Abstract
The movement from the subjects during in vivo confocal Raman spectral measurements could change the measurement volume, leading to non-specific signals and inaccurate interpretation of the acquired spectrum. Here we introduce a generally applicable method that includes (1) developing a multimodal system to achieve real-time monitoring of every spectral measurement with reflectance confocal microscopy (RCM) and multiphoton microscopy (MPM) imaging; (2) performing region-of-interest measurement by scanning an area of the tissue during spectral acquisition. The developed method has been validated by measuring different micro-structures of in vivo human skin. Our results demonstrated great consistency between RCM images and confocal Raman spectra. The superior quality of the images and spectra allows us to derive blood flow velocity and blood glucose level. We believe this method is valuable for realizing accurate microscopic spectral measurement and have great potential to be adapted into clinic to achieve non-invasive measurement of important biological parameters.
Similar content being viewed by others
Introduction
Confocal Raman spectroscopy, which is a noninvasive optical technique, could provide micron-level resolution and depth-resolved biochemical information of in vivo biological tissues. This technique has been applied to study a number of in vivo health related phenomenon of human tissues, such as noninvasive assessment of human corneal hydration1, estimation of stratum corneum thickness2 and monitoring drug penetration depth inside the skin3. Confocal Raman spectroscopy has also been proved to have potentials in helping early diagnosis of a variety of cancers4,5,6,7. With the permissible laser power followed by ANSI standard8, the usual acquisition time for a confocal Raman spectrum from biological tissue can be as long as 80s9. During the tens of seconds of acquisition time of each spectrum, the acquired confocal Raman spectrum will very likely contain information from out-of-target area due to movement of the subject. Reflectance confocal microscopy (RCM) imaging, which is capable of optical sectioning, has been considered to guide the spectral measurements. There have been some studies on combining reflectance confocal imaging with confocal Raman spectroscopy, but the imaging channel and the spectral acquisition are not from the same excitation laser source and the wavelengths are different10. Therefore, registration between the confocal image and the spot/area of the Raman spectral measurement could be problematic. Moreover, most of the Raman spectral measurements were performed as single-point measurements, which require turning off the optical scanner to generate a still beam. Even co-registration of the sampling locations of the imaging and spectral channels are performed, with the movement from the subject during in vivo measurement, the confocal Raman spectrum will not necessarily come from the targeted point inside the tissue, but from an integrated area according to the random movement range from the subject11. In addition, RCM imaging is only able to provide morphological information based on refractive index variations, while multiphoton microscopy (MPM), which employs two-photon fluorescence (TPF) and second-harmonic-generation (SHG) images, has been proven to provide complementary information comparing with reflectance confocal imaging and could be better utilized as a diagnostic tool12.
Therefore, our objective is to develop a novel but generally applicable method that is able (1) to achieve co-registered reflectance confocal microscopy imaging and confocal Raman spectral measurements using the same laser; (2) to perform well defined region-of-interest (ROI) confocal Raman spectral measurements without turning off the scanner and under the imaging guidance of reflectance confocal microscopy; and (3) to integrate multiphoton microscopy to enhance the imaging capability. We validated our method by performing ROI confocal Raman spectral measurement under the guidance of reflectance confocal microscopy imaging and multiphoton microscopy on skin in vivo. We hope our result will setup the trend of performing accurate micro-spectroscopic measurements under advanced microscopy imaging guidance for other researchers.
Results
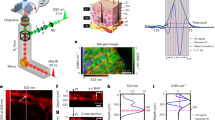
A multimodal spectroscopic and imaging system has been developed as shown in Figure 1. Briefly, a 785 nm continuous-wave (cw) diode laser is used for both confocal Raman spectral measurement and cw reflectance confocal microscopy (cwRCM) imaging. The laser beam is first directed to an optical scanning system consisting of a resonance scanner and a galvanometer scanner and then is focused by a 60X (NA = 1.0) water-immersion microscope objective onto human skin. A polarization beamsplitter (PBS) along with a quarter waveplate is used to direct the descanned reflectance confocal signals to an avalanche photodiode (APD) module with a 30 μm pinhole in front. A dichroic beamsplitter is used to direct the Raman signals to a 50 μm fiber connected with a Raman spectrometer. A femtosecond (fs) Ti:Sappire laser is used for multiphoton microscopic imaging. The fs laser beam is scanned by the same scanner and focused onto the skin by the same microscope objective. The TPF and SHG signals are collected by a photomultiplier tube to generate integrated SHG/TPF images (SHG + TPF imaging mode12) and the descanned fs reflectance confocal microscopy (fsRCM) signals will be directed by another PBS onto an APD with a 50 μm pinhole. A flip mirror is used to switch between the Raman excitation cw laser and the MPM excitation fs laser and the two beam paths have been optimized to be the same. To reduce involuntary body movement, double sided tape and a metal ring were used to magnetically mate the skin with the imaging objective. Water was used in between the microscope objective and the skin surface for refractive index matching. The Raman laser power incident on the skin was 27 mW and the fs laser power incident on the skin was adjusted to be less than 40 mW using a half-waveplate/polarizer combination at the laser exit. The imaging field of view (FOV) can vary from 10 μm × 10 μm to 300 μm × 300 μm. The study was approved by the University of British Columbia Research Ethics Board (#H96-70499). Informed consent was obtained from the volunteer subject.
The validation experiment that we performed was to measure different micro-structures of a cherry angioma lesion in vivo on the upper arm of an Asian male volunteer. A dermoscopic image was first taken with DinoLite (AD4013T-TVW, AnMo Electronics Corporation, Hsinchu, Taiwan) to record the clinical appearance of the lesion as shown in Figure 2(a). Both fsRCM and SHG + TPF imaging channels were used to locate a blood vessel inside the cherry anigoma lesion as our 1st example ROI with an imaging FOV of 300 μm × 300 μm. The flip mirror was then flipped on to allow cwRCM imaging and confocal Raman spectral acquisition. The 300 μm × 300 μm FOV cwRCM image is shown in Figure 2(b). Then, the FOV of the cwRCM imaging channel was shrunk to only cover the ROI shown in Figure 2(c), which is the same area shown as the red square in Figure 2(b). A confocal Raman spectrum was then taken under the guidance of the cwRCM imaging. A demonstration video of a process of finalizing the ROI for spectral measurement can be found in Supplementary Video 1 (a more complicated case than the case of figure 2). The acquisition time for one spectrum is 20 s. The acquired confocal Raman spectrum is shown in Figure 2(d). The accompanied cwRCM images were also recorded as a video during the confocal Raman spectral measurement which can be found in Supplementary Video 2. The movement from the volunteer can still be seen, but clear blood structures are shown in the video during the whole acquisition period, indicating that the confocal Raman spectrum is valid from an ROI completely and always within the blood vessel itself. Strong peaks from hemoglobin (752 cm−1), glucose (1123 cm−1 and 1343 cm−1) and protein (940 cm−1 and 1665 cm−1) can be found on the Raman spectrum. As examples, we have rejected Raman measurement results where the videos show that there are moments when the ROI had moved outside of the blood vessel.
In vivo confocal Raman spectra of blood in a cherry angioma acquired under guidance of reflectance confocal imaging and multiphoton microscopy imaging.
(a) Dermoscope image of the cherry angioma lesion. The plastic layer with a 2 mm punched hole are visible in the image, which was utilized to stabilize the skin and to target the lesion; (b) cwRCM image of the blood vessel, image field of view (FOV) is 300 μm × 300 μm; (c) cwRCM image extracted from a video (Supplementary Video 2) of the blood vessel of the region of interest (ROI) from the area of the red square in (b), the image FOV is 100 μm × 100 μm; (d) confocal Raman spectrum of the blood vessel of the area shown in (c) and (b) as red squares, the exposure time is 20 s.
To validate the method further, surrounding fiber structures and cellular structures were selected as our 2nd and 3rd example ROI, respectively. Figure 3(a) and Figure 3(e) were first taken at a FOV of 300 μm × 300 μm from the SHG + TPF and fsRCM imaging channels, respectively. The cwRCM images shown in Figure 3(b) and Figure 3(f) were 150 μm × 150 μm FOV and were from the green squares labeled in Figure 3(a) and Figure 3(e). The confocal Raman spectra of the fiber structures and cells (shown in Figure 3(d) and Figure 3(h), respectively) were acquired from the region with a FOV of 60 μm × 60 μm shown in Figure 3(c) and Figure 3(g) as red squares. Strong Raman peaks at 856 cm−1, 937 cm−1 and 1454 cm−1 representing collagen and elastin are shown in the confocal Raman spectrum in Figure 3(d). Raman peaks of cell nuclei (722 cm−1 and band 1325–1330 cm−1), protein (1450 cm−1), phenylalanine (1003 cm−1) and lipids (1655 cm−1) can be found in the confocal Raman spectrum shown in Figure 3(h). Tentative Raman peak assignments are listed in Table 1.
In vivo confocal Raman spectra of cells and fibers surrounding a cherry angioma acquired under guidance of reflectance confocal and multiphoton imaging.
(a) SHG + TPF image of the fiber structures surrounding a blood vessel of the cherry angioma lesion, image FOV is 300 μm × 300 μm; (b) zoom-in cwRCM image of the area shown in the green square of (a), image FOV is 150 μm × 150 μm; (c) cwRCM image showing the ROI from the area of the red square in (b), the image FOV is 60 μm × 60 μm; (d) confocal Raman spectrum of the fiber structures of the red square area shown in (c) and (b), the exposure time is 20 s; (e) fsRCM image of the cellular structures surrounding a blood vessel of the cherry angioma lesion, image FOV is 300 μm × 300 μm; (f) zoom-in cwRCM image of the area shown in the green square of (e), image FOV is 150 μm × 150 μm; (g) cwRCM image showing the ROI from the area of the red square in (f), the image FOV is 60 μm × 60 μm; (h) confocal Raman spectrum of the cellular structures of the area shown in (f) and (g), the exposure time is 20 s.
The fsRCM and cwRCM images clearly show blood flow inside each vessel and single blood cells can be identified. We found that the blood flow velocity inside the cherry angioma lesion varies significantly from vessel to vessel. Therefore, the high quality cwRCM images from the blood vessel allow us to derive the blood flow velocity by simply tracking a single blood cell. For example, Figure 4 shows 4 consecutive frames from a blood flow video (Supplementary Video 1, frame19–22). The targeted blood cell labeled with a yellow arrow in Figure 4(b) (frame f20)was not observed in Figure 4(a) (frame f19), but its trajectory can be seen in the next two frames (f20 and f21) shown in Figure 4(c) and Figure 4(d). The distance that the cell moved from f21 to f22 was 0.03 mm and the time difference between each frame is 1/15 s. Therefore, the velocity can be calculated out as 0.45 mm/s.
Interestingly, the glucose peaks (1124 cm−1 and 1343 cm−1) can be clearly found in the confocal Raman spectra that we acquired at the blood vessel (Figure 2(d)). To explore whether we could noninvasively detect blood glucose level changes, an in vivo spectrum from the blood vessel of the cherry angioma lesion was first taken with an integration time of 20 s as “zero-point” reference. The volunteer was then asked to drink a standard glucose liquid (75 g glucose tolerance test beverage, Thermo Fisher Scientific Inc., Waltham, MA) within 5 min. Confocal Raman spectra were taken every 15 min for the next 105 min time. The highest glucose level was found between 45 to 60 min after the volunteer took the glucose liquid, with ~10% increase compared with zero-point reference. The change of glucose level is plotted over time by taking ratio of the area under the glucose peak at 1124 cm−1 and the area under the protein peak at 1450 cm−1, which is considered as a stable Raman signal in the blood, not affected by glucose concentration (Figure 5).
Discussion
We believe that the method that we demonstrated is a powerful tool to perform accurate region-of-interest confocal Raman spectral measurements under the guidance of reflectance confocal and multiphoton microscopic imaging. This is especially critical when performing measurements on in vivo subjects because being able to evaluate the movement level during the measurement is the key factor for accurate interpretation of the acquired spectral data. We did find that sometimes there were severe movements during the measurement, the spectrum then contained contributions from outside of the ROI. In this case, we will simply re-take the measurement one more time to make sure the level of movement from the subject is small enough to be negligible. However, if there is no real-time monitoring from an imaging modality, the qualitative or quantitative analysis based on the spectral data will result in inaccurate conclusions. For example, when using confocal Raman spectroscopy to study skin cancer, a spectral measurement which should target on cancer cells only, may contain both normal and cancer cells due to the movement from patients and the only way to exclude the invalid spectral data is the real time imaging guidance. Therefore, we believe that the imaging guidance is crucial for in vivo biological micro-Raman spectral measurements.
The flexibility of changing the area of ROI from 300 μm × 300 μm to 10 μm × 10 μm can easily allow us targeting on micro-structures of different sizes. With the help from multiphoton microscopic imaging, both morphological and functional information of biological tissues can be obtained. For our specific experiment on the cherry angioma measurements, SHG and TPF signals were not separated because our focus was not to target on separating collagen from elastin fibers. But with simple instrument modification12, SHG and TPF signals can be well separated and be used for measuring specific fiber structures in the dermis.
We also found that reflectance confocal imaging is very good for imaging blood flow in vivo. When deriving the blood flow velocity, we did not consider the factor that the blood cell may not exactly move within the screen plane. The reason is that the travel distance between the 2 consecutive frames was relatively short and the difference of the blood flow direction may not be very crucial. We believe that with further improvement on imaging penetration depth, the capability of deriving blood flow may be applicable to noninvasively analysis of blood related diseases.
It is very interesting to find clear glucose peaks in the confocal Raman spectra of blood with greatly reduce interferences from other tissue components. Although the idea of using Raman spectroscopy to noninvasively derive glucose level is not new22, our measurements may provide a few novel and valuable insights. First, under confocal imaging guidance, we can make sure that the glucose information is only from the blood vessel, not elsewhere in the tissue. This improves the accuracy of noninvasively measure the blood glucose level. Secondly, the Raman spectral quality of our measurement is significantly improved compared with results shown in previous literatures. This is probably due to our confocal measurement geometry, precise blood vessel targeting under imaging guidance and the high sensitivity of our spectrometer. Thirdly, other important, micro-scale localized biochemical information such as the level of oxyhemoglobin and deoxyhemoglobin may also be derived using our method.
In conclusion, our method of real time monitoring in vivo confocal Raman spectral measurements with reflectance confocal and multiphoton imaging could provide more accurate spectral interpretations. We have validated our method by measuring different skin micro-structures on in vivo volunteer. The demonstrated ROI method is valuable for precise in vivo spectral measurement and the imaging modalities allows us noninvasively obtaining information of the blood flow velocity. In addition, the special non-invasive method of accurately obtaining blood glucose level is novel and practical to be adapted into clinical use. Therefore, we believe our developed method can be generally applied into various types of in vivo micro-spectroscopy measurements. Researchers should perform micro-spectroscopy measurement carefully under advanced microscopy imaging guidance in order to generate accurate spectral interpretation of the biological, physiological and biomedical phenomena under investigation.
Methods
Multimodal spectroscopy and imaging system
The Raman excitation laser (I0785SA0100B-TK, Innovative Photonic Solutions, Monmouth Junction, NJ), which is a 785 nm continuous-wave (cw) diode laser, has been used for both the confocal Raman spectral measurement and the cw reflectance confocal microscopy (cwRCM) imaging. The Raman laser first passes a spatial filter system with a pinhole size of 30 μm and then is directed to an optical scanning system consisting of an 8 kHz resonance scanner for the fast axis and a galvanometer scanner for the slow axis. The laser beam is then focused by a 60X (NA = 1.0) water-immersion microscope objective (LUMPLFLN60X/W, Olympus Canada, Markham, Ontario) onto human skin. A polarization beamsplitter (PBS) along with a quarter waveplate is used to direct the descanned reflectance confocal signals to an avalanche photodiode (APD) module (C10508, Hamamatsu Corp., Bridgewater, NJ) with a 30 μm pinhole in front. A dichroic beamsplitter is used to direct the Raman signals to a 50 μm core-sized fiber connected with a home-made Raman spectrometer (spectrograph: HoloSpec-f/2.2-NIR, Kaiser, Ann Arbor, MI; CCD detector: Spec-10 : 100 BR/LN, Princeton Instruments, Trenton, NJ).
A femtosecond (fs) Ti:Sappire laser (Chameleon, Coherent Inc., Santa Clara, California), which has a tunable wavelength range of 720–950 nm, is used for multiphoton microscopy imaging. The fs laser beam is first pulsewidth-compressed by an optical compressor and expanded using two lenses. The fs laser beam is then scanned by the same scanner and focused onto the skin by the same microscope objective. The TPF and SHG signals are collected by a photomultiplier tube (PMT, active area 3.7 mm × 13 mm, > 15% quantum efficiency between 200 and 650 nm, H9433MOD-03, Hamamatsu Corp., Bridgewater, New Jersey) to generate SHG + TPF images and the descanned fs reflectance confocal (fsRCM) signals are directed by another PBS onto an APD with a 50 μm pinhole. A flip mirror is used to switch between the Raman laser and the fs laser and the two beam paths have been optimally aligned to be the same. To reduce involuntary body movement, double sided tape and a metal ring were used to magnetically mate the skin with the imaging objective. Water was used in between the microscope objective and the skin surface. The Raman laser power incident on the skin was 27 mW and the fs laser power incident on the skin was adjusted to be less than 40 mW using a half-waveplate/polarizer combination at the laser exit.
Data processing
For Raman spectra analysis, the fluorescence background was removed using the Vancouver Raman Algorithm which was developed by our group and based on an iterative polynomial fitting method23. To plot the blood glucose level over time, the background-subtracted Raman spectra were first derived. Then, the ratio of the area under the glucose peak at 1124 cm−1 and the area under the protein peak at 1450 cm−1 was calculated as the y-axis. This specific protein peak was chosen because it is considered as a stable Raman signal in the blood, not affected by glucose concentration.
Change history
11 October 2013
A correction has been published and is appended to both the HTML and PDF versions of this paper. The error has not been fixed in the paper.
References
Bauer, N. J. C., Hendrikse, F. & March, W. F. In vivo confocal Raman spectroscopy of the human cornea. Cornea 18, 483–488 (1999).
Egawa, M., Hirao, T. & Takahashi, M. In vivo estimation of stratum corneum thickness from water concentration profiles obtained with Raman spectroscopy. Acta. Derm. Venereol. 87, 4–8 (2007).
Caspers, P. J., Williams, A. C., Carter, E. A., Edwards, H. G. M., Barry, B. W., Bruining, H. A. & Puppels, G. J. Monitoring the Penetration Enhancer Dimethyl Sulfoxide in Human Stratum Corneum in Vivo by Confocal Raman Spectroscopy. Pharm. Res. 19, 1577–1580 (2002).
Lieber, C. A., Majumder, S. K., Ellis, D. L., Billheimer, D. D. & Mahadevan-Jansen, A. In Vivo Nonmelanoma Skin Cancer Diagnosis Using Raman Microspectroscopy. Laser. Surg. Med. 40, 461-467 (2008).
Shafer-Peltier, K. E., Haka, A. S., Fitzmaurice, M., Crowe, J., Myles, J., Dasari1, R. R. & Feld, M. S. Raman microspectroscopic model of human breast tissue: implications for breast cancer diagnosis in vivo. J. Raman Spectrosc. 33, 552–563 (2002).
Chan, J. W., Taylor, D. S., Zwerdling, T., Lane, S. T., Ihara, K. & Huser, T. Micro-Raman spectroscopy detects individual neoplastic and normal hematopoietic cells. Biophys. J. 90, 648–656 (2006).
Mordechai, S., Sahu, R. K., Hammody, Z. et al. Possible common biomarkers from FTIR microspectroscopy of cervical cancer and melanoma. J. Microsc. 215, 86–91 (2004).
White, I. J. & Dederich, H. D. American National Standard for Safe Use of Lasers, ANSI Z 136.1–2007. Laser Institute of America:Orlando (2007).
Zhang, G., Moore, D. J., Flach, C. R. & Mendelsohn, R. Vibrational microscopy and imaging of skin: from single cells to intact tissue. Anal Bioanal Chem 387, 1591–1599 (2007).
Caspers, P. J., Lucassen, G. W. & Puppels, G. J. Combined in vivo confocal Raman spectroscopy and confocal microscopy of human skin. Biophys. J. 85, 572–580 (2003).
Arrasmith, C. L., Patil, C. A., Dickensheets1, D. L. & Mahadevan-Jansen, A. A MEMS based handheld confocal microscope with Raman spectroscopy for in-vivo skin cancer diagnosis. Proc. of SPIE 7169, 71690N1–71690N8 (2009).
Wang, H., Lee, A. M. D., Frehlick, Z., Lui, H., McLean, D. I., Tang, S. & Zeng, H. Perfectly registered multiphoton and reflectance confocal video rate imaging of in vivo human skin. J. Biophoton. 10.1002/jbio.201200067 (2012).
Binoy, J., Abraham, J. P., Joe, I. H., Jayakumar, V. S., Petit, G. R. & Nielsen, O. F. NIR-FT Raman and FT-IR spectral studies and ab initio calculations of the anti-cancer drug combretastatin-A4. J Raman Spectrosc. 35, 939–946 (2004).
Deng, J. L., Wei, Q., Zhang, M. H., Wang, Y. Z. & Li, Y. Q. Study of the effect of alcohol on single human red blood cells using near-infrared laser tweezers. J. Raman Spectrosc. 36, 257–261 (2005).
Cheng, W., Liu, M., Liu, H. & Lin, S. Micro-Raman spectroscopy used to identify and grade human skin pilomatrixoma. Microsc. Res. Techniq. 68, 75–79 (2005).
Stone, N., Kendall, C., Smith, J., Crow, P. & Barr, H. Raman spectroscopy for identification of epithelial cancers. Faraday Discuss. 126, 141–157 (2004).
Robichaux Viehoever, A., Anderson, D., Jansen, D. & Mahadevan-Jansen, A. Organotypic Raft Cultures as an Effective In Vitro Tool for Understanding Raman Spectral Analysis of Tissue. Photochem. Photobiol. 78, 517–524 (2003).
Krafft, C., Neudert, L., Simat, T. & Salzer, R. Near infrared Raman spectra of human brain lipids. Spectrochim. Acta. A 61, 1529–1535 (2005).
Lau, D. P., Huang, Z., Lui, H., Man, C. S., Berean, K., Morrison, M. D. & Zeng, H. Raman spectroscopy for optical diagnosis in normal and cancerous tissue of the nasopharynx-Preliminary findings. Laser. Surg. Med. 32, 210–214 (2003).
Gniadecka, M., Wulf, H., Mortensen, N., Nielsen, O. & Christensen, D. Diagnosis of basal cell carcinoma by Raman spectroscopy. J. Raman Spectrosc. 28, 125–129 (1997).
Huang, Z., McWilliams, A., Lui, H., McLean, D. I., Lam, S. & Zeng, H. Near infrared Raman spectroscopy for optical diagnosis of lung cancer. Int. J. Cancer 107, 1047–1052 (2003).
Barman, F. I., Kong, C. R., Singh, G. P., Dasari, R. R. & Feld, M. S. Accurate Spectroscopic Calibration for Noninvasive Glucose Monitoring by Modeling the Physiological Glucose Dynamics. Anal. Chem. 82, 6104–6114 (2010).
Zhao, J., Lui, H. I., McLean, D. I. & Zeng, H. Automated Autofluorescence Background Subtraction Algorithm for Biomedical Raman Spectroscopy. Appl. Spectrosc. 61, 1225–1232 (2007).
Acknowledgements
This work was funded by the Canadian Dermatology Foundation, the VGH & UBC Hospital Foundation, the BC Hydro Employees Community Services Fund and the BC Cancer Agency. A. M. D. Lee acknowledges the Michael Smith Foundation for Health Research (MSFHR) and the CIHR Skin Research Training Centre for postdoctoral funding. H. Wang acknowledges CIHR Skin Research Training Centre for doctoral funding.
Author information
Authors and Affiliations
Contributions
For this publication, HequnWang designed the study, partially constructed the system, performed all the measurements, analyzed the data, prepared all the figures and movies and wrote the manuscript. Anthony Lee helped in developing the system and provided feedbacks to the manuscript. Haishan Zeng helped in study design. Harvey Lui, David McLean and Haishan Zeng provided essential comments to the manuscript.
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Electronic supplementary material
Supplementary Information
Supplementary Information
Supplementary Information
Supplementary Video 1
Supplementary Information
Supplementary Video 2
Rights and permissions
This work is licensed under a Creative Commons Attribution-NonCommercial-NoDerivs 3.0 Unported License. To view a copy of this license, visit http://creativecommons.org/licenses/by-nc-nd/3.0/
About this article
Cite this article
Wang, H., Lee, A., Lui, H. et al. A Method for accurate in vivo micro-Raman spectroscopic measurements under guidance of advanced microscopy imaging. Sci Rep 3, 1890 (2013). https://doi.org/10.1038/srep01890
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/srep01890
This article is cited by
-
Rapid and label-free screening of echinococcosis serum profiles through surface-enhanced Raman spectroscopy
Analytical and Bioanalytical Chemistry (2020)
-
Raman spectroscopic detection of high-grade cervical cytology: Using morphologically normal appearing cells
Scientific Reports (2018)
-
Digital and Computational Imaging in Pathology
Current Pathobiology Reports (2017)
-
Measurement of human serum albumin concentration using Raman spectroscopy setup
Optical and Quantum Electronics (2016)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.