Abstract
Abnormal activity of oncogenic and tumor-suppressor signaling pathways contributes to cancer and cancer risk in humans. Transcriptional dysregulation of these pathways is commonly associated with tumorigenesis and the development of cancer. Genetic and epigenetic alterations may mediate dysregulated transcriptional activity. One of the most important epigenetic alternations is the non-coding regulatory element, which includes both enhancers and super-enhancers (SEs). SEs, characterized as large clusters of enhancers with aberrant high levels of transcription factor binding, have been considered as key drivers of gene expression in controlling and maintaining cancer cell identity. In cancer cells, oncogenes acquire SEs and the cancer phenotype relies on these abnormal transcription programs driven by SEs, which leads to cancer cells often becoming addicted to the SEs-related transcription programs, including prostate cancer. Here, we summarize recent findings of SEs and SEs-related gene regulation in prostate cancer and review the potential pharmacological inhibitors in basic research and clinical trials.
Similar content being viewed by others
The hallmarks of cancer, including sustaining proliferation, activating invasion, metastasis, and aberrant replicative immortality, are closely connected to cancer-specific regulatory mechanisms of gene expression1. Prostate cancer (PCa) is one of the leading causes of cancer-related deaths in men worldwide2,3. Although clinically beneficial treatment options for localized PCa include surgery, radiotherapy, and androgen ablation therapy, there is basically no cure for metastatic castration-resistant PCa (CRPC)4,5,6. Therefore, it is most necessary than ever to further understand the crucial regulators within the PCa and develop new therapies7.
The enhancer is a class of regulatory DNA sequence that defines the genetic regulatory circuitry8. It increases the promoter activity to activate target genes through specific transcription factors (TF)9. For cancer-type-specific gene expression, enhancer closely interact with the promoter, over either short or long distances, independent of the corresponding orientation and position concerning the transcription start sites (TSS)10,11. Enhancers often contain conserved recognition DNA sites for RNA polymerase, TFs, and co-activators and function as binding platforms10,11,12. Accumulated evidence reveals that enhancers are bound by epigenetic modifications, such as mono-methylation at H3 lysine 4 (H3K4me1) and acetylation at H3 lysine 27(H3K27ac), generally13,14. In addition, the three-dimensional structure of enhancers to TSS affects the interaction activity and expression output, with the distance varying from less than 10 Kb to more than 1 Mb15,16.
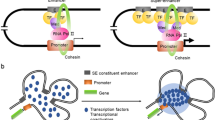
Super-enhancer (SE) is defined as large clusters of enhancers spanning across a long-range region of genomic DNA that drives stronger transcriptional activation ability than individual enhancers4. Similar to the typic enhancer, SE incorporates modelized regulation mechanisms. Specific TFs bind to SE to trigger promoter-enhancer interaction, mediated by chromatin looping, to load the SE to the cognate promoter. Then the basal machinery is recruited to initiate the downstream transcription activity (depicted in Fig. 1). The SE was first proposed in mouse embryonic stem cells (mESCs), by chromatin immunoprecipitation (ChIP)-sequencing analysis of active histone marker (H3K27ac) and other TFs17. The ROSE (Rank ordering of super-enhancers) algorithm is designed to search SEs by locating genomic proximity for grouping elements to a putative target gene17,18. Generally accepted models of SEs are considered to be large clusters of regulatory elements (after over 20 Kb) binding with dense transcriptional coactivators, such as BRD4 and CDK7, and with high potential to activate target gene expression output19 (summarized in Table 1). As shown in mESCs, pluripotency genes like OCT4, SOX2, and NANOG are all controlled and activated by SEs, given the concept that SEs can drive specific gene expression that controls and defines the cell identity and engages in cell-type-specific biological processes20. It is worth noting that the expression level of SE-related genes is significantly higher than that of the typical enhancer-control genes, which has been widely validated in a variety of cancer types21. On the other hand, this addiction makes SEs and SE-related genes as potential therapeutic targets and diagnosis22.
The most cutting-edge research has revealed that membrane-less organelles, such as the nucleolus, stress granule, processing body, and nuclear speckles form subcellular compartments to facilitate signaling transduction and transcriptional regulation by liquid–liquid phase separation23,24,25. In essence, the SE can be described as a co-assembly of high-density transcription factors, co-factors, chromatin regulators, non-coding RNA, and RNA polymerases26,27. Computer simulations and experimental validations indicate that, in the context of the number and valence of interacting components and the affinity of the interaction between transcription factors and nucleic acids, phase separations play essential roles in the SE assembly and function26,28. Inhibition of the activity of SE function highlights sensitivity and vulnerability in cancers along with cancer-specific oncogene downregulation, which could be a possible therapeutic target for cancer treatment29. SE complexes are dynamic, highly enriched condensates, particularly sensitive to SE-related inhibition26,28. The essential components of SE are dysregulated in PCa, such as increased MED1 phosphorylation at T1457 in a CDK7-dependent manner in metastatic CRPC and enzalutamide-resistant cells. Bromodomain-containing proteins (BRDs) are overexpressed in CRPC and show particular prognostic value in PCa.
Given the cancer-specific SE complexes in PCa, we summarize the critical transcriptional regulation and therapeutic targets of the SEs and SE-associated regulator proteins and provide promising future directions for SEs in the prostate cancer community (depicted in Fig. 2).
Proposed mode of the mechanism of super-enhancer-mediated action in prostate cancer. H3K27ac, acetylation of histone 3 lysine 27; AR, androgen receptor; BRD2/3/4, bromodomain containing protein 2/3/4; CDK7, cyclin dependent kinase 7; CDK9, cyclin dependent kinase 9; ERG, erythroblast transformation-specific-related gene; MED1, mediator complex subunit 1; RNA pol II, RNA polymerase II.
SE-related protein BET/BRD4 in prostate cancer
The bromodomain and extra-terminal domain (BET) family proteins act as “readers” of acetylated histones, and are important transcriptional regulators. BRD2, BRD3, BRD4, and BRDT (bromodomain testis-specific protein) are part of the BET family30. BRDT is most commonly expressed in germ cells, while the other three are ubiquitously expressed. BRD2, BRD3, BRD4, and BRDT share an extra-terminal domain and conserved N-terminal bromodomains (BD1 and BD2). BRD4, as a chromatin reader, recognizes acetylated histones and facilitates the transcription activation by propelling the recruitment of the positive transcription elongation factor P-TEFB31. In particular, BRD4 shows a density binding activity in SE and drives the cell-identical gene expression32. In PCa, AR-positive or AR-signaling-component cell lines (VCaP, LNCaP, and 22RV1) are selectively sensitive to BRD4 inhibition, but not in AR-negative cell lines (PC3 and DU145)33,34. BRD4 has been found that sequence-specific DNA-binding TFs may physically interact with it in a gene-specific manner, such as AR35. Mechanically, BRD4 physically interacts with the N-terminal domain of AR, and BRD4 colocalizes with AR at AR target loci to drive AR-mediated gene transcription. For the BMPR1B gene, AR-and BRD4-associated binding signals in the enhancers and SEs are significantly induced by dihydrotestosterone (DHT) treatment. The expression levels of BMPR1B are corroborating with the ChIP-seq data. Moreover, almost half of the known BRD-containing proteins are related to PCa, which also contribute the main chromatin-related processes and changes in PCa, beyond BRD436. These BRD-containing proteins (such as ATAD2, BRD8, CREBBP, and KTM2A) perform a wide range of downstream functions by recognizing acetylated histones, also present to be TFs, AR co-activators or methyltransferases37,38,39. Therefore, BRD-containing proteins are important transcriptional regulators that initiate chromatin restructuring beyond the SE function.
AR signaling remains the most common resistance mechanism in most CRPC patients40. On one hand, BRD4, by functioning downstream of AR signaling, appears to be effectively blocking the oncogenic drivers of PCa and less likely to be bypassed by acquired treatment resistance of AR therapy. BRD4 inhibition preferentially blocks both BRD4 and AR recruitment (the BRD4 and AR cistrome) to target loci on a genome-wide scale and leads to defects in transcriptional elongation. The acquisition of BRD4-associated SEs leads to prompt expression of key oncogenic genes, including TMPRSS2-ETS, KLK3, and BMPR1B in PCa, especially CRPC. Urbanucci et al. demonstrated that the transcription of certain BET proteins (BRD2 and BRD4), and ATAD2, a recently reported common coactivator of AR, may be induced by androgen41. The upregulated AR leads to a positive loop that boosts the expression of BRDs to expand the AR cistrome by increasing chromatin accessibility. On the other hand, BET inhibitors can also operate via mechanisms other than AR signaling. Shah et al. demonstrated that BET inhibitors resensitized drug-resistant tumors to enzalutamide by inhibiting the glucocorticoid receptor42. Cai et al. showed that the BRD4 inhibitor, JQ1, significantly inhibited androgen-independent growth of CRPC cells in vitro and in vivo by blocking AR-V7 chromatin binding and transcriptional programs co-activated by AR-V7 and ZFX in addition to the canonical AR signaling43.
SE-related protein CDK7 in prostate cancer
The mammalian cyclin-dependent kinases (CDKs) contain subfamilies with specific functions related to cell cycle (CDK1, CDK2, CDK4, CDK6) and transcriptional regulation (CDK7, CDK8, CDK9, CDK12, and CDK19)44,45. CDK7 is a ubiquitously expressed kinase that activates the cell cycle controlling through phosphorylation of CDKs and plays a key role in transcription regulation46. The mRNA and protein expression levels of CDK7 alone showed no significant change in benign cells and PCa cells47. However, CDK7 activity affects chromatin modification by promoting the recruitment of histone methyltransferases SETD1A/B and SETD2, which are phosphorylated by the C-terminal domain48,49. CDK7 also phosphorylates a range of TFs, including p53 and nuclear hormone receptors (RAR, AR, ER, etc.), to promote the transcription activation activity and subsequent protein-degradation50,51,52,53. Studies show that CDK7 inhibition can transiently block the functional activity and expression of the oncogenic drivers (AR, ETS, MYC, and E2F) in prostate cancer, and all these changes require the involvement of MED1 as a cofactor47,54,55,56. CDK7 activates MED1 via ligand-specific phosphorylation, and the IDRs (intrinsically disordered region) of MED1 form phase-separation properties to be recruited to the AR-bound SEs, resulting in the high-density assembly of the SE-related transcription apparatus. CDK7 inhibition has been effectively tested in several aggressive cancer types, including MYCN-amplified neuroblastoma, T-cell acute lymphoblastic leukemia, triple-negative breast cancer, and small-cell lung cancer50,57,58,59,60. The CDK7-related SEs mediate the recruitment of AR and RNA polymerase II to boost the expression of a host of target genes, known as the “Achilles cluster” genes, thus becoming transcriptional addictive and sensitive to CDK7 inhibition50.
SE-related protein ERG in prostate cancer
The erythroblast transformation-specific (ETS) family proteins are the essential transcription factors necessary for cell-type-specific lineage differentiation and expression patterns61. ERG protein is the master transcription factor for endothelial, hematopoietic, and luminal cell differentiation62,63,64,65. It interacts with other transcription factors by forming complexes to establish the cell-type-specific patterns. In PCa, ERG expression and rearrangement is per se not a strong prognostic biomarker and only relevant in the context of a specific molecular subtype66,67,68. Gerke et al. found that it was crucial to determine the prognostic value with other biomarkers, such as RRM2 and TYMS, when applying the ERG status to predict the outcomes69. ChIP-seq data show ERG binds to the vast majority of SEs in VCaP (a TMPRSS2-EGR fusion-positive cell line) cell65. The SE-associated lineage-specific machinery of ERG is linked with the TFs like FOXA1 and HOXB13, which play important roles in prostate-cancer-specific gene expression. Mechanically, ERG increases the SEs activity partially through the BRG1-associated chromatin remodeling complex to establish accessible chromatin of SEs and transcriptional regulation. In summary, ERG drives the prostate-cancer-specific lineage genes by regulating SEs. TMPRSS2-ERG structural rearrangements occur in close to 50% of PCa patients and contribute to the ERG overexpression70. The proposed model of ERG overexpression was previously thought to be driven by TMPRSS2 promoter hijacking. Ken et al. showed that TMPRSS2, along with the rearranged ERG allele, formed an expanded SE71. The expanded SE still contains cis-regulatory elements and extends into the ERG locus, which promotes ERG overexpression. This work firstly confirms that the expansion of the SE region after chromosomal rearrangements could positively drive the target gene expression. As for ERG, it synergistically regulates by physically interacting with prostate-cancer-specific regulators AR, HOXB13, and FOXA1. The regulatory landscape difference between TMPRSS2-ERG fusion and non-fusion PCa types may depend on the SE-related ERG-specific transcriptional profile, including activated NOTCH pathway. In light of the present findings, we conclude that ERG contributes to the SE-driven oncogenic transcriptional addiction, and that SEs lead to the overexpression of the ERG gene, leading to subsequent overexpression of ERG-target genes that drive the development of PCa.
Other SE-associate factors in prostate cancer
Abundant evidence shows that the transcriptional regulatory regions of SEs are practically enriched with cancer-related single nucleotide polymorphisms (SNPs), resulting in dysregulation of target genes and contribution to cancer development72,73,74. Chen et al. demonstrated that high enrichment of PCa-specific risk variants in SE regions, particularly in the disease-specific regulatory regions and the DNA regulatory elements, may lead to prostate carcinogenesis75. O-GlcNAc transferase (OGT), a glycosyltransferase, catalyzes the addition of a single O-GlcNAc sugar to serine and threonine residues76. OGT, as a major metabolic integration point in human cells, is also involved in the hexosamine biosynthetic pathway (HBP) to increase O-GlcNAcylation-modified nuclear proteins77. RNA polymerase II is reported to be the most prominent O-GlcNAcylation-modified protein that regulates the formation of the transcriptional pre-initiation complex formation. Also, increased OGT expression has been found in many cancers, including prostate cancer, where high O-GlcNAc protein levels are associated with poor clinical prognosis. Harri et al. showed that OGT regulated SE-dependent transcription through chromatin compaction78. Over 70% of the SE-related genes are related to the OGT chromatin mark, and the OGT inhibition significantly decreases the SE-related mRNA expression. The activity of OGT is required for the expression of MYC-target mitotic proteins. Thus, OGT could be an effective target for MYC-addicted PCa.
AR is a predominant target for PCa, owing to the functional AR signaling in the early-and late-stage PCa. Simon et al. demonstrated that R1881-activated AR bound at a comparable number of SE sites in VCaP cells, and indicated that AR attributed to the SE-associated action79. The androgen-regulated SE-target genes are associated with cell-proliferation-associated gene sets, stem cell properties, and hallmark AR signaling, such as KLK2, KLK3, TMPRRSS2, and FKBP5. In addition, abundant evidence shows that AR also tightly cross-talks with other factors80. In low androgen environments or AR-overexpression status, AR promotes the expression of some SE-complex-associated proteins, such as BRD441. AR closely interacts with the BRDs in the N-terminal domain (NTD) instead of the ligand-binding domain (LBD), since the NTD was essential for the transcriptional activity of AR81. Alternative splicing variants of the AR that lack LBD are overexpressed in patients who are resistant to enzalutamide or the androgen synthesis inhibitor abiraterone acetate82. For AR mutants (full-length AR with point-mutated forms and AR with lack of LBD splice variants and nonsense mutants), such as AR-V7, AR T878A, and AR H875Y, some BET inhibitors (PFI-1 and BETi) could reduce these mutants’ expression by regulating RNA processing and reducing alternative splicing83,84. In this regard, we suspect that the formation of AR-bound-SE may be partly affected and the cistrome of AR mutants could redistribute while retaining some binding sites. The current inhibitors may still be effective for AR mutants, but the extent of the effect still needs to be specifically evaluated.
The forkhead box A1 (FOXA1) transcription factor plays a pivotal role for the development and differentiation of several endoderm-derived organs, including prostate85. FOXA1 directly binds to and de-compacts condensed chromatin to increase accessibility of the binding sites for partnering nuclear hormone receptors, including estrogen receptor and AR86. Lupien et el. showed that FOXA1 functionally collaborated with AR and was predominantly recruited to the AR-regulated enhancers and SEs to establish a lineage-specific program in PCa87. FOXA1 plays a key role in prostate tumorigenesis by reprogramming the AR cistrome to new binding sites and driving the transformation of normal prostate epithelial cells88. Besides, FOXA1 also interacts directly with AR89. Considering the AR-bound-SE complexes, the alterations in FOXA1 may impact subsequent effects on the AR cistrome and lineage-specific programs in PCa.
Promising pharmacological targets
SEs drive PCa cells into becoming addicted to dysregulated transcriptional programs mediated by BRD4, CDK7, ERG, and other factors, but also become a powerful rationale for therapeutic interventions. Targeting SE may disrupt the dysregulated networks of the oncogenic functions, and some small molecule inhibitors and blockers have been tested to selectively target PCa cells. We summarize these inhibitors and their mechanisms as below and an overview description in Table 2.
First, BET proteins have been targeted by JQ1 in preclinical models in vitro and in vivo90. JQ1 exhibits a high binding affinity to the bromodomain pocket and displaces BRD4 from the active chromatin in most SE sites and is believed to act predominantly on BRD4, BRD2, BRD3, and, BRDT. In acute myeloid leukemia (AML) and myeloma, pre-clinical models and clinical trials already show the BRD4 inhibition induces strong suppression of tumor progression91. In CRPC xenograft mouse models, BRD4 inhibition (JQ1) is more effective than direct AR antagonism (enzalutamide)33,84. This novel approach can be used to synergistically block the oncogenic drivers in advanced PCa for better treatments. BRD4 contains two conserved bromodomains, BD1 and BD2. Dual-bromodomain BET inhibitors are designed to competitively inhibit the binding of the BD1 and BD2, such as OTX015, CPI-0610, and ABBV-07592,93,94,95,96. However, in some monotherapy clinical trials, dose-limiting adverse events, such as reduced numbers of thrombocytes in the blood and some gastrointestinal toxicity, are limited the clinical activity. Fairre et al. proposed a highly potent and selective inhibitor of the BD2 domain by a medicinal chemistry campaign named ABBV-74497. ABBV-744 selectively maintains high activity in PCa cell lines and xenografts and has a lower toxicity than ABBV-075. Further analyses demonstrate that ABBV-744 displaces BRD4 from AR-bound SE sites and disrupts the AR-target transcriptional programs. The successful development of the preclinical compound JQ1 for BET inhibition has also enabled several compounds of BET inhibitors (ABBV-075, ABBV-744, GSK525762, ZEN003694, and GS-5829) to successfully enter the clinical trials.
THZ1, a covalent inhibitor of CDK7, shows the ability to suppress the CDK7-dependent phosphorylation activity to achieve clinical activity58. The inhibition of CDK7 by THZ1 is related to the global transcriptional downregulation at high dose levels, but studies have found that cancer cell lines are sufficiently sensitive to lower doses of THZ1. Further reports indicate that THZ1 may selectively target SE-driven transcription programs, including MYC-dependent transcription amplification and the expression of other cancer-specific oncogenic TFs and signaling molecules98,99. Rasool et al. demonstrated that THZ1 attenuated the AR-signaling and maintained efficacy in CRPC and enzalutamide-resistant PCa cells47. Also, CDK7 selective inhibitors have been developed, such as SY1365 and CT7001, and evaluated in clinical trials in other advanced solid tumors100,101.
ERG overexpression is observed in a large group of primary PCa and CRPC. A highly selective small-molecule inhibitor of ERG, NSC139021, inhibits the growth of ERG-positive cancer cells102. Another small-molecule inhibitor, YK-4-279, reduces the ERG-positive PCa patient-derived xenograft growth103.
Advances in SE profiling and chromatin landscape profiling make it possible for identifying essential SE regulators and SE-target genes in cancers104,105. Also, the relevant information will feed to the pharmacologic industry for therapeutic interventions.
Future directions
Super-enhancers are a large cluster of active transcriptional enhancers and are rich in enhancer-related chromatin features. Compared to typical enhancers, SEs are larger, exhibit a higher density of TFs, and are often associated with critical lineage-specific genes.
In PCa, fundamental components of the SE complexes, such as BDR4 and ERG, densely bind to the enhancer and SE elements to promote tumorigenesis and tumor growth. In advanced CRPC patients, the SE activity created by these transcription factors remains stable and evolves to be more specific, which makes it possible that SEs may be promising therapeutic targets47,97,106,107.
Several pre-clinical and clinically relevant studies targeting SEs have been successfully carried out in various tumors, including PCa21. BRD4 inhibition has been validated and investigated in leukemia, lymphoma, myeloma, neuroblastoma, breast cancer, prostate cancer, and other cancer types. Compared to castration-sensitive status, advanced CRPC patients exhibit an AR deregulation status41. Evidence shows that BRDs (BRD2, BRD4, and ATAD2) are prognostic markers that are overexpressed in CRPC. AR-deregulation-mediated BRDs upregulation intensifies the SE activity and feeds back a positive loop for AR chromatin binding. In other words, AR deregulation forms a transcription addiction status by enhancing the SE activities. To define the BET inhibition response in CRPC, a ten-gene signature, BROMO-10, was designed and used to guide patient selection for combinatorial trials of the BET inhibition against other agents. Remarkably, the BROMO-10 signature still needs to be refined and evaluated for AR signaling status and BRDs expression as well. Using this method, we can design specific gene signatures to evaluate the efficacy of other inhibitors and screen patients for potential therapeutic benefits. CDK7 inhibition has shown surprising therapeutic effects in preclinical models without evident systemic toxicity, which gives us high hopes for further human clinical trials108. Besides, it has been reported that SEs tend to undergo double-strand breaks and are therefore susceptible to deficiencies in cellular DNA-repair mechanisms. This shows that the combination of endocrine therapy and drugs targeting DNA damage repair will improve the anti-tumor efficiency, and clinical studies have already been conducted. Strikingly, Ma et al. found that SEs reorganization was tightly linked to drug resistance109. In repetitive cisplatin-treated cancer cells, the developmental transcription factor ISL1 invokes an unconventional trans-differentiation identity via SE reorganization to escape the drug-induced near-to-death status and facilitate tumor colonization. This may raise the question of how to effectively target cancer cells by avoiding the acquisition resistance in a SE-reorganization manner. The inhibition of transcription factors, such as SOX10 and ISL1, might potentially blockade the SE reorganization beyond BRDs or CDK7 inhibitors.
However, researchers have discovered that SE-related inhibitors might not have therapeutic effects in certain PCa types110,111. PCa cell lines and organoids from individuals with SPOP mutations show therapeutic resistance to cell growth arrest and apoptosis induced by BRD4 inhibitors112,113. The resistance to BRD4 inhibition in SPOP-mutant PCa can be overcome by combining with AKT inhibitors, and SPOP mutations may be used as biomarkers to guide the choice of treatment options for PCa patients, including those with urgent needs seeking precision medicine, and to determine whether the treatment is valid or not114,115. Moreover, it is worth noting that targeting SEs for cancer can cause side effects that cannot be ignored, because blocking SEs may also suppress specific SE-dependent tumor suppressor genes that are associated with cancer risk, such as cancer cell death116,117. Therefore, before using SEs as a therapeutic approach for the treatment of specific cancers, we urgently need to do more detailed research and make effective decisions.
References
Hanahan, D. & Weinberg, R. A. Hallmarks of cancer: the next generation. Cell 144, 646–674 (2011).
Litwin, M. S. & Tan, H. J. The diagnosis and treatment of prostate cancer: a review. JAMA 317, 2532–2542 (2017).
Shafi, A. A., Yen, A. E. & Weigel, N. L. Androgen receptors in hormone-dependent and castration-resistant prostate cancer. Pharmacol. Therapeutics 140, 223–238 (2013).
Tian, J. Y., Guo, F. J., Zheng, G. Y. & Ahmad, A. Prostate cancer: updates on current strategies for screening, diagnosis and clinical implications of treatment modalities. Carcinogenesis 39, 307–317 (2018).
Damber, J. E. & Aus, G. Prostate cancer. Lancet 371, 1710–1721 (2008).
Nakazawa, M., Paller, C. & Kyprianou, N. Mechanisms of therapeutic resistance in prostate cancer. Curr. Oncol. Rep. 19, 13 (2017).
Labbé, D. P. & Brown, M. Transcriptional regulation in prostate cancer. Cold Spring Harbor Perspect. Med. 8, https://doi.org/10.1101/cshperspect.a030437 (2018).
Tippens, N. D., Vihervaara, A. & Lis, J. T. Enhancer transcription: what, where, when, and why? Genes Dev. 32, 1–3 (2018).
Zabidi, M. A. & Stark, A. Regulatory enhancer-core-promoter communication via transcription factors and cofactors. Trends Genet. 32, 801–814 (2016).
Sur, I. & Taipale, J. The role of enhancers in cancer. Nat. Rev. Cancer 16, 483–493 (2016).
Grimmer, M. R. & Farnham, P. J. Can genome engineering be used to target cancer-associated enhancers? Epigenomics 6, 493–501 (2014).
Nizovtseva, E. V., Todolli, S., Olson, W. K. & Studitsky, V. M. Towards quantitative analysis of gene regulation by enhancers. Epigenomics 9, 1219–1231 (2017).
Reiter, F., Wienerroither, S. & Stark, A. Combinatorial function of transcription factors and cofactors. Curr. Opin. Genet. Dev. 43, 73–81 (2017).
Plank, J. L. & Dean, A. Enhancer function: mechanistic and genome-wide insights come together. Mol. cell 55, 5–14 (2014).
Robson, M. I., Ringel, A. R. & Mundlos, S. Regulatory landscaping: how enhancer-promoter communication is sculpted in 3D. Mol. Cell 74, 1110–1122 (2019).
Gorkin, D. U., Leung, D. & Ren, B. The 3D genome in transcriptional regulation and pluripotency. Cell Stem Cell 14, 762–775 (2014).
Whyte, W. A. et al. Master transcription factors and mediator establish super-enhancers at key cell identity genes. Cell 153, 307–319 (2013).
Lovén, J. et al. Selective inhibition of tumor oncogenes by disruption of super-enhancers. Cell 153, 320–334 (2013).
Gryder, B. E. et al. Chemical genomics reveals histone deacetylases are required for core regulatory transcription. Nat. Commun. 10, 3004 (2019).
Hnisz, D. et al. Super-enhancers in the control of cell identity and disease. Cell 155, 934–947 (2013).
Thandapani, P. Super-enhancers in cancer. Pharmacol. Therapeutics 199, 129–138 (2019).
He, Y., Long, W. & Liu, Q. Targeting super-enhancers as a therapeutic strategy for cancer treatment. Front. Pharmacol. 10, 361 (2019).
Tang, L. Liquid phase separation. Nat. Methods 16, 18 (2019).
Hyman, A. A., Weber, C. A. & Jülicher, F. Liquid-liquid phase separation in biology. Annu. Rev. Cell Dev. Biol. 30, 39–58 (2014).
Alberti, S. & Dormann, D. Liquid-liquid phase separation in disease. Annu. Rev. Genet. 53, 171–194 (2019).
Hnisz, D., Shrinivas, K., Young, R. A., Chakraborty, A. K. & Sharp, P. A. A phase separation model for transcriptional control. Cell 169, 13–23 (2017).
Wang, X., Cairns, M. J. & Yan, J. Super-enhancers in transcriptional regulation and genome organization. Nucleic Acids Res. 47, 11481–11496 (2019).
Sabari, B. R. et al. Coactivator condensation at super-enhancers links phase separation and gene control. Science 361, https://doi.org/10.1126/science.aar3958 (2018).
Baumgart, S. J. & Haendler, B. Exploiting epigenetic alterations in prostate cancer. Int. J. Mol. Sci. 18, https://doi.org/10.3390/ijms18051017 (2017).
Shi, J. & Vakoc, C. R. The mechanisms behind the therapeutic activity of BET bromodomain inhibition. Mol. Cell 54, 728–736 (2014).
Devaiah, B. N. et al. BRD4 is a histone acetyltransferase that evicts nucleosomes from chromatin. Nat. Struct. Mol. Biol. 23, 540–548 (2016).
Lee, J. E. et al. Brd4 binds to active enhancers to control cell identity gene induction in adipogenesis and myogenesis. Nat. Commun. 8, 2217 (2017).
Asangani, I. A. et al. Therapeutic targeting of BET bromodomain proteins in castration-resistant prostate cancer. Nature 510, 278–282 (2014).
Lochrin, S. E., Price, D. K. & Figg, W. D. BET bromodomain inhibitors–a novel epigenetic approach in castration-resistant prostate cancer. Cancer Biol. Ther. 15, 1583–1585 (2014).
Donati, B., Lorenzini, E. & Ciarrocchi, A. BRD4 and cancer: going beyond transcriptional regulation. Mol. Cancer 17, 164 (2018).
Urbanucci, A. & Mills, I. G. Bromodomain-containing proteins in prostate cancer. Mol. Cell. Endocrinol. 462, 31–40 (2018).
Grasso, C. S. et al. The mutational landscape of lethal castration-resistant prostate cancer. Nature 487, 239–243 (2012).
Garcia-Carpizo, V., Ruiz-Llorente, S., Sarmentero, J., González-Corpas, A. & Barrero, M. J. CREBBP/EP300 bromodomain inhibition affects the proliferation of AR-positive breast cancer cell lines. Mol. Cancer Res. 17, 720–730 (2019).
Jiang, C. Y. et al. MiR-185 attenuates androgen receptor function in prostate cancer indirectly by targeting bromodomain containing 8 isoform 2, an androgen receptor co-activator. Mol. Cell. Endocrinol. 427, 13–20 (2016).
Dai, C., Heemers, H. & Sharifi, N. Androgen signaling in prostate cancer. Cold Spring Harbor Perspect. Med. 7, https://doi.org/10.1101/cshperspect.a030452 (2017).
Urbanucci, A. et al. Androgen receptor deregulation drives bromodomain-mediated chromatin alterations in prostate cancer. Cell Rep. 19, 2045–2059 (2017).
Shah, N. et al. Regulation of the glucocorticoid receptor via a BET-dependent enhancer drives antiandrogen resistance in prostate cancer. Elife 6, https://doi.org/10.7554/eLife.27861 (2017).
Cai, L. et al. ZFX mediates non-canonical oncogenic functions of the androgen receptor splice variant 7 in castrate-resistant prostate cancer. Mol. Cell 72, 341–354. e346 (2018).
Malumbres, M. Cyclin-dependent kinases. Genome Biol. 15, 122 (2014).
Malumbres, M. & Barbacid, M. Mammalian cyclin-dependent kinases. Trends Biochem. Sci. 30, 630–641 (2005).
Asghar, U., Witkiewicz, A. K., Turner, N. C. & Knudsen, E. S. The history and future of targeting cyclin-dependent kinases in cancer therapy. Nat. Rev. Drug Discov. 14, 130–146 (2015).
Rasool, R. U. et al. CDK7 inhibition suppresses castration-resistant prostate cancer through MED1 inactivation. Cancer Discov. 9, 1538–1555 (2019).
Ebmeier, C. C. et al. Human TFIIH kinase CDK7 regulates transcription-associated chromatin modifications. Cell Rep. 20, 1173–1186 (2017).
Lu, H. et al. Phase-separation mechanism for C-terminal hyperphosphorylation of RNA polymerase II. Nature 558, 318–323 (2018).
Wang, Y. et al. CDK7-dependent transcriptional addiction in triple-negative breast cancer. Cell 163, 174–186 (2015).
Rochette-Egly, C., Adam, S., Rossignol, M., Egly, J. M. & Chambon, P. Stimulation of RAR alpha activation function AF-1 through binding to the general transcription factor TFIIH and phosphorylation by CDK7. Cell 90, 97–107 (1997).
Bour, G. et al. Cyclin H binding to the RARalpha activation function (AF)-2 domain directs phosphorylation of the AF-1 domain by cyclin-dependent kinase 7. Proc. Natl Acad. Sci. USA 102, 16608–16613 (2005).
Keriel, A., Stary, A., Sarasin, A., Rochette-Egly, C. & Egly, J. M. XPD mutations prevent TFIIH-dependent transactivation by nuclear receptors and phosphorylation of RARalpha. Cell 109, 125–135 (2002).
Russo, J. W., Nouri, M. & Balk, S. P. Androgen receptor interaction with mediator complex is enhanced in castration-resistant prostate cancer by CDK7 phosphorylation of MED1. Cancer Discov. 9, 1490–1492 (2019).
Chymkowitch, P., Le May, N., Charneau, P., Compe, E. & Egly, J. M. The phosphorylation of the androgen receptor by TFIIH directs the ubiquitin/proteasome process. EMBO J. 30, 468–479 (2011).
Han, Y. et al. Triptolide inhibits the AR signaling pathway to suppress the proliferation of enzalutamide resistant prostate cancer cells. Theranostics 7, 1914–1927 (2017).
Tee, A. E. et al. Combination therapy with the CDK7 inhibitor and the tyrosine kinase inhibitor exerts synergistic anticancer effects against MYCN-amplified neuroblastoma. Int. J. Cancer https://doi.org/10.1002/ijc.32936 (2020).
Kwiatkowski, N. et al. Targeting transcription regulation in cancer with a covalent CDK7 inhibitor. Nature 511, 616–620 (2014).
Christensen, C. L. et al. Targeting transcriptional addictions in small cell lung cancer with a covalent CDK7 inhibitor. Cancer Cell 26, 909–922 (2014).
Zhang, H. et al. CDK7 inhibition potentiates genome instability triggering anti-tumor immunity in small cell lung cancer. Cancer cell 37, 37–54. e39 (2020).
Sizemore, G. M., Pitarresi, J. R., Balakrishnan, S. & Ostrowski, M. C. The ETS family of oncogenic transcription factors in solid tumours. Nat. Rev. Cancer 17, 337–351 (2017).
Mounir, Z. et al. TMPRSS2:ERG blocks neuroendocrine and luminal cell differentiation to maintain prostate cancer proliferation. Oncogene 34, 3815–3825 (2015).
Zhang, J. et al. Deregulation of DUX4 and ERG in acute lymphoblastic leukemia. Nat. Genet. 48, 1481–1489 (2016).
Sugimura, R. et al. Haematopoietic stem and progenitor cells from human pluripotent stem cells. Nature 545, 432–438 (2017).
Kalna, V. et al. The transcription factor erg regulates super-enhancers associated with an endothelial-specific gene expression program. Circ. Res 124, 1337–1349 (2019).
Adamo, P. & Ladomery, M. R. The oncogene ERG: a key factor in prostate cancer. Oncogene 35, 403–414 (2016).
Falzarano, S. M. & Magi-Galluzzi, C. ERG protein expression as a biomarker of prostate cancer. Biomark. Med. 7, 851–865 (2013).
Clark, J. P. & Cooper, C. S. ETS gene fusions in prostate cancer. Nat. Rev. Urol. 6, 429–439 (2009).
Gerke, J. S. et al. Integrative clinical transcriptome analysis reveals TMPRSS2-ERG dependency of prognostic biomarkers in prostate adenocarcinoma. Int. J. Cancer 146, 2036–2046 (2020).
Weier, C. et al. Nucleotide resolution analysis of TMPRSS2 and ERG rearrangements in prostate cancer. J. Pathol. 230, 174–183 (2013).
Kron, K. J. et al. TMPRSS2-ERG fusion co-opts master transcription factors and activates NOTCH signaling in primary prostate cancer. Nat. Genet. 49, 1336–1345 (2017).
Cong, Z. et al. The SNP of rs6854845 suppresses transcription via the DNA looping structure alteration of super-enhancer in colon cells. Biochem. Biophys. Res. Commun. 514, 734–741 (2019).
Oldridge, D. A. et al. Genetic predisposition to neuroblastoma mediated by a LMO1 super-enhancer polymorphism. Nature 528, 418–421 (2015).
Kandaswamy, R. et al. Genetic predisposition to chronic lymphocytic leukemia is mediated by a BMF super-enhancer polymorphism. Cell Rep. 16, 2061–2067 (2016).
Chen, H. et al. Genetic associations of breast and prostate cancer are enriched for regulatory elements identified in disease-related tissues. Hum. Genet. 138, 1091–1104 (2019).
Hanover, J. A., Chen, W. & Bond, M. R. O-GlcNAc in cancer: an oncometabolism-fueled vicious cycle. J. Bioenerg. Biomembr. 50, 155–173 (2018).
Ong, Q., Han, W. & Yang, X. O-GlcNAc as an integrator of signaling pathways. Front. Endocrinol. 9, 599 (2018).
Itkonen, H. M. et al. High OGT activity is essential for MYC-driven proliferation of prostate cancer cells. Theranostics 9, 2183–2197 (2019).
Baumgart, S. J. et al. Darolutamide antagonizes androgen signaling by blocking enhancer and super-enhancer activation. Mol. Oncol. https://doi.org/10.1002/1878-0261.12693 (2020).
Baumgart, S. J., Nevedomskaya, E. & Haendler, B. Dysregulated transcriptional control in prostate cancer. Int. J. Mol. Sci. 20, https://doi.org/10.3390/ijms20122883 (2019).
Wong, Y. N., Ferraldeschi, R., Attard, G. & de Bono, J. Evolution of androgen receptor targeted therapy for advanced prostate cancer. Nat. Rev. Clin. Oncol. 11, 365–376 (2014).
Coutinho, I., Day, T. K., Tilley, W. D. & Selth, L. A. Androgen receptor signaling in castration-resistant prostate cancer: a lesson in persistence. Endocr. Relat. Cancer 23, T179–t197 (2016).
Hupe, M. C. et al. The BET-inhibitor PFI-1 diminishes AR/AR-V7 signaling in prostate cancer cells. World J. Urol. 37, 343–349 (2019).
Welti, J. et al. Targeting bromodomain and extra-terminal (BET) family proteins in castration-resistant prostate cancer (CRPC). Clin. Cancer Res. 24, 3149–3162 (2018).
Gao, N. et al. Forkhead box A1 regulates prostate ductal morphogenesis and promotes epithelial cell maturation. Development 132, 3431–3443 (2005).
Hankey, W., Chen, Z. & Wang, Q. Shaping chromatin states in prostate cancer by pioneer transcription factors. Cancer Res. 80, 2427–2436 (2020).
Lupien, M. et al. FoxA1 translates epigenetic signatures into enhancer-driven lineage-specific transcription. Cell 132, 958–970 (2008).
Pomerantz, M. M. et al. The androgen receptor cistrome is extensively reprogrammed in human prostate tumorigenesis. Nat. Genet. 47, 1346–1351 (2015).
Yang, Y. A. & Yu, J. Current perspectives on FOXA1 regulation of androgen receptor signaling and prostate cancer. Genes Dis. 2, 144–151 (2015).
Filippakopoulos, P. et al. Selective inhibition of BET bromodomains. Nature 468, 1067–1073 (2010).
Zuber, J. et al. RNAi screen identifies Brd4 as a therapeutic target in acute myeloid leukaemia. Nature 478, 524–528 (2011).
Boi, M. et al. The BET bromodomain inhibitor OTX015 affects pathogenetic pathways in preclinical B-cell tumor models and synergizes with targeted drugs. Clin. Cancer Res. 21, 1628–1638 (2015).
Asangani, I. A. et al. BET bromodomain inhibitors enhance efficacy and disrupt resistance to AR antagonists in the treatment of prostate cancer. Mol. Cancer Res. 14, 324–331 (2016).
Siu, K. T. et al. Preclinical activity of CPI-0610, a novel small-molecule bromodomain and extra-terminal protein inhibitor in the therapy of multiple myeloma. Leukemia 31, 1760–1769 (2017).
Bui, M. H. et al. Preclinical characterization of BET family bromodomain inhibitor ABBV-075 suggests combination therapeutic strategies. Cancer Res. 77, 2976–2989 (2017).
Faivre, E. J. et al. Exploitation of castration-resistant prostate cancer transcription factor dependencies by the novel BET inhibitor ABBV-075. Mol. Cancer Res. 15, 35–44 (2017).
Faivre, E. J. et al. Selective inhibition of the BD2 bromodomain of BET proteins in prostate cancer. Nature 578, 306–310 (2020).
Chipumuro, E. et al. CDK7 inhibition suppresses super-enhancer-linked oncogenic transcription in MYCN-driven cancer. Cell 159, 1126–1139 (2014).
Durbin, A. D. et al. Selective gene dependencies in MYCN-amplified neuroblastoma include the core transcriptional regulatory circuitry. Nat. Genet. 50, 1240–1246 (2018).
Hu, S. et al. Discovery and characterization of SY-1365, a selective, covalent inhibitor of CDK7. Cancer Res. 79, 3479–3491 (2019).
Sava, G. P., Fan, H., Coombes, R. C., Buluwela, L. & Ali, S. CDK7 inhibitors as anticancer drugs. Cancer Metastasis Rev. https://doi.org/10.1007/s10555-020-09885-8 (2020).
Mohamed, A. A. et al. Identification of a small molecule that selectively inhibits ERG-positive cancer cell growth. Cancer Res. 78, 3659–3671 (2018).
Winters, B. et al. Inhibition of ERG activity in patient-derived prostate cancer xenografts by YK-4-279. Anticancer Res. 37, 3385–3396 (2017).
Wei, Y. et al. SEA: a super-enhancer archive. Nucleic Acids Res. 44, D172–D179 (2016).
Jiang, Y. et al. SEdb: a comprehensive human super-enhancer database. Nucleic Acids Res. 47, D235–d243 (2019).
Nagasawa, M. et al. Long non-coding RNA MANCR is a target of BET bromodomain protein BRD4 and plays a critical role in cellular migration and invasion abilities of prostate cancer. Biochem. Biophys. Res. Commun. 526, 128–134 (2020).
McClurg, U. L. et al. Molecular mechanism of the TP53-MDM2-AR-AKT signalling network regulation by USP12. Oncogene 37, 4679–4691 (2018).
Chou, J., Quigley, D. A., Robinson, T. M., Feng, F. Y. & Ashworth, A. Transcription-associated cyclin-dependent kinases as targets and biomarkers for cancer therapy. Cancer Discov. 10, 351–370 (2020).
Ma, Q. et al. Super-enhancer redistribution as a mechanism of broad gene dysregulation in repeatedly drug-treated cancer cells. Cell Rep. 31, 107532 (2020).
Janouskova, H. et al. Opposing effects of cancer-type-specific SPOP mutants on BET protein degradation and sensitivity to BET inhibitors. Nat. Med. 23, 1046–1054 (2017).
Dai, X., Wang, Z. & Wei, W. SPOP-mediated degradation of BRD4 dictates cellular sensitivity to BET inhibitors. Cell Cycle 16, 2326–2329 (2017).
Dai, X. et al. Prostate cancer-associated SPOP mutations confer resistance to BET inhibitors through stabilization of BRD4. Nat. Med. 23, 1063–1071 (2017).
Bradley, C. A. Prostate cancer: BET inhibitors - SPOP right there! Nat. Rev. Cancer 17, 574–575 (2017).
Zhang, P. et al. Intrinsic BET inhibitor resistance in SPOP-mutated prostate cancer is mediated by BET protein stabilization and AKT-mTORC1 activation. Nat. Med. 23, 1055–1062 (2017).
Yan, Y. et al. The novel BET-CBP/p300 dual inhibitor NEO2734 is active in SPOP mutant and wild-type prostate cancer. EMBO Mol. Med 11, e10659 (2019).
Zhang, J. et al. The CREBBP acetyltransferase is a haploinsufficient tumor suppressor in B-cell lymphoma. Cancer Discov. 7, 322–337 (2017).
Ding, Y. et al. Ikaros tumor suppressor function includes induction of active enhancers and super-enhancers along with pioneering activity. Leukemia 33, 2720–2731 (2019).
Acknowledgements
This work was supported by National Natural Science Foundation of China (grand No. 82072851, 81872100, and 81772756) and Natural Science Foundation of Tianjin (18PTLCSY00030 and 19JCYBJC24900).
Author information
Authors and Affiliations
Contributions
X.C. and Q.M. contributed equally to this manuscript, and are considered as “co-first authors”. X.C. wrote section on SE-related proteins, promising pharmacological targets, future directions, and edited manuscript. Q.M. co-wrote introduction and abstract, SE-related proteins and edited manuscript. Z.S. co-wrote introduction and abstract, edited manuscript. N.Y. co-wrote introduction and abstract, edited manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Chen, X., Ma, Q., Shang, Z. et al. Super-enhancer in prostate cancer: transcriptional disorders and therapeutic targets. npj Precis. Onc. 4, 31 (2020). https://doi.org/10.1038/s41698-020-00137-0
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41698-020-00137-0
This article is cited by
-
KLF7 regulates super-enhancer-driven IGF2BP2 overexpression to promote the progression of head and neck squamous cell carcinoma
Journal of Experimental & Clinical Cancer Research (2024)
-
The PENGUIN approach to reconstruct protein interactions at enhancer-promoter regions and its application to prostate cancer
Nature Communications (2023)
-
Targeting ACE2-BRD4 crosstalk in colorectal cancer and the deregulation of DNA repair and apoptosis
npj Precision Oncology (2023)
-
Super-enhancer-associated gene CAPG promotes AML progression
Communications Biology (2023)
-
Transcription associated cyclin-dependent kinases as therapeutic targets for prostate cancer
Oncogene (2022)