected in peripheral blood of patients. Oncotarget 8(1), 1416–1428 (2017).
Azam, F. et al. Performance status assessment by using ECOG (Eastern Cooperative Oncology Group) score for cancer patients by oncology healthcare professionals. Case Rep. Oncol. 12(3), 728–736 (2019).
Ellingson, B. M., Wen, P. Y. & Cloughesy, T. F. Modified criteria for radiographic response assessment in glioblastoma clinical trials. Neurotherapeutics 14(2), 307–320 (2017).
Thery, C. et al. Minimal information for studies of extracellular vesicles 2018 (MISEV2018): A position statement of the International Society for Extracellular Vesicles and update of the MISEV2014 guidelines. J. Extracell. Vesicles 7(1), 1535750 (2018).
Momen-Heravi, F. et al. Impact of biofluid viscosity on size and sedimentation efficiency of the isolated microvesicles. Front. Physiol. 3, 162 (2012).
Ibanez de Caceres, I. et al. Tumor cell-specific BRCA1 and RASSF1A hypermethylation in serum, plasma, and peritoneal fluid from ovarian cancer patients. Cancer Res. 64(18), 6476–6481 (2004).
Vera-Puente, O. et al. MAFG is a potential therapeutic target to restore chemosensitivity in cisplatin-resistant cancer cells by increasing reactive oxygen species. Transl. Res. 200, 1–17 (2018).
Altman, D. G., McShane, L. M., Sauerbrei, W. & Taube, S. E. Reporting Recommendations for Tumor Marker Prognostic Studies (REMARK): Explanation and elaboration. PLoS Med. 9(5), e1001216 (2012).
Pascual, J. et al. ESMO recommendations on the use of circulating tumour DNA assays for patients with cancer: A report from the ESMO Precision Medicine Working Group. Ann. Oncol. 33(8), 750–768 (2022).
Landis, J. R. & Koch, G. G. The measurement of observer agreement for categorical data. Biometrics 33(1), 159–174 (1977).
McHugh, M. L. Interrater reliability: The kappa statistic. Biochem. Med. (Zagreb) 22(3), 276–282 (2012).
Revilla-Pacheco, F. et al. Extent of resection and survival in patients with glioblastoma multiforme: Systematic review and meta-analysis. Medicine (Baltimore) 100(25), e26432 (2021).
Molinaro, A. M. et al. Association of maximal extent of resection of contrast-enhanced and non-contrast-enhanced tumor with survival within molecular subgroups of patients with newly diagnosed glioblastoma. JAMA Oncol. 6(4), 495–503 (2020).
Brown, T. J. et al. Association of the extent of resection with survival in glioblastoma: A systematic review and meta-analysis. JAMA Oncol. 2(11), 1460–1469 (2016).
Jusue-Torres, I., Lee, J., Germanwala, A. V., Burns, T. C. & Parney, I. F. Effect of extent of resection on survival of patients with glioblastoma, IDH-wild-type, WHO Grade 4 (WHO 2021): Systematic review and meta-analysis. World Neurosurg. 171, e524–e532 (2023).
Caccese, M. et al. Definition of the prognostic role of MGMT promoter methylation value by pyrosequencing in newly diagnosed IDH wild-type glioblastoma patients treated with radiochemotherapy: A large multicenter study. Cancers (Basel) 14(10), 2425 (2022).
Leighl, N. B. et al. Clinical utility of comprehensive cell-free DNA analysis to identify genomic biomarkers in patients with newly diagnosed metastatic non-small cell lung cancer. Clin. Cancer Res. 25(15), 4691–4700 (2019).
Simonelli, M. et al. Cerebrospinal fluid tumor DNA for liquid biopsy in glioma patients’ management: Close to the clinic?. Crit. Rev. Oncol. Hematol. 146, 102879 (2020).
Mattox, A. K., Yan, H. & Bettegowda, C. The potential of cerebrospinal fluid-based liquid biopsy approaches in CNS tumors. Neuro Oncol. 21(12), 1509–1518 (2019).
Segura-Collar, B., Mata-Martinez, P., Hernandez-Lain, A., Sanchez-Gomez, P. & Gargini, R. Blood–brain barrier disruption: A common driver of central nervous system diseases. Neuroscientist 28(3), 222–237 (2022).
Lucien, F. et al. MIBlood-EV: Minimal information to enhance the quality and reproducibility of blood extracellular vesicle research. J. Extracell. Vesicles 12(12), e12385 (2023).
Majchrzak-Celinska, A. et al. Detection of MGMT, RASSF1A, p15INK4B, and p14ARF promoter methylation in circulating tumor-derived DNA of central nervous system cancer patients. J. Appl. Genet. 54(3), 335–344 (2013).
Lavon, I., Refael, M., Zelikovitch, B., Shalom, E. & Siegal, T. Serum DNA can define tumor-specific genetic and epigenetic markers in gliomas of various grades. Neuro Oncol. 12(2), 173–180 (2010).
Liu, B. L. et al. Quantitative detection of multiple gene promoter hypermethylation in tumor tissue, serum, and cerebrospinal fluid predicts prognosis of malignant gliomas. Neuro Oncol. 12(6), 540–548 (2010).
Weaver, K. D., Grossman, S. A. & Herman, J. G. Methylated tumor-specific DNA as a plasma biomarker in patients with glioma. Cancer Investig. 24(1), 35–40 (2006).
Balana, C. et al. O6-methyl-guanine-DNA methyltransferase methylation in serum and tumor DNA predicts response to 1,3-bis(2-chloroethyl)-1-nitrosourea but not to temozolamide plus cisplatin in glioblastoma multiforme. Clin. Cancer Res. 9(4), 1461–1468 (2003).
Mut, M. et al. Extracellular-vesicle-based cancer panels diagnose glioblastomas with high sensitivity and specificity. Cancers (Basel) 15(15), 3782 (2023).
Allenson, K. et al. High prevalence of mutant KRAS in circulating exosome-derived DNA from early-stage pancreatic cancer patients. Ann. Oncol. 28(4), 741–747 (2017).
Bernard, V. et al. Circulating nucleic acids are associated with outcomes of patients with pancreatic cancer. Gastroenterology. 156(1), 108-118 e4 (2019).
Manda, S. V. et al. Exosomes as a biomarker platform for detecting epidermal growth factor receptor-positive high-grade gliomas. J. Neurosurg. 128(4), 1091–1101 (2018).
Skouras, P., Gargalionis, A. N. & Piperi, C. Exosomes as novel diagnostic biomarkers and therapeutic tools in gliomas. Int. J. Mol. Sci. 24(12), 10162 (2023).
Greco, F. et al. Longitudinal bottom-up proteomics of serum, serum extracellular vesicles, and cerebrospinal fluid reveals candidate biomarkers for early detection of glioblastoma in a murine model. Molecules 26(19), 5992 (2021).
Gerlinger, M. et al. Intratumor heterogeneity and branched evolution revealed by multiregion sequencing. N. Engl. J. Med. 366(10), 883–892 (2012).
Bagley, S. J. et al. Clinical utility of plasma cell-free DNA in adult patients with newly diagnosed glioblastoma: A pilot prospective study. Clin. Cancer Res. 26(2), 397–407 (2020).
Fontanilles, M. et al. Cell-free DNA and circulating TERT promoter mutation for disease monitoring in newly-diagnosed glioblastoma. Acta Neuropathol. Commun. 8(1), 179 (2020).
Muralidharan, K. et al. TERT promoter mutation analysis for blood-based diagnosis and monitoring of gliomas. Clin. Cancer Res. 27(1), 169–178 (2021).
Zavridou, M. et al. Prognostic significance of gene expression and DNA methylation markers in circulating tumor cells and paired plasma derived exosomes in metastatic castration resistant prostate cancer. Cancers (Basel) 13(4), 780 (2021).
Maire, C. L. et al. Genome-wide methylation profiling of glioblastoma cell-derived extracellular vesicle DNA allows tumor classification. Neuro Oncol. 23(7), 1087–1099 (2021).
Acknowledgements
The authors thank Morote translation services for their assistance with language editing and the HULP-IdiAPZ Biobank.
Disclaimer
The information provided in this study is included in a patent application process (EP19382299.6); therefore, it must be treated, solely and exclusively, based on the purposes of this paper and should not be published if it does not respond to the purpose thereof.
Funding
This study was funded by Instituto de Salud Carlos III (ISCIII) and co-funded by the European Union. Grants numbers: PI18/000050; PI21/000145; DTS20/000029; JR17/000016; JR21/000003; FI19/000061; CIBERONC-CB16/12/0295. MICINN (PLEC2021-08034, PID19-104644RB-I00), GHETTI2021 and Instituto de Salud Carlos III (ISCIII), FORT23/00006, del Programa FORTALECE del Ministerio de Ciencia e Innovación.
Author information
Authors and Affiliations
Contributions
Conceptualization: RRA, ADL, RLL, JFG, GM, JDC, IIC. Methodology: RRA (sample collection and processing), JCF (sample collection and processing), OP (sample collection and processing), MB (sample collection and processing), CRA (data analysis), ILG (data analysis), TR (sample collection and processing), RMV (sample collection and processing), IER (pathologic diagnosis), VM (patient recruitment and clinical data collection), PY (patient recruitment), NCF (sample collection and processing), EDM (sample collection and processing), CVS (patient recruitment and clinical data collection), MLGG (patient recruitment and clinical data collection), JDC (patient recruitment and clinical data collection). Funding acquisition: IIC, JDC. Project administration: RRA, JDC, IIC. Supervision: RRA, ADL, GM, IIC. Writing original draft: RRA, IIC. All authors reviewed the manuscript. Institutional consent form was signed.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Rosas-Alonso, R., Colmenarejo-Fernández, J., Pernía, O. et al. Evaluation of the clinical use of MGMT methylation in extracellular vesicle-based liquid biopsy as a tool for glioblastoma patient management. Sci Rep 14, 11398 (2024). https://doi.org/10.1038/s41598-024-62061-8
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-024-62061-8
Keywords
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.