Abstract
Body-based biomolecular sensing systems, including wearable, implantable and consumable sensors allow comprehensive health-related monitoring. Glucose sensors have long dominated wearable bioanalysis applications owing to their robust continuous detection of glucose, which has not yet been achieved for other biomarkers. However, access to diverse biological fluids and the development of reagentless sensing approaches may enable the design of body-based sensing systems for various analytes. Importantly, enhancing the selectivity and sensitivity of biomolecular sensors is essential for biomarker detection in complex physiological conditions. In this Review, we discuss approaches for the signal amplification of biomolecular sensors, including techniques to overcome Debye and mass transport limitations, and selectivity improvement, such as the integration of artificial affinity recognition elements. We highlight reagentless sensing approaches that can enable sequential real-time measurements, for example, the implementation of thin-film transistors in wearable devices. In addition to sensor construction, careful consideration of physical, psychological and security concerns related to body-based sensor integration is required to ensure that the transition from the laboratory to the human body is as seamless as possible.
Key points
-
Glucose sensors traditionally dominate the commercial sensing market, but sensors for alternative analytes could advance personalized health care in coming decades.
-
Body-based biomolecular sensors, including wearable, implantable and ingestible sensors, provide simple and continuous access to user biomolecular data through various biological fluids.
-
Continuous monitoring requires kinetically favourable receptors and sensing mechanisms capable of detecting analytes without user intervention.
-
In addition to sensor efficacy, body-based systems require careful consideration of physical, psychological and security concerns related to device use and data handling.
Similar content being viewed by others
Introduction
Biomolecular sensing is a rapidly growing interdisciplinary field. By combining engineering, chemistry, biology, physics, medicine and computational data analytics, sensors have been developed for the real-time monitoring of various physiological conditions. However, in spite of the identification of new sensing mechanisms and unique biomolecular targets, there remains a gap between the number of laboratory-developed sensors and commercialized products. Despite major advances in analyte targeting, signal amplification and sensor portability, it remains challenging to match the practicality and performance of simple enzymatic glucose sensors in systems that detect other analytes. Glucose sensors use specific enzymes (for example, glucose oxidase and glucose dehydrogenase) capable of producing highly amplified responses (for example, through high catalytic turnover) in a continuous manner, which is not easily achievable for other small-molecule, protein and nucleic-acid analytes. The development and adoption of body-based biomolecular sensing platforms (for example, wearables and implantables) requires continuous-sensing mechanisms with sensitivities and specificities comparable to those of glucose sensors. Therefore, the continued exploration of these body-based platforms is key to expanding biomolecular analysis beyond simple metabolite detection, to enable comprehensive physiological monitoring and inform clinical decision-making.
Historically, biological sensors or biosensors were named for their incorporation of a biological component to recognize and detect analytes. These biorecognition elements were necessary because the intricacy and selectivity of biological macromolecules was difficult to imitate. However, advances in the development of synthetic recognition elements, such as molecularly imprinted polymers and artificial enzymes, have enabled sensors to achieve selectivity comparable to that of their natural counterparts1. At present, we use the term biomolecular sensor to more generally describe all sensors aimed at detecting biomolecules or other biologically relevant molecules, shifting the focus away from the origin of the selected recognition component, and towards the more relevant nature of the analyte being detected.
In terms of composition, biomolecular sensing systems typically comprise four main components: the analyte, which is the biomolecule being detected; the recognition element, which interacts with the analyte and provides some degree of selectivity; the transducer, which converts the analyte-recognition element interaction into a readable signal; and the analysis system, which interprets the received signals. In addition to these physical components, there are several key terms that are vital to understanding and comparing sensing schemes: the limit of detection (LoD) describes the lowest analyte concentration that can be consistently detected by a sensor; sensitivity, though often erroneously used interchangeably with LoD, refers to the ability of a sensor to differentiate between similar input concentrations; selectivity describes the ability of a sensor to selectively target certain analytes; specificity, which serves as an ultimate form of selectivity, describes the ability of a sensor to target a single analyte; linear range describes the analyte concentration range over which a sensor produces a proportional response; and physiological range describes the physiologically relevant concentration range of the analyte, which ideally falls within the linear range of the sensor.
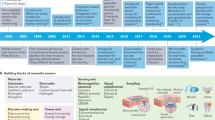
The concept of biomolecular sensing existed millennia prior to any understanding of the molecular concept (Fig. 1), with the analysis of urine being conducted in civilizations as old as Sumer (~4000 BC)2. The most considerable and lasting of these findings was the association between polyuria and sweet-tasting urine, recorded by the Indian physician Sushruta (~600 BC)3. Over the next two millennia, it was not uncommon for individuals referred to as water tasters to diagnose diabetes-like illness by tasting urine and assessing its sweetness4. The connection between diabetes and sweet urine was rediscovered and made mainstream in Europe by Thomas Willis (~1679)3. Over the next few centuries, scholars discovered that sugar was responsible for the sweetness of diabetic urine and established chemical tests to determine its presence5. These tests, including Trommer’s test, Fehling’s test and Benedict’s test, exploited sugar-assisted reduction of copper species to produce colorimetric changes and diagnose diabetes3,5. Although these tests were rather non-specific, they can be seen as the predecessors of modern biomolecular sensors.
Timeline of major events regarding the evolution of biomolecular sensing, including the first evidence of urine analysis2; the first connection between sweet urine and diabetes-like illness3; the rediscovery of the connection between sweet urine and diabetes in Europe3; the development of primitive metal salt glucose tests3,5; the first incorporation of a biological recognition element into a colorimetric biomolecular sensor8; the first proposed electrochemical biomolecular sensor6; the first commercial, portable glucose sensor9; the first commercial electrochemical glucose sensor12; and the US Food and Drug Administration (FDA) approval of the first continuous glucose monitor13.
The first modern biomolecular sensor is typically attributed to Leland Clark for his 1962 proposal6 to incorporate glucose oxidase enzymes into his previously developed oxygen electrode7. However, the Miles–Ames laboratory also made substantial contributions with the development of a colorimetric glucose oxidase test strip in 1957 (ref. 8). In 1970, the same laboratory released the first portable blood glucose meter, a landmark achievement in portable biomolecular analysis9. Clark eventually commercialized his electrochemical glucose monitor in 1975 (ref. 10). In the 1980s, as glucose monitoring became more mainstream, many alternative colorimetric glucose systems made their way to market11. Although it was difficult to miniaturize them, the first portable electrochemical glucose monitor eventually reached the market in 1987 and quickly dominated its colorimetric competitors12. This landmark led to market-wide adoption of electrochemical sensing, with most companies replacing their colorimetric devices with electrochemical ones that are still on the market today. Finally, in 1999, the first continuous glucose monitor received US Food and Drug Administration (FDA) approval13, sparking interest in the development of body-based sensing platforms.
The twenty-first century has brought with it a new wave of body-based sensing systems, with many large life sciences companies marketing their own continuous glucose monitoring system. While glucose measurement remains at the forefront of sensor innovation, the development of commercial sensors for alternative analytes has lagged behind, with the exception of a few other metabolites (for example, cholesterol). However, there is an abundance of other biochemical indicators (for example, proteins and DNA) that can provide invaluable information on physiological status. As sensor development continues towards new body-based platforms capable of long-term measurements, the number of analytes that can be reliably detected will hopefully continue to expand and allow these systems to realize their diagnostic potential.
This Review summarizes emerging biomolecular sensors, tackling two main challenges. The first challenge is the transition of sensors from laboratory-based assays to body-based systems to develop self-contained biomolecular sensors that are wearable, implantable or consumable and can easily be incorporated into everyday life. The second challenge is to expand biomolecular sensors beyond glucose detection, to realize practical and commercial devices for a broader range of relevant physiological markers (Box 1). To overcome these challenges requires biomolecular sensing approaches that are more sensitive, specific and amenable to continuous monitoring than ever before. Here we highlight many of the cutting-edge strategies expected to facilitate the development of new sensing devices. In addition, we explore relevant considerations regarding biological fluid compatibility and sensor design to ensure seamless lab-to-body transition.
Strategies for signal amplification
Sensitivity, specificity and adaptability to new analytes are the major challenges for biomolecular analysis14. Therefore, many amplification strategies have been developed to enable lower LoDs and higher sensitivities. These strategies can be implemented at any level of sensor development to help to achieve practical physiological monitoring.
Intracellular sensors
Intracellular sensors go beyond secreted (for example, insulin) and membrane-bound biomarkers (for example, viral surface proteins) to provide localized information at the cellular level (for example, quantifying mRNA expression in breast cancer cells and detecting overexpression of cytosolic cathepsin B in adenocarcinoma cells)15,16. They are also sensitivity enhancers, given the small size of the cell and the fact that intracellular concentrations often surpass those in biological fluids. These sensors allow single-cell diagnosis by probing cytoplasmic regions and enable access to diverse high-concentration biomarkers (Fig. 2a). Intracellular analysis is typically achieved by either direct insertion of nanostructures into the cytosol to probe16 or extract17 its contents, or indirect intake and expression of a genetic sensing framework; for example, through reporter gene activation upon interaction with intracellular human immunodeficiency virus (HIV) proteins18. These intracellular approaches have the potential to considerably amplify cell-localized biological phenomena (such as cytoplasmic protein overexpression) for diagnostic purposes.
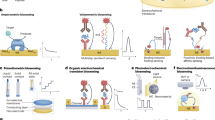
a, Intracellular sensing allows access to new, high-concentration biomarkers. b, Surrogate biomarkers can provide alternative detection pathways for trace analytes. c, Synthetic biomarkers can be introduced to the body to amplify biomolecular processes and enable their detection. d, Nanozymes and DNAzymes provide alternative catalytic routes for analyte monitoring. e, Mass transport advances, including analyte-guiding nanochannels, superhydrophobic transport metamaterials and analyte mixing micromotors increase the rate of interaction between an analyte and its recognition element. f, Organic electrochemical transistors serve as powerful amplifiers for biomolecular interactions. g, Reporter multimerization enhances the signal output from affinity interactions. h, Debye length-manipulating approaches, including structure-switching receptors, nanostructure-mediated electron transfer and membrane-mediated Debye extension bypass traditional Debye limitations to extend transducer influence and increase signal generation. i, Computational approaches, combined with multiplexed analysis, may allow health state amplification for predictive medicine. Part c is adapted from ref. 22, Springer Nature Limited. Part e (analyte-guiding nanochannel) is adapted from ref. 39, Springer Nature Limited. Part e (superhydrophobic transport metamaterials) is adapted from ref. 40, Springer Nature Limited. Part f is adapted from ref. 44, Springer Nature Limited. Part h (structure-switching receptor) from Nakatsuka, N. et al. Aptamer-field-effect transistors overcome Debye length limitations for small-molecule sensing. Science 362, 319–324 (2018)51. Adapted with permission from AAAS. Part h (nanostructure-mediated electron transfer) is adapted from ref. 53, CC BY 4.0 (https://creativecommons.org/licenses/by/4.0/). Part h (membrane-mediated Debye extension) is adapted from ref. 54, Springer Nature Limited.
Surrogate biomarkers
Sensor development has long focused on the assessment of biological conditions through directly related molecular targets, such as targeting viral surface proteins to detect a virus. Yet there is an abundance of additional targets or surrogate biomarkers (Fig. 2b) that are less intuitive but still correlate with physiological conditions19. Surrogate markers may also be present at higher concentrations than the direct target, and can thus amplify a biochemical signal. Advances in the understanding of molecular pathways can reveal new biological relationships and help to access a broader library of health indicators. For example, precursor or metabolite molecules can be targeted, such as tyrosine as precursor to norepinephrine — a neurological indicator20. Another approach is the targeting of indirectly related proteins that show high condition correlation, for example, measuring neurofilament light chain protein as an indicator of inflammatory brain demyelination21. These simple methods can provide precise indirect estimates of trace molecules or hidden biological events.
Synthetic biomarkers
The pool of endogenous biomarkers is large, but it is not limitless. Alternatively, synthetic biomarkers (Fig. 2c) that are designed to interact with biomolecules can be introduced into the body (for example, through injection) to indicate and amplify physiological conditions (for example, by reacting with an overexpressed enzyme)22,23. Synthetic biomarkers can have various configurations; however, all of them contain two essential components: a recognition region (for example, substrates24 and promoter sequences25) to target the biochemical process and a reporter region (for example, volatile organic compounds24 and genes25) to signal its presence. Synthetic markers can bypass common endogenous biomarker issues (for example, short half-lives and low concentrations), and detect previously inaccessible or subtle health states. For example, volatile reporter molecule-linked peptides injected into mice are cleaved by neutrophil elastase, to indicate its elevated activity, which is associated with several illnesses (such as pulmonary bacterial infections and alpha-1 antitrypsin deficiency)24.
Catalyst-mediated amplification
Enzymes are considered the ideal recognition element owing to their excellent selectivity and natural catalytic ability; however, they are restricted by the analytes they can target. Therefore, the discovery and fabrication of alternative catalysts (Fig. 2d) is important for universal high-sensitivity sensing.
Nanozymes are nanomaterials that possess catalytic ability and can replace enzymes in biomolecular sensing. These include nanoparticles26, metal–organic frameworks (MOFs)27, and many other nanomaterials28 capable of catalysing biochemical reactions and generating amplified biomolecular responses. For example, peroxidase-like MOFs capable of catalysing o-phenylenediamine to 2,2-diaminoazobenzene enable sensitive detection of Staphylococcus aureus with a LoD of only 6 colony-forming units (CFU) ml–1 (ref. 29). Additionally, single-atom nanozymes have atomically dispersed metal active sites that maximize catalytic efficiency, allow for tunable selectivity, and enable reaction kinetics similar to that of natural enzymes1,30,31. Moreover, computational approaches and artificial intelligence can be applied to the discovery of artificial catalysts (for example, superoxide-dismutase nanozymes and carbon dioxide electrocatalysts)32,33.
DNA catalysts, or DNAzymes, can also amplify biomolecular signals and offer alternative catalytic routes for sensing. DNAzymes catalyse various reactions, including RNA cleavage, RNA ligation and alkyne–azide cycloaddition34,35,36. These reactions can be implemented in a variety of ways to detect metal ions, small molecules, nucleic acids and proteins through a combination of a catalytic event and a released reporter molecule (for example, cleaving of a dye-conjugated RNA sequence). For example, DNAzymes selected to react with specific bacteria lysates can detect bacteria in single digits per millilitre in unprocessed blood samples37,38. In addition, the complementary nature of DNA within DNAzymes allows for programmable capture and release of the substrate, and easy integration with other functional nucleic acids (for example, extension of the DNAzyme sequence to include an aptamer sequence that can bind the analyte and provide additional selectivity)34.
Overcoming mass transport limitations
The sensitivities and LoDs of biomolecular sensors do not rely on the bulk concentrations of analyte in solution but on the localized amount of analyte interacting with the recognition component of the sensor. Therefore, maximizing the analyte-recognition element interactions is important to ensure sensitive and fast detection. Several strategies have been developed to overcome the limitations caused by slow mass transport of molecules and thus slow diffusion (Fig. 2e) to improve biomolecular recognition and to amplify early signal responses. These strategies include designing synthetic nanochannels to maximize transport efficiency39, using superhydrophobic surfaces to control molecule localization40 and using molecular micromotors to mix solutions and accelerate mass transport41. In addition, DNA nanostructures can improve surface probe distribution and reduce probe crowding, allowing for enhanced analyte association42. Microfluidic approaches for sample collection and transport can maximize mass transport efficiency, in addition to being compatible with wearable and implantable devices43.
Organic electrochemical transistors
Organic electrochemical transistors (OECTs) (Fig. 2f) can act as signal-amplifying components in biomolecular sensing and other bioelectronic devices44. OECTs contain three main properties which, when combined, set them apart from other field-effect transistors. First, they possess an organic semiconductor channel (such as poly(3,4-ethylenedioxythiophene) polystyrene sulfonate, PEDOT:PSS) that bridges the source and drain electrodes. Second, they possess an electrolyte between the channel and gate electrode. Third and most importantly, they allow for full penetration of electrolyte ions into the semiconductor channel45. Because semiconductor doping is reliant on ion penetration, OECTs can generate very large drain currents with small gate voltages, which makes them excellent signal amplifiers for biomolecular interactions44,46. For example, an interdigitated OECT sensor for adenosine triphosphate (ATP) achieves a LoD of 10 pM, which is nearly four orders of magnitude better than an amperometric transduction approach using the same recognition element47. The inexpensive fabrication costs of OECTs, as well as their capacity for flexible electronic integration, make these components attractive candidates for wearable and implantable devices48.
Reporter multimerization
A simple method of amplifying biomolecular interactions at the transduction level is to increase the amount of signal generated by each analyte. Typically, affinity interactions result in a 1:1 ratio between signal and interaction. However, this ratio can be increased through simple incorporation of multiple reporter molecules (Fig. 2g) into the recognition element complex. For example, incorporation of multiple methylene blue redox markers into a C-reactive protein sensor enhances signal output and enables a LoD of 1 pM, which is considerably lower than that of previous electrochemical and optical approaches using single reporter molecules49. Alternatively, secondary reporter complexes in the form of oligonucleotide systems or reporter-labelled nanomaterials (for example, dendrimers) may be used to produce amplified responses50. However, many of these multimerization amplification strategies are designed for use in more traditional immunoassays with externally added reporter molecules, and thus may not be compatible with body-based biomolecular sensing. This encourages the development of methods that focus on multimerization of recognition elements and require no external reagents, which are more compatible with continuous in vivo detection.
Overcoming Debye limitations
The screening of surface-bound charges by mobile ions beyond the Debye length, which is the effective sensing range, is a key obstacle in the development of high-sensitivity electronic sensors51. Although this issue is often avoided through alterations to the buffer solution (for example, decreased ionic strength), this approach is unrealistic for biofluid analysis52. Thus, several strategies have been introduced to overcome Debye length limitations and to amplify biomolecular interactions (Fig. 2h). Among these strategies is inducing near-electrode conformational changes to signal distant interactions51, using nanostructured materials to accelerate electron transfer53, and employing membrane-encapsulated ion-free water layers to extend the Debye length54.
Multiplexing and molecular data fusion
Multiplexing or using multiple markers allows the amplification of changes in physiological signals by increasing the number of biomolecular targets. Thus, sampling of multiple molecular avenues (for example, inflammatory, hormonal and metabolic) provides a more comprehensive health assessment55. The fusion and analysis of multiple molecular data (Fig. 2i) through artificially intelligent approaches, such as machine learning, can expand the capability of sensors to identify and diagnose physiological conditions faster, earlier and with more accuracy than techniques relying on single analyte analysis56,57,58. In addition, multiplexed controls (for example, additional sensors with non-specific recognition elements) allow better insight into sensor stability by providing separate channels to monitor sensor degradation.
Strategies for selectivity improvement
The incorporation of biological recognition elements that selectively detect biologically relevant molecules into molecular sensing approaches can improve selectivity6,8. Proteins, DNA and synthetic alternatives (such as polymers and nanoparticles) can be applied as recognition elements for specific analytes59,60. Although enzymes are ideal recognition elements owing to their high specificity and natural signal amplification through high catalytic turnover, most biomolecular targets do not have specific enzyme catalysts and thus require alternative recognition approaches. Robust affinity recognition elements (affinity receptors) that induce highly specific association interactions with their target are necessary to develop body-based biomolecular sensors. These recognition elements should be able to withstand the presence of high concentration interferants in native biofluids, remain stable during daily wear and tear, and possess the resolution and dynamic capabilities to capture physiologically relevant changes in biomarker levels. Here, we discuss the current state of affinity recognition elements, explore strategies to overcome non-specific binding and biofouling, and analyse the compatibility of various recognition elements for in vivo real-time monitoring.
Natural and artificial affinity receptors
Antibodies are the most popular biological affinity receptors employed in sensors (Fig. 3a). They are generated in vivo in response to specific immunostimulation to selectively bind targets under physiological conditions. Antibodies have high binding affinities, selectivity and sensitivity to detect biomarkers at picomolar to nanomolar concentrations. However, antibodies are fragile, highly sensitive to temperature and pH changes, exhibit batch-dependent variation, and have high production costs, which greatly limits their use in bioelectronic applications59. Antibody mimetics with high ligand-binding affinity can be designed in the form of antibody fragments59 (for example, nanobodies61) and engineered proteins (for example, avimers62, affimers63 and affibodies64). Compared to antibodies, these engineered receptors are smaller in size, have higher thermal stability, and can be engineered as multimers to increase avidity59,61. In addition, in vitro technologies such as phage display and ribosome display enable rapid, efficient and inexpensive selection of high-affinity protein-based binders65,66. However, although they are effective, these engineered alternatives often fail to outcompete antibodies in assay performance; this is probably due to their inability to mimic the glycosylation patterns and post-translational modifications of antibodies, which confer additional selectivity.
a, Classes of available recognition elements (protein, DNA and synthetic) including both traditional and emerging receptors. b, Strategies for prevention of non-specific binding (NSB). c, Recognition-element-based strategies for continuous monitoring include catalyst-mediated detection, kinetic control of analyte binding and release, and receptor regeneration following analyte attachment. koff, dissociation rate constant; kon, association rate constant; SB, specific binding; SOMAmer, slow off-rate modified aptamer; ssDNA, single-stranded DNA; ssRNA, single-stranded RNA.
Functional nucleic acids are promising affinity recognition element alternatives to antibodies. Aptamers are single-stranded nucleic acids generated in vitro through the systematic evolution of ligands by exponential enrichment (SELEX), in which sequences are selected to bind to a specific target, ranging from metal ions to whole cells67,68. SELEX allows the generation of affinity receptors for a broad range of targets, including toxic, non-immunogenic and small-molecule biomarkers that are difficult to target with antibodies69,70. Aptamers are particularly promising for implementation in body-based sensors owing to their simple and inexpensive mass synthesis, minimal batch-to-batch variation, high thermal stability, non-immunogenic properties and their amenability to site-specific chemical modification. As such, aptamers have been demonstrated in several wearable and implantable sensors (for example, in continuous cortisol- and serotonin-monitoring systems)71,72.
However, aptamers are inherently prone to nonspecific binding owing to their intrinsic negative charge and the large variety of RNA- and DNA-binding proteins present in body fluids73. In addition, they are susceptible to nucleases in biological fluids and sensitive to changes in environmental conditions, such as pH, temperature and salt concentration. Alternatively, chemically modified aptamers with non-natural nucleic acids can have low nonspecific binding, and improved stability and binding affinity (such as SOMAmers74 modified to have slow analyte dissociation)75. SELEX approaches run in biological fluids or directly in live animals can isolate aptamers that maintain high performance under physiological conditions76,77. Additionally, strategies to resist nuclease degradation such as using chemical alterations or surface coatings78,79 can minimize probe loss.
Synthetic recognition elements that are not based on biological monomers (such as molecularly imprinted polymers60 and supramolecular systems80) also show promise as selective receptors for biomolecular sensors.
Nonspecific binding
A key challenge in the translation of biomolecular sensors is nonspecific binding, which can occur on both the sensor surface (biofouling) and the recognition element (cross-reactivity)73. Biofouling is driven by the accumulation of cells, proteins or other species on the sensor surface through nonspecific interactions. This adsorption process hinders the diffusion of analytes to the sensor and leads to signal loss over time81. Cross-reactivity arises from recognition elements that interact with molecules other than the analyte of interest to produce false-positive responses. These interferants may have affinities close to that of the analyte, or they may be present in such high concentrations that they associate to a considerable degree at equilibrium. In biological fluids, high protein content compared to that of the analyte can cause substantial nonspecific binding that may appear as false positive signals73. Strategies and approaches, such as using protective coatings and exploiting kinetic differences between analyte and interferant must therefore be developed to minimize these extraneous interactions (Fig. 3b).
Generally, anti-fouling strategies aim to either fill the gaps in a sensing surface using a blocking agent (for example, bovine serum albumin (BSA) and casein) or to coat the entire sensing surface with an anti-fouling layer (for example, poly(ethylene glycol) and zwitterionic polymers such as derivatized polycarboxybetaine)73,82,83. More complex anti-fouling layers, based on bioinspired materials (for example, slippery liquid-infused porous surfaces inspired by pitcher plants)84, hydrogels (for example, polyacrylamide copolymer gels)85,86 and nanocomposites (for example, cross-linked BSA impregnated with conducting nanomaterials)87,88 also show fouling resistance in vivo. Additionally, methods have been developed to distinguish analyte binding from nonspecific interferent binding. One such approach exploits the affinity differences between analyte and interferant to physically remove weakly bound entities with an external force, for example by using an alternating electric field89. This approach allows active removal of interfering species, such as nonspecific IgG without ejection of the target HER2 protein. The inclusion of a reference or control sensor with a non-specific receptor (for example, scrambled aptamer sequence71) is another way to differentiate between real and non-specific signals. Alternatively, the molecular recognition approach itself can confer some resistance to nonspecific interactions. Sensing mechanisms that rely on an analyte-triggered event (for example, structure-switching aptamers51) are less prone to nonspecific signal interference than sensors that rely on bulk property changes. Similarly, stochastic sensing approaches (for example, solid-state nanopores90) that can computationally distinguish between analyte and interferant are highly resistant to nonspecific behaviour.
Continuous real-time monitoring
As an alternative to single-use sensors, body-based biomolecular sensors require sensor regeneration and reliability. Thus, sensors must be able to detect analytes using reversible molecular interactions, ensure minimal time between readings and provide continuous data with negligible hysteresis. Continuous monitoring is challenging, especially with high-affinity recognition elements that exhibit slow dissociation kinetics91; however, there are several solutions to overcoming such limitations (Fig. 3c). A receptor with fast binding kinetics (for example, an aptamer selected for high on/off rate) is ideal for continuous monitoring because it allows rapid equilibration with the surrounding fluid92. However, as these rapid kinetics edge closer to real-time monitoring, this temporal resolution often comes at the cost of sensor LoD. Therefore, biomolecular sensors that employ such recognition elements must also use highly sensitive transduction mechanisms (for example, transistor-based approaches) to compensate for sensitivity loss93. In addition, a proper fluid-sampling system (for example, microfluidic integration) is desirable to provide effective and rapid transport of the biological fluid over the sensor to ensure a reproducible, timely and accurate signal, along with negligible sample contamination and carryover.
Real-time monitoring can also be achieved through the regeneration of recognition elements after each measurement, using either chemical, thermal or electrochemical approaches94. Chemical regeneration methods rely on surface and fluid property alterations through the application of acids, bases or other solvents. However, these approaches can damage biological receptors and are not compatible with body-based biomolecular analysis. Thermal regeneration methods are more feasible for body-based sensing, relying on localized warming using miniaturized heating elements, and are particularly promising for nucleic-acid-based receptors95. Finally, electrochemical regeneration approaches employ electrical pulses or other waveforms to manipulate affinity interactions without risking damage to receptors96.
Approaches for continuous monitoring
Continuous monitoring requires complementary sensing approaches that enable real-time detection in biological fluids97. Most available biomolecular sensing methods rely on secondary reporters (for example, sandwich assays), indirect reporters (for example, steric hindrance assays) or chromatographic systems (for example, lateral flow assays). However, such techniques are incompatible with body-based biomolecular sensing owing to their dependence on external manipulation and washing steps. Some approaches avoid the need for such external interventions by combining recognition and reporting steps into one process. However, these methods typically rely on specialized receptors with intrinsic activity such as enzyme catalysts that are only available for a small subset of analytes. Thus, the development of universal reagentless platforms that offer versatility in target analysis, and employ self-contained reagent-free mechanisms, is crucial for the widescale adoption of body-based sensors.
Electrochemical aptamer-based sensors
Electrochemical aptamer-based (E-AB) sensors (Fig. 4a) allow reagentless, label-free detection of small molecules and proteins98. E-AB sensors use electrode-bound aptamers with terminal redox reporters to recognize and bind analytes. Analyte detection is achieved through target-induced proximity changes between the redox reporter and the electrode surface, which alters the received Faradaic current. E-AB sensors have been optimized to establish a detailed theoretical framework for their molecular interactions99, and enable their implementation in in vivo systems99,100,101. Moreover, E-AB sensors have excellent temporal resolution upon sequential in vivo measurements, for example, for the detection of plasma phenylalanine in rats102. Furthermore, E-AB molecular probes can be incorporated into implantable devices to offer non-invasive continuous monitoring. For example, an E-AB microelectrode sensor is used for the continuous measurement of doxorubicin in mouse tumour tissue103. Although E-AB approaches expand analyte diversity for continuous monitoring, they are restricted by the availability of aptamer sequences that can bind relevant targets and undergo sufficient conformational changes between their unbound and bound states.
a, Electrochemical aptamer-based sensors rely on aptamer binding-induced proximity changes between a redox reporter and an electrode. b, Electrochemical DNA sensors use hairpin DNA structures to detect DNA through binding-induced redox reporter proximity changes. c, DNA scaffold sensors rely on steric hindrance from small receptor-bound analytes to alter redox readout. d, Protein scaffold sensors use conformational changes in redox-tagged protein receptors to detect bound analytes. e, Nanoscale molecular pendulum sensors exploit hydrodynamic differences between bound and unbound probes to temporally separate redox readout. f, Impedimetric sensors rely on interfacial capacitance changes to detect analyte binding. g, Thin-film transistors use interaction-mediated alterations in current flow to detect analyte–receptor binding. Part a is reproduced with permission from Xiao, Y., Lubin, A. A., Heeger, A. J. & Plaxco, K. W. Label-free electronic detection of thrombin in blood serum by using an aptamer-based sensor. Angew. Chem. Int. Edn Engl. 44, 5456–5459 (2005)98. Copyright Wiley-VCH GmbH. Part b is adapted with permission from Fan, C., Plaxco, K. W. & Heeger, A. J. Electrochemical interrogation of conformational changes as a reagentless method for the sequence-specific detection of DNA. Proc. Natl Acad. Sci. USA 100, 9134–9137 (2003)104. Copyright (2003) National Academy of Sciences, USA. Part e is adapted from ref. 117, Springer Nature Limited.
Electrochemical DNA sensors
DNA is an important biomarker for the diagnosis of disease and surveillance of physiological conditions. Electrochemical DNA (E-DNA) sensors (Fig. 4b) operate in a similar way to E-AB sensors, that is, through target-induced proximity changes between a terminal redox reporter and an electrode. However, instead of aptamers, E-DNA sensors rely on DNA hairpin probes that hybridize to nucleic acid analytes104. Alternatively, E-DNA approaches can use strand displacement techniques105. E-DNA sensors can be implemented into in vivo systems, for example, for the detection of pM-level DNA in whole blood106. However, these sensors are often susceptible to false-positive results owing to probe degradation and non-specific interactions. In addition, ultralow concentrations of circulating DNA in physiological matrices complicates the commercial adaptation of these approaches107. Ongoing research aims to overcome these challenges and enable in vivo monitoring through signal amplification and advanced probe design (for example, through the incorporation of multiple redox reporters per probe)108.
DNA-scaffold-based sensors
DNA-scaffold-based sensors enable reagentless detection of proteins, nucleic acids and small molecules, relying on DNA frameworks (Fig. 4c). These sensors employ a variety of small recognition elements such as peptides that are anchored to electrodes by DNA sequences. In addition, these systems typically contain redox reporter molecules within the DNA scaffold that enable electrochemical readout109,110,111,112,113. DNA-scaffold-based detection is achieved through differential access to the electrode surface of bound versus unbound probes, often through analyte-mediated steric hindrance, which leads to differences in faradaic current. DNA-scaffold-based sensors can detect analytes in complex biological fluids, for example, HIV and syphilis antibodies in human serum110,113, making them a promising chemical approach for body-based sensing. Even though these sensors work well for small receptors, they are incompatible with larger recognition elements such as antibodies owing to slow diffusion and low baseline currents. Thus, analytes that lack corresponding small receptors, such as many large proteins, are difficult to detect with this approach. Using protein-binding DNA can be a solution for this challenge114, but these methods are also limited by the availability of such recognition elements.
Protein-scaffold-based sensors
Protein-scaffold-based sensors (Fig. 4d) operate on principles similar to those of their DNA counterparts, enabling reagentless detection through use of amino acid scaffolds115,116. Like DNA scaffolds, protein scaffolds contain an electroactive reporter that confers electrochemical activity. However, protein-based approaches do not typically involve a separate recognition element, with the protein scaffold itself acting as the analyte receptor. Upon binding of the protein scaffold to its corresponding analyte, conformational changes facilitate the alteration of faradaic current received at the electrode. Protein-scaffold-based sensors can enable real-time measurements in biological fluids, for example, for the detection of Fyn kinase peptide targets in whole blood115. However, their dependence on protein-based receptors greatly limits the range of analytes they can detect.
Molecular pendulum-based sensors
Nanoscale molecular pendulums (NMPs) allow reagentless electrochemical sensing for continuous biomarker analysis117 (Fig. 4e). NMPs are comprised of two hybridized DNA strands, one of which contains a terminal ferrocene redox reporter and the other a terminal antibody. Upon application of a sufficiently positive potential, the negatively charged NMPs descend towards the electrode surface in a motion resembling that of an inverted pendulum. When the terminal ends of the NMPs fall close enough to the electrode surface, ferrocene oxidizes and produces a measurable faradaic current. Detection with NMPs occurs through differential rates of ferrocene oxidation, where hydrodynamic differences between bound and unbound antibodies dictate the rate of pendulum descent. This method allows detection of proteins and peptides (for example, troponin T, brain natriuretic peptide and the interleukin IL-6) in a variety of biological fluids, including blood, sweat, and saliva117. NMPs can also detect SARS-CoV-2 proteins and whole viruses in untreated clinical saliva samples118. However, further theoretical development and optimization is required prior to the adoption of NMP sensors in biomedical devices.
Impedance spectroscopy-based sensors
Electrochemical impedance spectroscopy (EIS) (Fig. 4f) is a powerful technique for measuring interfacial properties related to electrode-localized molecular recognition events119. Many EIS sensors rely on faradaic impedance, and thus require addition of external redox probes. Alternatively, non-faradaic impedance approaches enable reagentless detection without the need for sample pretreatment120. Given the detail-rich nature of impedance measurements, these non-faradaic methods use alternative parameters (for example, double-layer capacitance121) to monitor analyte interaction with receptor-functionalized surfaces. Non-faradaic methods allow the detection of nucleic acids, proteins and many other analytes continuously in complex biological fluids (for example, detection of insulin in serum and detection of miRNAs in plasma)122,123. However, because of their dependence on interfacial properties, impedance-based sensors are particularly susceptible to surface biofouling; thus, further research into anti-fouling strategies is required to apply these techniques in body-based systems87.
Thin-film-transistor-based systems
Thin-film transistors (Fig. 4g) are a subset of field-effect transistors (FETs) that can be integrated into body-based sensors, showing high sensitivity, great flexibility and capacity for real-time continuous sensing124. Similar to impedimetric approaches, FETs rely on interfacial property alterations and thus do not require external reporter molecules to indicate molecular interactions. Promising semiconductors for thin-film transistors include organic materials, such as PEDOT:PSS45,46, carbon allotropes125 and metal oxides71. Although these materials differ in their individual properties, for example in their conductivity, they can all facilitate continuous monitoring of a variety of analyte types in complex biological fluids. For example, FETs are used for the detection of amino-terminal pro-brain natriuretic peptide (NT-proBNP) in human serum and SARS-CoV-2 RNA in nasopharyngeal samples46,126,127. In addition, thin-film transistors can be implemented in wearable and implantable systems for small molecule sensing in vivo (for example, wearable cortisol monitoring in human sweat and serotonin monitoring in mice using an implanted neural probe)71,72. However, similar to EIS-based sensors, thin-film transistors are susceptible to biofouling and environmental degradation128.
Alternative transduction approaches
Besides electrochemical and electronic detection, optical approaches can be applied for signal transduction. Here, the distance between a fluorophore and quencher are exploited, using optical analogues of E-AB sensors129, E-DNA sensors128, DNA/protein-scaffold-based sensors130 and NMP sensors131. Advances in fibre optics and nanophotonics have enabled optical body-based biomolecular sensors132,133.
The biological fluid frontier
A variety of bodily fluids can be assessed for biomolecular analysis (Table 1; Fig. 5). Ideally, a biological fluid should be non-invasively collected or sampled, contain diverse analytes, reflect physiologically relevant biomarker concentrations, and capture transient fluctuations in targeted biomarkers. In addition, biological fluid sampling should be compatible with day-to-day life, such that sensors can take readings without the need for user handling.
Approaches to incorporating body-based sensors into everyday life, showing frequently used biological fluids and specific applications for these fluids (apart from general applications that multiple fluids can be used for), and existing commercial products that can house these fluid-sampling devices.
Urine
Urine is a reliable fluid for at-home biomolecular sensing (for example, pregnancy tests) owing to its non-invasive and easy collection. Because of its direct derivation from blood, urine contains a wealth of molecular information, including metabolites, DNA, RNA and proteins, that tend to correlate well with blood concentrations134. In addition, urine does not typically undergo proteolytic degradation, as with other blood-derived fluids, greatly extending protein lifetime and stability135. However, biomolecules tend to exist in urine at much lower concentrations than in blood, which can complicate detection134,135. Furthermore, the long filtration process of urine production reflects analyte concentrations over long time frames, which makes analysis of transient conditions difficult136. The use of urine in biomolecular sensors is rare, but it still may be advantageous over other fluids in diagnosing reproductive cancers135, sexually transmitted diseases137 and other urogenital conditions. Urine has also been explored as a potential fluid for wearable sensor analysis (for example, in diapers)138.
Blood
Blood is the most information-rich biological fluid in the body owing to its ubiquitous involvement in homeostasis and transport. Because of the diverse biomolecular composition of blood, blood analysis has long served as the gold standard for physiological diagnosis. Many other body fluids, including interstitial fluid (ISF), saliva, tears and urine are derived from blood, making it an excellent reference for alternative fluid comparison. Blood is being explored as a potential fluid for in vivo body-based sensors139; however, its sampling tends to be invasive and risks systemic infections. In addition, blood contains high concentrations of cells and proteins, which can cause biofouling and decrease the stability and reliability of sensors over time. Although blood still serves as an excellent fluid for laboratory-based analysis and sensor comparison, blood derivatives are favoured in the development of body-based sensors.
Saliva
Saliva can be easily collected or stimulated140, and has a diverse molecular composition. Although saliva is 99% water, it also contains an abundance of small molecules, proteins, DNA, and other molecular signatures that can be used to monitor physiology and diagnose disease141. Saliva is derived from blood plasma; however, its biochemical composition has little correlation with that of whole blood142. Therefore, the use of salivary markers to monitor physiology requires independent validation and testing. Saliva–blood glucose comparisons show decent similarity, with lag times of approximately 15 minutes143. Body-based sensing wearables for the analysis of saliva include mouthguards and pacifiers that can continuously monitor small molecules144,145. Unlike physiological markers, exogenous analytes, such as those from bacteria or viruses, are typically used in presence-based detection, which makes saliva an ideal testing fluid for infectious diseases141,146.
Sweat
Sweat can be non-invasively collected for physiological monitoring147. Small-molecule concentrations in sweat have demonstrated good correlation with blood, with a reported lag time of 10 minutes for glucose148. However, although wearable sweat sensors have been developed for ions and other small molecules20,55,149, sweat-based detection of proteins and other large molecules remains a challenge150. Protein concentrations in sweat correlate with blood concentrations in some instances151, but have remained difficult to reliably detect owing to their trace concentrations, often unexplored physiological correlation, and a lack of biomarker-focused sweat proteomic analyses147,152. Despite these hurdles, protein sensing in sweat has had some success (for example, in the detection of cytokines153,154) and has the potential to advance considerably as the physiological distribution of proteins in sweat becomes clearer150.
Sweat collection and sampling is straightforward as its secretion can be stimulated through heat, stress, chemicals or iontophoresis in addition to passive release147,155. However, the partitioning of analytes into sweat varies with the different stimulation approaches155. Furthermore, sweat composition is largely dependent on sweat rate, which influences the degree of analyte dilution155. Ionic strength differences among sweat collection conditions alter the electric double layer of electronic surfaces, which complicates detection schemes that rely on stable Debye parameters51,52. Sweat-based wearables can be incorporated into everyday life in the form of watches, tattoos or any other product in contact with skin156.
Tears
Tears have gained attention as a non-invasive fluid for biomolecular analysis owing to their external secretion and diverse biochemical composition, including metabolites, small molecules and proteins157. Although tear composition can differ substantially from that of blood, many tear-based analytes correlate closely with their corresponding blood concentrations158. Tear analyte concentrations lag behind those of blood, with a recorded lag time of approximately 13 minutes for glucose159. Advances in flexible electronics have allowed the development of fully integrated contact lenses that can sample tears without user intervention158 for continuous monitoring of tear glucose160,161. However, tear-based sensors face several challenges, including small sample volumes, fast evaporation rates and variations in tear secretion rates and composition depending on their source of stimulation162,163,164.
Interstitial fluid
ISF is a promising fluid for biomolecular analysis, owing to its compositional similarity to blood and potential for non-invasive collection165. Analyte partitioning between blood and ISF occurs mainly through continuous capillaries owing to high capillary densities, low flow rates and the ability to exchange even very large proteins (up to 1 MDa). Therefore, many analytes pass easily between these two fluids and have near-equal concentrations in both155. However, larger analytes (for example, proteins >10 kDa) typically exist at lower concentrations in ISF owing to their reliance on more complex transport pathways, with their ISF concentrations varying inversely with the logarithm of their molecular weight166. Analyte changes in blood are also detected quickly in ISF, with the lag time for glucose being approximately 5–6 minutes behind that of blood167. Although ISF-based continuous glucose monitors have been commercialized (for example, Dexcom’s G6 and Abbott’s FreeStyle Libre continuous glucose monitoring system168), there remains a demand for robust and simple ISF collection approaches with high extraction efficiency and resistance to potential contamination155. Microneedle patches, containing arrays of micrometre-sized needles, can be used to sample large volumes of ISF non-invasively169,170. Although ISF-based glucose sensing has been extensively optimized, fundamental research into ISF analyte partitioning, physiological correlation and lag times will be needed to allow the detection of other analytes171.
Breath
Breath contains volatile organic compounds, proteins, fatty acids and germ particles that can provide valuable information on physiology and disease172. Although the diversity and quantity of biologically relevant molecules in breath is lower than in other biological fluids, biomolecular breath sensors can be applied to detect small molecules, proteins and viruses172,173,174. In addition, synthetic biomarkers (for example, protease-responsive nanoparticles) may enable breath-based detection of various physiological conditions (for example, alpha-1 antitrypsin deficiency)24. Breath analysis can also be implemented in wearable sensors (such as masks) that can directly sample analytes from exhaled air132.
Digestive fluids
Digestive fluids may be analysed by consumable electronics (for example, diagnostic pills). The molecular biomarkers within digestive fluids are still being explored, but contain an abundance of biochemical information relating to health (for example, gut microbiota)175 and disease (for example, digestive system cancers)176. Unlike other bodily fluids, digestive fluids are characterized by high acidity and abundant enzymes, requiring highly resilient sensors177. Optical and electrochemical sensors can continuously monitor analytes within these harsh environments (for example, through amperometric detection of glucose in gastric and intestinal fluids178) and demonstrate success in animal models (for example, detecting gastrointestinal bleeding in swine using a wireless optical readout capsule specific for haem)179. Such consumable biomolecular sensors may improve access to and analysis of the gastrointestinal tract180.
Outlook
Biomolecular sensing has drawn substantial attention during the ongoing SARS-CoV-2 pandemic, with the rapid development and implementation of viral antigen tests demonstrating the power of such sensors to provide easy access to health information. Whereas glucose monitoring has traditionally dominated the biomolecular sensing landscape, body-based bioelectronic devices have sparked immense interest in the development of continuous sensors for alternative analytes that can operate in many body fluids180.
The development of body-based biomolecular sensors requires flexible and biocompatible electronic materials that can seamlessly interface with soft biological tissues and that can be manufactured using inexpensive, facile and easily scalable methods20,181. Materials such as conductive hydrogels and laser-engraved graphene can serve as sensing platforms20,182. Additionally, plastics and organic materials can serve as substrates and semiconductors benefiting from cost-efficiency, flexibility and potential biodegradability183. Metals that are fluid at room temperature (for example, gallium) also hold great promise for soft electronics development184. Application-dependent material considerations are also essential in body-based sensor development; for example, wearable sensors that can be integrated into clothing as smart textiles are non-intrusive and allow dispersed units to communicate easily185,186. Moreover, implantable sensors made with biofilm-resistant materials are resilient to internal physiological conditions187, and consumable sensors made with edible, biosafe materials are safe for user ingestion188. In addition, materials that can harvest biochemical or biomechanical energy (for example, biofuel cells189, triboelectric generators190 and piezoelectric generators191), store energy (for example, supercapacitors192), and enable multi-sensor communication (for example, surface-plasmon-like metamaterials193) will likely prove vital in the evolution of body-based platforms.
However, the clinical translation of biomolecular sensors will require proof of safety. Sensors must be sanitized prior to implementation194, have low component toxicity195 and minimal immunogenicity196. Furthermore, sensors must produce minimal irritation and stimuli (for example, loud sounds and bright lights) and they should be comfortable to wear197. Moreover, devices must be resistant to cyberattacks or other intrusive events198 and ensure user data protection199.
Body-based sensing systems will also advance sensing applications, such as in personalized healthcare to provide tailored diagnoses, interventions and management plans through access to real-time, patient-specific biomolecular data. Using this technology, it may be possible to identify effective drugs for different illnesses200, monitor and optimize drug dosages201, and detect early signs of disease. In addition, body-based sensors can provide preventative information, through metabolic and nutritional profiles, for example, to improve patient health and avoid disease202. Body-based systems also show promise in the development of advanced closed-loop systems, such as an artificial pancreas device capable of monitoring and delivering insulin, for the treatment of diabetes203. In addition to diabetes, these closed-loop systems may allow the treatment of various health conditions related to biomolecular deficiencies (for example, neural chemical imbalances and iron-deficiency anaemia).
The next generation of biomolecular sensors aims to advance biomolecular analysis by using advanced body-based systems to detect diverse analytes in complex biological environments. However, there are outstanding challenges regarding adequate signal amplification (for example, the need for enhanced sensitivity and flexible LoDs), practical continuous measurements (for example, the need for regenerable sensing approaches), biological fluid composition (for example, the need for strategies to overcome interferences in complex media), and device integration (for example, the need for reliable and practical sensors) that must be addressed prior to widespread adoption.
Citation diversity statement
We acknowledge that papers authored by scholars from minoritized groups are systematically under-cited. Here, we have made every attempt to reference relevant papers in a manner that is equitable in terms of racial, ethnic, gender and geographical representation.
References
Ji, S. et al. Matching the kinetics of natural enzymes with a single-atom iron nanozyme. Nat. Catal. 4, 407–417 (2021).
Armstrong, J. A. Urinalysis in Western culture: a brief history. Kidney Int. 71, 384–387 (2007).
Karamanou, M., Protogerou, A., Tsoucalas, G., Androutsos, G. & Poulakou-Rebelakou, E. Milestones in the history of diabetes mellitus: the main contributors. World J. Diabetes 7, 1–7 (2016).
Thomas, M. C., Jandeleit-Dahm, K. & Bonnet, F. Beyond glycosuria: exploring the intrarenal effects of SGLT-2 inhibition in diabetes. Diabetes Metab. 40, S17–S22 (2014).
Guthrie, D. W. & Humphreys, S. S. Diabetes urine testing: an historical perspective. Diabetes Educ. 14, 521–525 (1988).
Clark, L. C. Jr & Lyons, C. Electrode systems for continuous monitoring in cardiovascular surgery. Ann. NY Acad. Sci. 102, 29–45 (1962). This article describes the first electrochemical sensing platform that incorporates a biorecognition element to provide selectivity for an analyte.
Clark, L. C., Kaplan, S., Matthews, E. C., Edwards, F. K. & Helmsworth, J. A. Monitor and control of blood oxygen tension and pH during total body perfusion. J. Thorac. Surg. 36, 488–496 (1958).
Free, A. H., Adams, E. C., Kercher, M. L., Free, H. M. & Cook, M. H. Simple specific test for urine glucose. Clin. Chem. 3, 163–168 (1957). This article describes the first time a biorecognition element was incorporated into a sensing mechanism, despite this feat being usually misattributed to Clark and Lyons (1962).
Mazzaferri, E. L., Skillman, T. G., Lanese, R. R. & Keller, M. P. Use of test strips with colour meter to measure blood-glucose. Lancet 295, 331–333 (1970).
Chua, K. S. & Tan, I. K. Plasma glucose measurement with the Yellow Springs glucose analyzer. Clin. Chem. 24, 150–152 (1978).
Clarke, S. F. & Foster, J. R. A history of blood glucose meters and their role in self-monitoring of diabetes mellitus. Br. J. Biomed. Sci. 69, 83–93 (2012).
Matthews, D. R. et al. Pen-sized digital 30-second blood glucose meter. Lancet 329, 778–779 (1987).
Ginsberg, B. H. The FDA panel advises approval of the first continuous glucose sensor. Diabetes Technol. Ther. 1, 203–204 (1999).
Kelley, S. O. et al. Advancing the speed, sensitivity and accuracy of biomolecular detection using multi-length-scale engineering. Nat. Nanotechnol. 9, 969–980 (2014).
Sero, J. E. & Stevens, M. M. Nanoneedle-based materials for intracellular studies. In Bio-Nanomedicine For Cancer Therapy 191–219 (Springer, 2021).
Chiappini, C. et al. Mapping local cytosolic enzymatic activity in human esophageal mucosa with porous silicon nanoneedles. Adv. Mater. 27, 5147–5152 (2015).
Cao, Y. et al. Nondestructive nanostraw intracellular sampling for longitudinal cell monitoring. Proc. Natl Acad. Sci. USA 114, E1866–E1874 (2017).
Siciliano, V. et al. Engineering modular intracellular protein sensor–actuator devices. Nat. Commun. 9, 1881 (2018).
Poste, G. Bring on the biomarkers. Nature 469, 156–157 (2011).
Yang, Y. et al. A laser-engraved wearable sensor for sensitive detection of uric acid and tyrosine in sweat. Nat. Biotechnol. 38, 217–224 (2020).
Weinhofer, I. et al. Neurofilament light chain as a potential biomarker for monitoring neurodegeneration in X-linked adrenoleukodystrophy. Nat. Commun. 12, 1816 (2021).
Kwong, G. A. et al. Synthetic biomarkers: a twenty-first century path to early cancer detection. Nat. Rev. Cancer 21, 655–668 (2021). This review describes the development of synthetic biomarkers, including their function at the physiological level, their design and how they advance biomolecular sensing.
Nishihara, T. et al. Beta-galactosidase-responsive synthetic biomarker for targeted tumor detection. Chem. Commun. 54, 11745–11748 (2018).
Chan, L. W. et al. Engineering synthetic breath biomarkers for respiratory disease. Nat. Nanotechnol. 15, 792–800 (2020).
Aalipour, A. et al. Engineered immune cells as highly sensitive cancer diagnostics. Nat. Biotechnol. 37, 531–539 (2019).
Chen, J. et al. Glucose-oxidase like catalytic mechanism of noble metal nanozymes. Nat. Commun. 12, 3375 (2021).
Liu, S. et al. Metal–organic frameworks and their derivatives as signal amplification elements for electrochemical sensing. Coord. Chem. Rev. 424, 213520 (2020).
Huang, Y., Ren, J. & Qu, X. Nanozymes: classification, catalytic mechanisms, activity regulation, and applications. Chem. Rev. 119, 4357–4412 (2019).
Hu, W.-C. et al. Ultrasensitive detection of bacteria using a 2D MOF nanozyme-amplified electrochemical detector. Anal. Chem. 93, 8544–8552 (2021).
Huang, L., Chen, J., Gan, L., Wang, J. & Dong, S. Single-atom nanozymes. Sci. Adv. 5, eaav5490 (2019).
Jiao, L. et al. Single-atom catalysts boost signal amplification for biosensing. Chem. Soc. Rev. 50, 750–765 (2021).
Wang, Z. et al. Accelerated discovery of superoxide-dismutase nanozymes via high-throughput computational screening. Nat. Commun. 12, 6866 (2021).
Zhong, M. et al. Accelerated discovery of CO2 electrocatalysts using active machine learning. Nature 581, 178–183 (2020).
McConnell, E. M. et al. Biosensing with DNAzymes. Chem. Soc. Rev. 50, 8954–8994 (2021).
Borggräfe, J. et al. Time-resolved structural analysis of an RNA-cleaving DNA catalyst. Nature 601, 144–149 (2022).
Liu, K., Lat, P. K., Yu, H.-Z. & Sen, D. CLICK-17, a DNA enzyme that harnesses ultra-low concentrations of either Cu+ or Cu2+ to catalyze the azide–alkyne ‘click’ reaction in water. Nucleic Acids Res. 48, 7356–7370 (2020).
Liu, M. et al. Programming a topologically constrained DNA nanostructure into a sensor. Nat. Commun. 7, 12074 (2016).
Kang, D.-K. et al. Rapid detection of single bacteria in unprocessed blood using integrated comprehensive droplet digital detection. Nat. Commun. 5, 5427 (2014).
Shen, J., Liu, G., Han, Y. & Jin, W. Artificial channels for confined mass transport at the sub-nanometre scale. Nat. Rev. Mater. 6, 294–312 (2021).
de Angelis, F. et al. Breaking the diffusion limit with super-hydrophobic delivery of molecules to plasmonic nanofocusing SERS structures. Nat. Photon. 5, 682–687 (2011).
Morales-Narváez, E., Guix, M., Medina-Sánchez, M., Mayorga-Martinez, C. C. & Merkoçi, A. Micromotor enhanced microarray technology for protein detection. Small 10, 2542–2548 (2014).
Lin, M. et al. Programmable engineering of a biosensing interface with tetrahedral DNA nanostructures for ultrasensitive DNA detection. Angew. Chem. 127, 2179–2183 (2015).
Zhang, P. et al. Ultrasensitive detection of circulating exosomes with a 3D-nanopatterned microfluidic chip. Nat. Biomed. Eng. 3, 438–451 (2019).
Rivnay, J. et al. Organic electrochemical transistors. Nat. Rev. Mater. 3, 17086 (2018). This review describes the evolution of organic electrochemical transistors, including their fabrication, various configurations and their ability to amplify biochemical signals.
Rivnay, J. et al. Organic electrochemical transistors with maximum transconductance at zero gate bias. Adv. Mater. 25, 7010–7014 (2013).
Pappa, A. M. et al. Direct metabolite detection with an n-type accumulation mode organic electrochemical transistor. Sci. Adv. 4, eaat0911 (2018).
Liang, Y., Wu, C., Figueroa-Miranda, G., Offenhäusser, A. & Mayer, D. Amplification of aptamer sensor signals by four orders of magnitude via interdigitated organic electrochemical transistors. Biosens. Bioelectron. 144, 111668 (2019).
Ersman, P. A. et al. All-printed large-scale integrated circuits based on organic electrochemical transistors. Nat. Commun. 10, 5053 (2019).
Jarczewska, M., Rębiś, J., Górski, Ł. & Malinowska, E. Development of DNA aptamer-based sensor for electrochemical detection of C-reactive protein. Talanta 189, 45–54 (2018).
Reiber, T., Zavoiura, O., Dose, C. & Yushchenko, D. A. Fluorophore multimerization as an efficient approach towards bright protein labels. Eur. J. Org. Chem. 2021, 2817–2830 (2021).
Nakatsuka, N. et al. Aptamer-field-effect transistors overcome Debye length limitations for small-molecule sensing. Science 362, 319–324 (2018).
Kesler, V., Murmann, B. & Soh, H. T. Going beyond the Debye length: overcoming charge screening limitations in next-generation bioelectronic sensors. ACS Nano 14, 16194–16201 (2020).
Fu, K. et al. Accelerated electron transfer in nanostructured electrodes improves the sensitivity of electrochemical biosensors. Adv. Sci. 8, 2102495 (2021).
Lee, D. et al. Ionic contrast across a lipid membrane for Debye length extension: towards an ultimate bioelectronic transducer. Nat. Commun. 12, 3741 (2021).
Gao, W. et al. Fully integrated wearable sensor arrays for multiplexed in situ perspiration analysis. Nature 529, 509–514 (2016).
Cui, F., Yue, Y., Zhang, Y., Zhang, Z. & Zhou, H. S. Advancing biosensors with machine learning. ACS Sens. 5, 3346–3364 (2020).
King, R. C. et al. Application of data fusion techniques and technologies for wearable health monitoring. Med. Eng. Phys. 42, 1–12 (2017).
Habib, C., Makhoul, A., Darazi, R. & Salim, C. Self-adaptive data collection and fusion for health monitoring based on body sensor networks. IEEE Trans. Ind. Inf. 12, 2342–2352 (2016).
Yu, X., Yang, Y.-P., Dikici, E., Deo, S. K. & Daunert, S. Beyond antibodies as binding partners: the role of antibody mimetics in bioanalysis. Annu. Rev. Anal. Chem. 10, 293–320 (2017).
Belbruno, J. J. Molecularly imprinted polymers. Chem. Rev. 119, 94–119 (2019).
Muyldermans, S. Nanobodies: natural single-domain antibodies. Annu. Rev. Biochem. 82, 775–797 (2013).
Silverman, J. et al. Multivalent avimer proteins evolved by exon shuffling of a family of human receptor domains. Nat. Biotechnol. 23, 1556–1561 (2005).
Tiede, C. et al. Affimer proteins are versatile and renewable affinity reagents. eLife 6, e24903 (2017).
Löfblom, J. et al. Affibody molecules: engineered proteins for therapeutic, diagnostic and biotechnological applications. FEBS Lett. 584, 2670–2680 (2010).
Bratkovič, T. Progress in phage display: evolution of the technique and its applications. Cell. Mol. Life Sci. 67, 749–767 (2010).
Bradbury, A. R. M., Sidhu, S., Dübel, S. & McCafferty, J. Beyond natural antibodies: the power of in vitro display technologies. Nat. Biotechnol. 29, 245–254 (2011).
Tuerk, C. & Gold, L. Systematic evolution of ligands by exponential enrichment: RNA ligands to bacteriophage T4 DNA polymerase. Science 249, 505–510 (1990).
Ellington, A. D. & Szostak, J. W. In vitro selection of RNA molecules that bind specific ligands. Nature 346, 818–822 (1990).
Qian, S., Chang, D., He, S. & Li, Y. Aptamers from random sequence space: accomplishments, gaps and future considerations. Anal. Chim. Acta 1196, 339511 (2022).
Yu, H., Alkhamis, O., Canoura, J., Liu, Y. & Xiao, Y. Advances and challenges in small-molecule DNA aptamer isolation, characterization, and sensor development. Angew. Chem. Int. Edn 60, 16800–16823 (2021).
Wang, B. et al. Wearable aptamer-field-effect transistor sensing system for noninvasive cortisol monitoring. Sci. Adv. 8, eabk0967 (2022). This article demonstrates a fully integrated body-based sensing platform for the continuous detection of cortisol using field-effect transistors and aptamer recognition elements.
Zhao, C. et al. Implantable aptamer-field-effect transistor neuroprobes for in vivo neurotransmitter monitoring. Sci. Adv. 7, eabj7422 (2021).
Frutiger, A. et al. Nonspecific binding — fundamental concepts and consequences for biosensing applications. Chem. Rev. 121, 8095–8160 (2021). This review provides a detailed analysis of non-specific binding, including its history, physiological causes and solutions to overcome its effects.
Gawande, B. N. et al. Selection of DNA aptamers with two modified bases. Proc. Natl Acad. Sci. USA 114, 2898–2903 (2017).
Yoshikawa, A. M. et al. Discovery of indole-modified aptamers for highly specific recognition of protein glycoforms. Nat. Commun. 12, 7106 (2021).
Wang, J. et al. Multiparameter particle display (MPPD): a quantitative screening method for the discovery of highly specific aptamers. Angew. Chem. Int. Edn Engl. 129, 762–765 (2017).
Zhou, J. & Rossi, J. Aptamers as targeted therapeutics: current potential and challenges. Nat. Rev. Drug. Discov. 16, 181–202 (2017).
Gerling, T., Kube, M., Kick, B. & Dietz, H. Sequence-programmable covalent bonding of designed DNA assemblies. Sci. Adv. 4, eaau1157 (2018).
Anastassacos, F. M., Zhao, Z., Zeng, Y. & Shih, W. M. Glutaraldehyde cross-linking of oligolysines coating DNA origami greatly reduces susceptibility to nuclease degradation. J. Am. Chem. Soc. 142, 3311–3315 (2020).
Biedermann, F. & Schneider, H.-J. Experimental binding energies in supramolecular complexes. Chem. Rev. 116, 5216–5300 (2016).
Bixler, G. D. & Bhushan, B. Biofouling: lessons from nature. Phil. Trans. R. Soc. A 370, 2381–2417 (2012).
Ostuni, E. et al. Self-assembled monolayers that resist the adsorption of proteins and the adhesion of bacterial and mammalian cells. Langmuir 17, 6336–6343 (2001).
Liu, S. & Guo, W. Anti-biofouling and healable materials: preparation, mechanisms, and biomedical applications. Adv. Funct. Mater. 28, 1800596 (2018).
Li, S. et al. Slippery liquid-infused microphase separation surface enables highly robust anti-fouling, anti-corrosion, anti-icing and anti-scaling coating on diverse substrates. Chem. Eng. J. 431, 133945 (2022).
Zhang, L. et al. Zwitterionic hydrogels implanted in mice resist the foreign-body reaction. Nat. Biotechnol. 31, 553–556 (2013).
Chan, D. et al. Combinatorial polyacrylamide hydrogels for preventing biofouling on implantable biosensors. Adv. Mater. 34, 2109764 (2022).
del Río, J. S., Henry, O. Y. F., Jolly, P. & Ingber, D. E. An antifouling coating that enables affinity-based electrochemical biosensing in complex biological fluids. Nat. Nanotechnol. 14, 1143–1149 (2019).
Timilsina, S. S. et al. Ultrarapid method for coating electrochemical sensors with antifouling conductive nanomaterials enables highly sensitive multiplexed detection in whole blood. Adv. Healthc. Mater. 11, 2102244 (2022).
Shiddiky, M. J. A., Vaidyanathan, R., Rauf, S., Tay, Z. & Trau, M. Molecular nanoshearing: an innovative approach to shear off molecules with AC-induced nanoscopic fluid flow. Sci. Rep. 4, 3716 (2014).
Xue, L. et al. Solid-state nanopore sensors. Nat. Rev. Mater. 5, 931–951 (2020).
Yang, B., Jiang, X., Fang, X. & Kong, J. Wearable chem-biosensing devices: from basic research to commercial market. Lab Chip 21, 4285–4310 (2021).
Fercher, C., Jones, M. L., Mahler, S. M. & Corrie, S. R. Recombinant antibody engineering enables reversible binding for continuous protein biosensing. ACS Sens. 6, 764–776 (2021).
Wilson, B. D. & Soh, H. T. Re-evaluating the conventional wisdom about binding assays. Trends Biochem. Sci. 45, 639–649 (2020).
Goode, J. A., Rushworth, J. V. H. & Millner, P. A. Biosensor regeneration: a review of common techniques and outcomes. Langmuir 31, 6267–6276 (2015).
Delport, F. et al. Real-time monitoring of DNA hybridization and melting processes using a fiber optic sensor. Nanotechnology 23, 065503 (2012).
Gilles, P. N., Wu, D. J., Foster, C. B., Dillon, P. J. & Chanock, S. J. Single nucleotide polymorphic discrimination by an electronic dot blot assay on semiconductor microchips. Nat. Biotechnol. 17, 365–370 (1999).
Clifford, A. et al. Strategies for biomolecular analysis and continuous physiological monitoring. J. Am. Chem. Soc. 143, 5281–5294 (2021).
Xiao, Y., Lubin, A. A., Heeger, A. J. & Plaxco, K. W. Label-free electronic detection of thrombin in blood serum by using an aptamer-based sensor. Angew. Chem. Int. Edn Engl. 44, 5456–5459 (2005).
Arroyo-Currás, N. et al. Real-time measurement of small molecules directly in awake, ambulatory animals. Proc. Natl Acad. Sci. USA 114, 645–650 (2017).
Dauphin-Ducharme, P., Ploense, K. L., Arroyo-Currás, N., Kippin, T. E. & Plaxco, K. W. Electrochemical aptamer-based sensors: a platform approach to high-frequency molecular monitoring in situ in the living body. Biomed. Eng. Technol. 2393, 479–492 (2022).
Ferguson, B. S. et al. Real-time, aptamer-based tracking of circulating therapeutic agents in living animals. Sci. Transl. Med. 5, 213ra165 (2013).
Idili, A., Gerson, J., Kippin, T. & Plaxco, K. W. Seconds-resolved, in situ measurements of plasma phenylalanine disposition kinetics in living rats. Anal. Chem. 93, 4023–4032 (2021).
Seo, J. W. et al. Real-time monitoring of drug pharmacokinetics within tumor tissue in live animals. Sci. Adv. 8, eabk2901 (2022). This article describes the real-time monitoring of doxorubicin in animal tumour tissue, demonstrating the effectiveness of reagentless sensing approaches in animal models.
Fan, C., Plaxco, K. W. & Heeger, A. J. Electrochemical interrogation of conformational changes as a reagentless method for the sequence-specific detection of DNA. Proc. Natl Acad. Sci. USA 100, 9134–9137 (2003).
Xiao, Y., Lubin, A. A., Baker, B. R., Plaxco, K. W. & Heeger, A. J. Single-step electronic detection of femtomolar DNA by target-induced strand displacement in an electrode-bound duplex. Proc. Natl Acad. Sci. USA 103, 16677–16680 (2006).
Li, C. et al. Design of DNA nanostructure-based interfacial probes for the electrochemical detection of nucleic acids directly in whole blood. Chem. Sci. 9, 979–984 (2018).
Yu, H. L. L., Maslova, A. & Hsing, I.-M. Rational design of electrochemical DNA biosensors for point-of-care applications. ChemElectroChem 4, 795–805 (2017).
Wang, M. et al. A reagentless triplex DNA junctions-based electrochemical DNA sensor using signal amplification strategy of CHA and tetraferrocene. Sens. Actuators B 358, 131496 (2022).
Ranallo, S., Porchetta, A. & Ricci, F. DNA-based scaffolds for sensing applications. Anal. Chem. 91, 44–59 (2019).
Parolo, C. et al. E-DNA scaffold sensors and the reagentless, single-step, measurement of HIV-diagnostic antibodies in human serum. Microsyst. Nanoeng. 6, 13 (2020).
Cash, K. J., Ricci, F. & Plaxco, K. W. An electrochemical sensor for the detection of protein–small molecule interactions directly in serum and other complex matrices. J. Am. Chem. Soc. 131, 6955–6957 (2009).
White, R. J. et al. Wash-free, electrochemical platform for the quantitative, multiplexed detection of specific antibodies. Anal. Chem. 84, 1098–1103 (2012).
Ogden, N. E., Kurnik, M., Parolo, C. & Plaxco, K. W. An electrochemical scaffold sensor for rapid syphilis diagnosis. Analyst 144, 5277–5283 (2019).
Ricci, F., Bonham, A. J., Mason, A. C., Reich, N. O. & Plaxco, K. W. Reagentless, electrochemical approach for the specific detection of double- and single-stranded DNA binding proteins. Anal. Chem. 81, 1608–1614 (2009).
Kurnik, M., Pang, E. Z. & Plaxco, K. W. An electrochemical biosensor architecture based on protein folding supports direct real-time measurements in whole blood. Angew. Chem. Int. Edn 59, 18442–18445 (2020).
Kang, D. et al. New architecture for reagentless, protein-based electrochemical biosensors. J. Am. Chem. Soc. 139, 12113–12116 (2017).
Das, J. et al. Reagentless biomolecular analysis using a molecular pendulum. Nat. Chem. 13, 428–434 (2021).
Yousefi, H. et al. Detection of SARS-CoV-2 viral particles using direct, reagent-free electrochemical sensing. J. Am. Chem. Soc. 143, 1722–1727 (2021).
Bertok, T. et al. Electrochemical impedance spectroscopy based biosensors: mechanistic principles, analytical examples and challenges towards commercialization for assays of protein cancer biomarkers. ChemElectroChem 6, 989–1003 (2019).
Flauzino, J. M. R. et al. Label-free and reagentless electrochemical genosensor based on graphene acid for meat adulteration detection. Biosens. Bioelectron. 195, 113628 (2022).
Cecchetto, J., Fernandes, F. C. B., Lopes, R. & Bueno, P. R. The capacitive sensing of NS1 Flavivirus biomarker. Biosens. Bioelectron. 87, 949–956 (2017).
Capaldo, P. et al. Circulating disease biomarker detection in complex matrices: real-time, in situ measurements of DNA/miRNA hybridization via electrochemical impedance spectroscopy. ACS Sens. 1, 1003–1010 (2016).
Luo, X., Xu, M., Freeman, C., James, T. & Davis, J. J. Ultrasensitive label free electrical detection of insulin in neat blood serum. Anal. Chem. 85, 4129–4134 (2013).
Kergoat, L., Piro, B., Berggren, M., Horowitz, G. & Pham, M.-C. Advances in organic transistor-based biosensors: from organic electrochemical transistors to electrolyte-gated organic field-effect transistors. Anal. Bioanal. Chem. 402, 1813–1826 (2012).
Hajian, R. et al. Detection of unamplified target genes via CRISPR–Cas9 immobilized on a graphene field-effect transistor. Nat. Biomed. Eng. 3, 427–437 (2019).
Wang, L. et al. Rapid and ultrasensitive electromechanical detection of ions, biomolecules and SARS-CoV-2 RNA in unamplified samples. Nat. Biomed. Eng. 6, 276–285 (2022).
Tai, T. Y. et al. Design and demonstration of tunable amplified sensitivity of AlGaN/GaN high electron mobility transistor (HEMT)-based biosensors in human serum. Anal. Chem. 91, 5953–5960 (2019).
Tyagi, S., Bratu, D. P. & Kramer, F. R. Multicolor molecular beacons for allele discrimination. Nat. Biotechnol. 16, 49–53 (1998).
Tuleuova, N. et al. Development of an aptamer beacon for detection of interferon-gamma. Anal. Chem. 82, 1851–1857 (2010).
Bai, Y., Shu, T., Su, L. & Zhang, X. Functional nucleic acid-based fluorescence polarization/anisotropy biosensors for detection of biomarkers. Anal. Bioanal. Chem. 412, 6655–6665 (2020).
Kruse, M. et al. Measuring influenza A virus and peptide interaction using electrically controllable DNA nanolevers. Adv. Mater. Technol. 7, 2101141 (2021).
Nguyen, P. Q. et al. Wearable materials with embedded synthetic biology sensors for biomolecule detection. Nat. Biotechnol. 39, 1366–1374 (2021).
Oh, S.-H. et al. Nanophotonic biosensors harnessing van der Waals materials. Nat. Commun. 12, 3824 (2021).
Aitekenov, S., Gaipov, A. & Bukasov, R. Review: Detection and quantification of proteins in human urine. Talanta 223, 121718 (2021).
Ploussard, G. & de La Taille, A. Urine biomarkers in prostate cancer. Nat. Rev. Urol. 7, 101–109 (2010).
Gray, M. et al. Implantable biosensors and their contribution to the future of precision medicine. Vet. J. 239, 21–29 (2018).
Soler, M. et al. Multiplexed nanoplasmonic biosensor for one-step simultaneous detection of Chlamydia trachomatis and Neisseria gonorrhoeae in urine. Biosens. Bioelectron. 94, 560–567 (2017).
Zhang, J. et al. A wearable self-powered biosensor system integrated with diaper for detecting the urine glucose of diabetic patients. Sens. Actuators B 341, 130046 (2021).
Lager, W. et al. Implantable electrocatalytic glucose sensor. Horm. Metab. Res. 26, 526–530 (1994).
Topkas, E., Keith, P., Dimeski, G., Cooper-White, J. & Punyadeera, C. Evaluation of saliva collection devices for the analysis of proteins. Clin. Chim. Acta 413, 1066–1070 (2012).
Zhang, C.-Z. et al. Saliva in the diagnosis of diseases. Int. J. Oral. Sci. 8, 133–137 (2016).
Bel’skaya, L. V., Sarf, E. A. & Kosenok, V. K. Age and gender characteristics of the biochemical composition of saliva: correlations with the composition of blood plasma. J. Oral Biol. Craniofac. Res. 10, 59–65 (2020).
Lin, C. et al. Toward the development of a glucose dehydrogenase-based saliva glucose sensor without the need for sample preparation. J. Diabetes Sci. Technol. 12, 83–89 (2018).
Arakawa, T. et al. Mouthguard biosensor with telemetry system for monitoring of saliva glucose: a novel cavitas sensor. Biosens. Bioelectron. 84, 106–111 (2016).
García-Carmona, L. et al. Pacifier biosensor: toward noninvasive saliva biomarker monitoring. Anal. Chem. 91, 13883–13891 (2019).
Kevadiya, B. D. et al. Diagnostics for SARS-CoV-2 infections. Nat. Mater. 20, 593–605 (2021).
Bariya, M., Nyein, H. Y. Y. & Javey, A. Wearable sweat sensors. Nat. Electron. 1, 160–171 (2018).
la Count, T. D., Jajack, A., Heikenfeld, J. & Kasting, G. B. Modeling glucose transport from systemic circulation to sweat. J. Pharm. Sci. 108, 364–371 (2019).
He, W. et al. Integrated textile sensor patch for real-time and multiplex sweat analysis. Sci. Adv. 5, eaax0649 (2019).
Pérez, D. & Orozco, J. Wearable electrochemical biosensors to measure biomarkers with complex blood-to-sweat partition such as proteins and hormones. Microchim. Acta 189, 127 (2022).
Cizza, G. et al. Elevated neuroimmune biomarkers in sweat patches and plasma of premenopausal women with major depressive disorder in remission: the P.O.W.E.R. study. Biol. Psychiat. 64, 907–911 (2008).
Qiao, L., Benzigar, M. R., Subramony, J. A., Lovell, N. H. & Liu, G. Advances in sweat wearables: sample extraction, real-time biosensing, and flexible platforms. ACS Appl. Mater. Interf. 12, 34337–34361 (2020).
Jagannath, B. et al. Novel approach to track the lifecycle of inflammation from chemokine expression to inflammatory proteins in sweat using electrochemical biosensor. Adv. Mater. Technol. 7, 2101356 (2022).
Jagannath, B. et al. Temporal profiling of cytokines in passively expressed sweat for detection of infection using wearable device. Bioeng. Transl. Med. 6, e10220 (2021).
Heikenfeld, J. et al. Accessing analytes in biofluids for peripheral biochemical monitoring. Nat. Biotechnol. 37, 407–419 (2019). This review discusses opportunities and challenges for biomolecular sensing in various biological fluids, with information on biological fluid generation, analyte partitioning and sampling technologies.
Kaya, T. et al. Wearable sweat sensors: background and current trends. Electroanalysis 31, 411–421 (2019).
You, J. et al. Tear fluid protein biomarkers. Adv. Clin. Chem. 62, 151–196 (2013).
Farandos, N. M., Yetisen, A. K., Monteiro, M. J., Lowe, C. R. & Yun, S. H. Contact lens sensors in ocular diagnostics. Adv. Healthc. Mater. 4, 792–810 (2015).
la Belle, J. T. et al. Self-monitoring of tear glucose: the development of a tear based glucose sensor as an alternative to self-monitoring of blood glucose. Chem. Commun. 52, 9197–9204 (2016).
Kim, J. et al. Wearable smart sensor systems integrated on soft contact lenses for wireless ocular diagnostics. Nat. Commun. 8, 14997 (2017).
Elsherif, M., Hassan, M. U., Yetisen, A. K. & Butt, H. Wearable contact lens biosensors for continuous glucose monitoring using smartphones. ACS Nano 12, 5452–5462 (2018).
Kim, J., Campbell, A. S., de Ávila, B. E. F. & Wang, J. Wearable biosensors for healthcare monitoring. Nat. Biotechnol. 37, 389–406 (2019).
Sempionatto, J. R., Jeerapan, I., Krishnan, S. & Wang, J. Wearable chemical sensors: emerging systems for on-body analytical chemistry. Anal. Chem. 92, 378–396 (2020).
Stuchell, R. N., Feldman, J. J., Farris, R. L. & Mandel, I. D. The effect of collection technique on tear composition. Invest. Ophthalmol. Vis. Sci. 25, 374–377 (1984).
Kim, Y. & Prausnitz, M. R. Sensitive sensing of biomarkers in interstitial fluid. Nat. Biomed. Eng. 5, 3–5 (2021).
Vermeer, B. J., Reman, F. C. & van Gent, C. M. The determination of lipids and proteins in suction blister fluid. J. Investig. Dermatol. 73, 303–305 (1979).
Basu, A. et al. Time lag of glucose from intravascular to interstitial compartment in humans. Diabetes 62, 4083–4087 (2013).
Soni, A. et al. A practical approach to continuous glucose monitoring (rtCGM) and FreeStyle Libre systems (isCGM) in children and young people with type 1 diabetes. Diabetes Res. Clin. Pract. 184, 109196 (2022).
Samant, P. P. et al. Sampling interstitial fluid from human skin using a microneedle patch. Sci. Transl. Med. 12, eaaw0285 (2020).
Samant, P. P. & Prausnitz, M. R. Mechanisms of sampling interstitial fluid from skin using a microneedle patch. Proc. Natl Acad. Sci. USA 115, 4583–4588 (2018).
Friedel, M. et al. Opportunities and challenges in the diagnostic utility of dermal interstitial fluid. Nat. Biomed. Eng. https://doi.org/10.1038/s41551-022-00998-9 (2023). This Perspective provides a detailed analysis of the opportunities and challenges related to interstitial-fluid-based sensing and provides a roadmap for future interstitial fluid sensor development.
Vasilescu, A., Hrinczenko, B., Swain, G. M. & Peteu, S. F. Exhaled breath biomarker sensing. Biosens. Bioelectron. 182, 113193 (2021).
Chen, H. et al. Automated in vivo nanosensing of breath-borne protein biomarkers. Nano Lett. 18, 4716–4726 (2018).
Ates, H. C. & Dincer, C. Wearable breath analysis. Nat. Rev. Bioeng. 1, 80–82 (2023).
McBurney, M. I. et al. Establishing what constitutes a healthy human gut microbiome: state of the science, regulatory considerations, and future directions. J. Nutr. 149, 1882–1895 (2019).
Melton, S. D., Genta, R. M. & Souza, R. F. Biomarkers and molecular diagnosis of gastrointestinal and pancreatic neoplasms. Nat. Rev. Gastroenterol. Hepatol. 7, 620–628 (2010).
Salama, R., Arshavsky-Graham, S., Sella-Tavor, O., Massad-Ivanir, N. & Segal, E. Design considerations of aptasensors for continuous monitoring of biomarkers in digestive tract fluids. Talanta 239, 123124 (2022).
Ruiz-Valdepeñas Montiel, V. et al. Direct electrochemical biosensing in gastrointestinal fluids. Anal. Bioanal. Chem. 411, 4597–4604 (2019).
Mimee, M. et al. An ingestible bacterial-electronic system to monitor gastrointestinal health. Science 360, 915–918 (2018).
Beardslee, L. A. et al. Ingestible sensors and sensing systems for minimally invasive diagnosis and monitoring: the next frontier in minimally invasive screening. ACS Sens. 5, 891–910 (2020).
Yang, Y. & Gao, W. Wearable and flexible electronics for continuous molecular monitoring. Chem. Soc. Rev. 48, 1465–1491 (2019).
Wang, L., Xu, T. & Zhang, X. Multifunctional conductive hydrogel-based flexible wearable sensors. TrAC Trends Anal. Chem. 134, 116130 (2021).
Someya, T., Bao, Z. & Malliaras, G. G. The rise of plastic bioelectronics. Nature 540, 379–385 (2016).
Park, Y.-G. et al. Liquid metal-based soft electronics for wearable healthcare. Adv. Healthc. Mater. 10, 2002280 (2021).
Libanori, A., Chen, G., Zhao, X., Zhou, Y. & Chen, J. Smart textiles for personalized healthcare. Nat. Electron. 5, 142–156 (2022).
Wang, L. et al. Functionalized helical fibre bundles of carbon nanotubes as electrochemical sensors for long-term in vivo monitoring of multiple disease biomarkers. Nat. Biomed. Eng. 4, 159–171 (2020).
Shah, S. R., Tatara, A. M., D’Souza, R. N., Mikos, A. G. & Kasper, F. K. Evolving strategies for preventing biofilm on implantable materials. Mater. Today 16, 177–182 (2013).
Kim, J. et al. Edible electrochemistry: food materials based electrochemical sensors. Adv. Healthc. Mater. 6, 1700770 (2017).
Yin, L. et al. A passive perspiration biofuel cell: high energy return on investment. Joule 5, 1888–1904 (2021).
Hinchet, R. et al. Transcutaneous ultrasound energy harvesting using capacitive triboelectric technology. Science 365, 491–494 (2019).
Ali, F., Raza, W., Li, X., Gul, H. & Kim, K.-H. Piezoelectric energy harvesters for biomedical applications. Nano Energy 57, 879–902 (2019).
Jeong, Y. R., Lee, G., Park, H. & Ha, J. S. Stretchable, skin-attachable electronics with integrated energy storage devices for biosignal monitoring. Acc. Chem. Res. 52, 91–99 (2019).
Hajiaghajani, A. et al. Textile-integrated metamaterials for near-field multibody area networks. Nat. Electron. 4, 808–817 (2021).
Chung, J., Sepunaru, L. & Plaxco, K. W. On the disinfection of electrochemical aptamer-based sensors. ECS Sens. Plus 1, 011604 (2022).
Zhang, Y., Bindra, D. S., Barrau, M.-B. & Wilson, G. S. Application of cell culture toxicity tests to the development of implantable biosensors. Biosens. Bioelectron. 6, 653–661 (1991).
Avula, M. N., Rao, A. N., McGill, L. D., Grainger, D. W. & Solzbacher, F. Foreign body response to subcutaneous biomaterial implants in a mast cell-deficient Kit(w-Sh) murine model. Acta Biomater. 10, 1856–1863 (2014).
Fang, Y.-M. & Chang, C.-C. Users’ psychological perception and perceived readability of wearable devices for elderly people. Behav. Inf. Technol. 35, 225–232 (2016).
Pycroft, L. & Aziz, T. Z. Security of implantable medical devices with wireless connections: the dangers of cyber-attacks. Expert Rev. Med. Devices 15, 403–406 (2018).
Sadowski, J., Viljoen, S. & Whittaker, M. Everyone should decide how their digital data are used — not just tech companies. Nature 595, 169–171 (2021).
Teymourian, H. et al. Wearable electrochemical sensors for the monitoring and screening of drugs. ACS Sens. 5, 2679–2700 (2020).
Goud, K. Y. et al. Wearable electrochemical microneedle sensor for continuous monitoring of Levodopa: toward Parkinson management. ACS Sens. 4, 2196–2204 (2019).
Sempionatto, J. R., Montiel, V. R.-V., Vargas, E., Teymourian, H. & Wang, J. Wearable and mobile sensors for personalized nutrition. ACS Sens. 6, 1745–1760 (2021).
Thabit, H. & Hovorka, R. Coming of age: the artificial pancreas for type 1 diabetes. Diabetologia 59, 1795–1805 (2016).
Biosensors market size by type (wearable, non-wearable), by technology (electrochemical, optical, thermal, piezoelectric), by medical application (blood glucose testing, cholesterol testing, blood gas analysis, pregnancy testing, drug discovery. Global Market Insights https://www.gminsights.com/industry-analysis/biosensors-market (2022).
Acknowledgements
The authors thank A. Jiao for her illustrative contributions to the manuscript.
Author information
Authors and Affiliations
Contributions
All authors contributed to the writing of the manuscript. C.D.F. and S.O.K. edited the manuscript.
Corresponding author
Ethics declarations
Competing interests
S.O.K. and H.Y. are cofounders and equity holders in Arma Biosciences, which is commercializing new sensing technologies.
Peer review
Peer review information
Nature Reviews Bioengineering thanks Chunhai Fan and the other, anonymous, reviewer(s) for their contribution to the peer review of this work.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Flynn, C.D., Chang, D., Mahmud, A. et al. Biomolecular sensors for advanced physiological monitoring. Nat Rev Bioeng 1, 560–575 (2023). https://doi.org/10.1038/s44222-023-00067-z
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s44222-023-00067-z
This article is cited by
-
Motion artefact management for soft bioelectronics
Nature Reviews Bioengineering (2024)
-
Anti-freezing Dough for Renewable and Reconfigurable Flexible Strain Sensors
Journal of Electronic Materials (2024)
-
Capacitive spectroscopy as transduction mechanism for wearable biosensors: opportunities and challenges
Analytical and Bioanalytical Chemistry (2024)
-
Breaking barriers in electrochemical biosensing using bioinspired peptide and phage probes
Analytical and Bioanalytical Chemistry (2024)
-
Wearable flexible microfluidic sensing technologies
Nature Reviews Bioengineering (2023)