Abstract
Knowledge on diet composition allows defining well-targeted conservation measures of large carnivores. Little is known about ecology of critically endangered Asiatic cheetah, especially the overall diet and its possible regional differences. We used cheetah scats, metabarcoding technique and microsatellite markers to assess the individual and overall diet composition of the species across its entire range in Asia. Cheetahs were primarily predating on mouflon; following by ibex, cape hare and goitered gazelle. Despite their high availability, small-sized livestock was never detected. Goitered gazelles were only detected in an area where the habitat is mainly flatlands. In hilly areas, mouflon was the most frequent prey item taken. Ibex was typically taken in rugged terrain, but mouflon was still the most frequently consumed item in these habitats. High consumption of mouflon in comparison to goitered gazelle suggests that human pressure on lowland habitats has possibly forced Asiatic cheetahs to occupy suboptimal habitats where gazelles are less abundant. The protection of flatlands and the removal of livestock from them are needed to ensure the long-term survival of Asiatic cheetah. The laboratory and bioinformatics pipelines used in this study are replicable and can be used to address similar questions in other threatened carnivores.
Similar content being viewed by others
Introduction
Intensive human activities have resulted in the decline of global biodiversity including mammals1,2. Large carnivores are one of the groups that are globally threatened by population decline, range contraction and habitat fragmentation3. Conservation measures grounded in scientific studies can reduce these negative effects and allow carnivore populations to recover. Availability of prey is one of the primary factors related to carnivore abundance4. Thus, precise knowledge on diet composition and prey availability are needed to define appropriate conservation decisions.
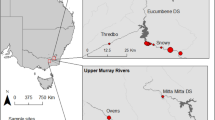
The cheetah (Acinonyx jubatus) is one of the most threatened carnivores at global level. Historically, it was widespread across Africa and southwestern Asia but it was extirpated from 91% of the former range due to habitat transformation, degradation and fragmentation, and depletion of its prey items5. The range decline was most severe in Asia: it was extirpated from almost all of its historical range and its distribution has been limited to the central plateau of Iran since the 1970s6. Within Iran, the Asiatic Cheetah (Acinonyx jubatus venaticus) has lost more than 70% of its suitable habitat over the past decades due to habitat transformation and loss of most of their main natural prey7. The population declined from over 200 individuals distributed across 44 areas in the mid-1970s to 50–100 individuals distributed in 14 areas in the 2000s8,9. Currently, the population is estimated at about 50 individuals distributed in three subpopulations located in the central plateau of Iran, encircling the Dasht-e Kavir desert (Fig. 1). The population is continuously declining and is divided into three subpopulations with an average distance of about 500 km between them: (1) Northern Subpopulation, (2) Southern Subpopulation and (3) Kavir Subpopulation; although since 2013, there have been no confirmed observations of Kavir Subpopulation10. Estimated boundaries of each subpopulation are shown by dashed lines in Fig. 1. The protected areas labelled in Fig. 1 constitute the last stronghold of the Asiatic cheetah and need an urgent boost in conservation efforts to prevent further population decline. The remaining Northern and Southern Subpopulations inhabit distinct habitat types. In general, the Northern Subpopulation occupies flat arid lowlands and the Southern Subpopulation ranges over mountainous deserts (Table 1). Consequently, prey abundance and availability are rather distinct in each of the regions, and potential regional differences in diet composition may be expected. As such, it is crucial to understand whether there are regional differences in diet composition and if diet in each region is reflecting prey abundance. Such information will allow defining conservation actions tailored to each region.
Current distribution of Cheetah in Iran (small inset) and location of subpopulations/protected areas from which samples were available for this study. The dashed lines depict the estimated boundaries of each subpopulation: Northern in the north, Southern in the south and Kavir in the west. Northern Subpopulation: (1) Touran Biosphere Reserve, (2) Miandasht WR and Zamen-e Ahoo NP; Southern subpopulation: (3) Naybandan WR; (4) Kamki Bahabad HPA, (5) Bafgh PA, (6) Ariz HPA, (7) Dareh Anjir WR. This figure was produced using ArcGIS (version 10.3.1 [www.esri.com/software/Arcgis]).
Most information on cheetah’s diet composition comes from Africa, where cheetahs have been reported to prey on a large variety of prey items, from large ungulates to small rodents11, preferring small to medium sized ungulates with body masses in the range of 15 to 65 kg, with the mode of 36 kg12,13. When these prey items are not available, such as outside birth peaks of ungulates or areas with lower ungulate population, cheetahs rely on smaller prey items such as hares (Leporidae)14,15. Cheetahs also predate on livestock, including sheep, goat and camel16,17, although they primarily consume wild prey15,18. Prey choice depends on prey abundance, presence of competitors, and cheetah’s sex19. Male coalitions are able to kill larger preys20, solitary females usually hunt smaller preys13,21, but when they have dependent cubs, they may take larger prey items22. In Iran, a few regional studies assessing cheetah’s diet reported jebeer (Gazella bennettii), goitered gazelle (Gazella subgutturosa), mouflon (Ovis vignei), ibex (Capra aegagrus), wild boar (Sus scrofa) and livestock (sheep, goat and dromedary) as main prey items23,24,25,26. Given that one of the most important cheetah refuges in central Iran, the Touran Biosphere Reserve, is inhabited by more than 70,000 livestock (sheep and goat) during the grazing season, knowing whether cheetahs regularly predate on livestock is vital to mitigate possible human-wildlife conflicts.
Diet analyses based on the morphological identification from scats are prone to errors regarding the identification of both, scat depositors and prey items27,28. Given that cheetahs start consuming their prey with muscle and viscera before chewing on skin and bones29, this can lead to biased estimations of consumed prey, e.g. to overestimate small sized prey and underestimate large sized prey30,31. Therefore, assessing cheetah diet using scat analyses, needs more accurate methods. Recent advances in DNA metabarcoding allow gathering accurate data on species’ diet, from bats to large mammals32,33,34. The method has been widely used in ecological studies, addressing the diet of carnivores35,36. Use of blocking primers can increase diet data acquisition by reducing amplifications from the predator37. The efficiency of using scat DNA metabarcoding for the analyses of cheetah diet using feeding trials was recently evaluated, reporting that it is possible to obtain diet information from 60-day old scats38. DNA metabarcoding from scat has been tested using wild cheetah scats in Africa and was reported to be a valuable complement to traditional dietary analysis methods, especially for items that might be missed by direct observation of killing events17. Patterns of prey selection may be different among individuals of the same species39,40 therefore individual identification and assessment of individual diet can provide important insights on population dynamics and conservation ecology41.
The main aim of this study is to provide accurate data on the foraging ecology of Asiatic cheetah based on scat DNA metabarcoding to support the definition of conservation measures, e.g. to identify the most important prey items for cheetah and boost their conservation. This is the first study assessing Asiatic cheetah diet using this approach and thus provides baseline methods and data that can be used as a guideline for future ecological surveys. This study aims to answer the following questions: What is the overall diet composition of Asiatic cheetahs? Is livestock consumed and do all individuals prey upon sheep and goats? Are there differences in diet between regions? Is diet composition correlated with habitat type and prey population size? Are there differences in individual dietary? The results of this study are intended to contribute to the conservation of the remaining critically endangered population of Asiatic cheetah in Iran.
Results
From 376 collected scat samples, 138 (37%) were confirmed to be from cheetah (Table 2). Other predators found were common fox (Vulpes vulpes; 22%), unidentified Felidae (15%), wolf (Canis lupus; 16%), caracal (Caracal caracal; 6%), porcupine (Hystrix indica; 1%), and striped hyaena (Hyaena hyaena; 1%). Yazd had the highest percentage of confirmed cheetah scats (66%), while Miandasht with 7% had the lowest, and in Naybandan and Touran, the percentage was 33% and 39%, respectively. Cheetah scats were most frequently confused with fox scats in all areas, except in Miandasht where they were most often confused with other felids and Canis lupus (30% and 36%, respectively). Identification to species level in many specimens of Felidae family was not possible, neither to discern whether Canis lupus samples were from wolf or dog (Canis lupus familiaris), as the metabarcodes chosen in this work do not disambiguate these taxa.
We were able to genotype 100 of the original 138 cheetah scat samples and to identify a total of 14 individual cheetahs (3 females and 11 males): 2 females and 5 males in Touran, 1 male in Miandasht, 2 males in Naybandan (of which, one was also identified in Touran) and 4 males and 1 female in Yazd. We assessed diet composition from 119 out of 138 scats. We were unable to genotype 38 scats and assess the diet of 18 scats, due to very low quality of samples. A total of 13 distinct food items (species or genus) were observed in these scats. The main food items were mouflon (67%), ibex (26%), Cape hare (Lepus capensis; 13%), goitered gazelle (3%), wild boar (3%) and dromedary camel (Camelus dromedarius) (3%) (Table 3). Other food items with low occurrence were Midday jird (Meriones meridianus; N = 2), house mouse (Mus musculus), common fox and food items from genus Jaculus and orders Galliformes, Squamata and Chiroptera (N = 1 for each). On average, each scat contained 1.2 food items. Touran exhibited the highest richness of food items (N = 10). Regional accumulation curve of species observed in the diet is presented in Supplementary Fig. 1. Details of food items consumed by each genotyped individual are provided in Supplementary Tables 1 and 2. Given the low number of female cheetahs detected (N = 4), it was not possible to statistically compare the diet variation between different sexes. Comparing the diet of males and females shows females tend to predate on smaller preys such as cape hare, although they are capable of hunting larger preys, especially in more rugged areas such as Yazd. Males tend to predate on larger preys, such as mouflon and ibex, and dromedary camel was taken only by males. The male coalition in Yazd had the higher percentage of preying ibex in comparison to the males from other areas. Inferences of individual dietary differences are presented in Supplementary Table 2.
In total and in each of the analysed areas, except Miandasht, mouflon was the most common food item found in scats, followed by Cape hare in Touran, dromedary camel in Naybandan, and ibex in Yazd (Table 3). Cape hare was the only food item that was observed in scats from all areas. Wild boar was detected a few times (total N = 4) in all areas except Naybandan. Despite being present in Touran and Miandasht, goitered gazelles were observed in the diet only in Miandasht, where they had the highest percentage of consumption (RFO = 57) jebeer was never observed in the analysed scats.
There were differences between prey abundance and prey consumed when analysing individual areas (Table 3). Mouflon was the most consumed food item in Touran, Naybandan and Yazd (RFO = 58, 71 and 52, respectively) and it was the food item with highest population size in Touran, while in Naybandan and Yazd, ibex has the highest population size. Ibex was observed only one time in scats from Naybandan, despite having larger population size than mouflon. In Yazd, where population of ibex is higher than mouflon, more scats contained ibex, but still the percentage of scats containing mouflon was higher (RFO = 52 for mouflon and 41 for ibex). Even though goitered gazelle have relatively large numbers in some parts of Touran, they were only observed in the diet of individuals from Miandasht. Visual comparison of relative percent consumption of different species to relative abundance of prey in each area is presented in Supplementary Fig. 2. Despite the large abundance of small-sized livestock, especially in the Northern Subpopulation, it was not found in any of the scats. Dromedary camel was rarely detected (total N = 4) in scats from Naybandan and Touran, where it is being kept as livestock.
Discussion
In this study, we collected scats from all cheetah populations within the extant distribution range of the species in Asia, identified some different cheetah individuals, and analysed their diet using non-invasive and metabarcoding methods. Our study shows that cheetahs tended to hunt mouflon in study sites with predominantly more hilly terrains and gazelles in study sites that were predominantly flatter; small livestock was not detected in their diet. These findings have implications for cheetah conservation, as discussed in the following sections.
Diet composition
We identified mouflon, ibex, cape hare, goitered gazelle, wild boar, and dromedary camel (in decreasing order of importance) as cheetah’s main food items. In comparison to other studies in Iran23,24,25,26, small livestock and jebeer were not detected in our results. This discrepancy could partially be due to misidentification of scat depositors in previous studies, as they were only based on the morphological identification of scats and could be partially due to low number of samples in this study, especially in Miandasht and Naybandan. The visual identification of faeces in diet composition studies has limitations, namely regarding the misidentification of scat depositors27,42 which can bias the results and misinform conservation managers28,43. For instance, Asiatic cheetah faeces used in the previous studies may have been confused with scats from leopard (Panthera pardus), wolf or hyaena, which are also present in the region (Table 2).
Overall, mouflon was the most consumed food item in Iran. Likewise, other studies also identified mouflon as the most consumed food item in the Southern Subpopulations23,25,26. In Africa, cheetahs mainly consume steppe-dwelling gazelles, such as impala (Aepyceros melampus) and Thomson’s gazelle (Eudorcas thomsonii)44,45. However, our results show that Asiatic cheetahs mainly take mouflon, which inhabits hilly and mountainous areas, rather than gazelles that inhabit plains. Given that Asiatic cheetahs are 30% smaller than South African ones46 and considering that preys such as Thomson’s gazelles are known as their preferred prey in Africa, it would be expected for jebeer and goitered gazelle to be the primary prey for the Asiatic cheetah. Indeed, previous studies conducted in 1970s to 2000s in Iran reported gazelles as cheetah’s primary prey item47,48,49, and recent studies found jebeer as the most preferred food item in Yazd-Naybandan25,26. Higher proportion of mouflon in the diet instead of the expected gazelles can be due to the severe declines in population range and size that cheetah experienced over the last decade, where most populations have been extirpated from their preferred plains, the preferred habitat of gazelles7.
We found that the diet composition apparently varies between geographic areas and these differences may be related with prey abundance and habitat characteristics. In some areas, cheetahs consume the most abundant prey, such as in Miandasht and Touran where they take goitered gazelle and mouflon, respectively. In other areas, the results shows that cheetah apparently consumes the second most abundant prey, such as in Naybandan and Yazd where they prey on mouflon rather than ibex (Table 3). Habitat characteristics are likely to interplay with prey availability: in comparison to ibex, mouflon inhabits less rugged areas, therefore in Naybandan, where rolling hills are more common than mountainous areas and TRI is lower than in Yazd, predation on ibex was only observed once. On the other hand, in Yazd where mountainous areas are the most common habitat type and TRI is higher than all the areas (Table 1), the proportion of ibex consumption was higher than in Naybandan (RFO = 41 in Yazd and 7 in Naybandan), similar to previous studies23,25,26. Although cheetahs are able to hunt successfully in various habitats12, their morphological adaption suggests that their primary habitat was open plains and that they evolved to hunt steppe-dwelling prey by chasing them in high speed, especially in open habitats13,50,51. In addition, old literature also suggested gazelles as their primary prey47,48,49. In Iran, when gazelles are unavailable, cheetahs need to hunt mouflon and ibex, which usually inhabit rugged mountains that are less suitable for cheetahs, where they might be even killed as competitors by leopards7,52. The absence of goitered gazelle in the scats analysed from Touran suggests that flat plains have become unsuitable for both cheetahs and gazelles, most likely due to poaching of gazelles and competition with livestock. Recent observations in Touran confirms that the goitered gazelles mainly occur in the vicinity of farmlands (HA personal communication), which are unsuitable for cheetahs due to disturbance associated with human settlements, transportation and livestock raising7. Although we sampled all known cheetah habitats in Yazd and Naybandan, jebeer was not detected in any of the analysed scats, possibly due to the severe decline in their population in recent years (Soofi et al., under review) and remarkable reduction in cheetah population size.
Small livestock was not detected in the diet analysed in our study, which contrasts with previous studies24,26, where domestic sheep and goat were detected as food items. This could be partially due to small sample size, especially in Miandasht and Naybandan. Despite large number of small livestock present in Touran and Miandasht and large herds of dromedary camel in Naybandan and Touran and smaller herds in Miandasht, sheep and goat were not detected in the analysed scats, and dromedary camel was detected only four times. Likewise, predation of Asiatic cheetahs on livestock was unreported in the old literature48,49. Therefore, the detection of small livestock as a food item in some recent studies24,25,26 may be the result of misidentification of scat depositors. In Africa, cheetahs prefer wild prey, but also consume small livestock, such as sheep and goat15. A possible reason for Asiatic cheetahs overlooking small livestock could be that livestock husbandry methods in Iran are an old practice where livestock herds are traditionally well protected by large guarding dogs that deter carnivores from approaching the herds53 and the smaller size of Asiatic cheetahs in comparison to southern African ones46. Still, the possibility of cheetahs occasionally consuming livestock is not fully rejected as there were evidences of predation on sheep in Chah-Shirin protected area (HA personal communication), suggesting that cheetahs may take small livestock in suboptimal habitats, with less wild prey54. As a matter of fact, we had only six samples from Miandasht which has the second highest livestock abundance, additional sampling may reveal consumption of livestock within protected areas. In addition, cheetahs may also consume dromedary, especially juveniles, as these are usually grazing freely with no shepherds or guarding dogs (B. Najafi personal communication).
Taken together, our results suggest that cheetahs in Iran tend to consume the most abundant wild prey, as long as the habitat characteristics make those prey items available. Moreover, our results can suggest that the recent decline in cheetah population10 and the shift in their distribution from flat areas to more unsuitable habitats7, such as hilly and mountainous habitats, may have induced also diet changes.
Implications for cheetah conservation
Our results have important implications for conservation of Asiatic cheetahs. To prevent Asiatic cheetahs from extinction, making the plains suitable for them again and to ensure their long-term survival, we propose as conservation action the removal of livestock from cheetah habitats in Touran and Miandasht. Livestock husbandry and agriculture is historically the main livelihood of local people in central plateau of Iran, as consequence of several crop and animal domestication events in Fertile Crescent55. Therefore, although these areas are under legal protection by DoE, local people still have traditional rights to graze their livestock. Buying out grazing rights in Touran and Miandasht has long been proposed as a solution to remove livestock pressure from cheetah habitats8, was recently emphasized as an urgent conservation action and the priority pastures have been identified10,56. Similarly, removal of dromedary camel should also be prioritized as their large numbers can have wide negative impact in quality of grazing lands. Removing livestock will improve the quality of grazing lands, decrease livestock-wild herbivores competition, decrease human-wildlife conflict and poaching, and eventually allow gazelles to reoccupy the plains57,58. Removal of livestock will ultimately enhance feeding opportunities for cheetahs, thus promoting their long-term survival44,59. In Naybandan and Yazd areas, protection of mountain ungulates should continue but at the same time, the conservation of plains should be boosted to recover the population of gazelles.
Sampling and barcoding constraints and innovations
The observed high confusion rate of scats with other predators (63%) emphasizes the importance of the genetic identification of scat depositors for complementing expert identification of scats in the field and for obtaining accurate data on the prey composition and predator community of each area. Several studies reported use of scat dogs as a complementary method to increase the probability of detecting scats in the field60. Using these dogs allows covering larger study areas and also decreasing laboratory costs by collecting scats from target species, and increasing the possibility of finding small scats from juveniles and females61. The problem of small number of samples size, especially in Miandasht and Naybandan can be overcome by using these dogs.
In our field work, we found scats from 11 male cheetahs but only from 3 females. The collection of scats in the field is usually biased towards male individuals, as they tend to mark their territories using scats29. Even though our samples were biased to male individuals, broad sampling (e.g., collecting both small and large-size scats) and sampling through line transects along previously known marking sites (instead of limiting our searches to marking sites) allowed us to obtain samples from four different females. The rate of identifying scat depositors’ species, and the individual and prey identifications using metabarcoding was high (98%, 72% and 87% respectively), which is probably due to low humidity of habitats, resulted in quick drying of scats and hence well preservation of scat’s DNA. The dry conditions of arid habitat make them very suitable for studies using non-invasive DNA.
The prey abundance data used in this study was obtained from annual census reports of Department of Environment. Although these data provide a coarse basis for estimation of prey abundance, it was not always collected with a standardize protocol. Therefore, for future studies, systematic sampling of prey abundance in all cheetah occurrence areas, preferably at the same time of sampling scats, is recommended to estimate prey abundance, which will allow estimating prey preference according to prey abundance. Systematic sampling of scats during several seasons to account for seasonality differences and retrieving all possible prey items and to prevent scat disappearance due to climate factors is also recommended38.
This was the first study using cheetah blocking primer to reduce predator amplifications from the diet analysis and better identify prey items. Although cheetah amplifications were not totally eliminated, they were decreased enough to allow obtaining more amplifications from the prey barcoding marker. The laboratory and bioinformatics pipeline presented in this study is an efficient and accurate tool to assess diet of large carnivores, especially in dry habitats. Using of molecular methods, allowed us to assess different ecological aspects of cheetah’s feeding ecology (e.g., sex and individual ID) which otherwise was not achievable. Therefore, we suggest to potential funders and donors to consider supporting molecular studies as a complementary part of field studies. To improve this pipeline, using additional markers to identify specimens of Felidae family to species level and to differentiate wolf and dog is recommended62,63 in order to have a better understanding of predator community of each area. Moreover, we recommend the development of a correction factor using feeding trials with captive animals to account for factors, such as meal size, prey species, feeding day, in order to obtain accurate data from the samples collected from the field.
Implications for carnivore conservation
Our study highlights the usefulness of using genomic techniques along with well-established methods of data collection (e.g., camera trapping) as a complement for answering ecological questions. These genomic methods allow complementing current field assessments and also to provide information on different ecological aspects that are important for wildlife conservation. They also help managers to make informed decisions so far difficult to base on traditional methods. For instance, scats collected from other predators give a better understanding of competitor community and how they may compete with each other over available prey; individual identification data can give insights on sexual and ontogenetic differences of tropic niches and even differences between individual hunting strategies. Management plans for conservation of large predators can be improved by understanding adequately the exact predator–prey interactions, and understanding problems that they may cause, such as predation on domestic animals64. Thus, these data can improve conservation plans for many endangered carnivores, such as leopard, snow leopard (Panthera unica), or tiger (Panthera tigris), by e.g. identifying problematic predators responsible for livestock killing65, assessing their diets, investigating levels of human-wildlife conflict, and providing suggestions to decrease these conflicts.
Methods
Study area
The study area covers the protected areas with confirmed cheetah observations in 2017, distributed in the central deserts of Iran. In this study, the Northern Subpopulation was further divided into Northern Subpopulation covering Touran National Park (NP), Protected Area (PA) and Wildlife Refuge (WR), (hereafter Touran); and North-eastern Subpopulation comprising Miandasht WR and Zamen-e Ahoo NP, (hereafter Miandasht). The Southern Subpopulation was further divided into South-eastern Subpopulation covering Naybandan WR; and South-western Subpopulation comprising Kamki Bahabad Hunting Prohibited Area (HPA), Bafgh PA, Ariz HPA and Dareh Anjir WR, (hereafter Yazd) (Fig. 1). These areas are mostly covered by desert and semi-desert habitats and exhibit regional differences in land-cover and prey population size and availability: (1) Touran, is mostly flat and rangelands are the most common ecosystem, it displays a variety of habitats including a saline river system, mountain systems with high peaks and vast sand areas, including moving dunes and salt flats contiguous to Dasht-e Kavir; (2) Miandasht, comprises flat plains interrupted by rolling hills and a seasonal river; (3) Naybandan, comprises mostly plains, with hilly and mountainous areas with high peaks, vast salt plains and moving sand dunes; and (4) Yazd, is the most mountainous area, comprises mostly mountainous habitat and rocky hills with some low and rolling hills66. Miandasht, with a maximum terrain roughness index (TRI) of 80.6 (SD = 11.4), is the only flat region. The other three regions contain more mountainous terrain and have higher TRI and altitude differences. Among them, the Yazd areas has the highest TRI difference (SD = 39.2) and highest mean altitude (1399 m). Consequently, prey populations are rather distinct in each of the regions. In terms of wild prey, the most abundant prey in Miandasht is goitered gazelle and in Touran is mouflon, while in Naybandan, the most abundant prey is ibex, followed by mouflon and jebeer. Finally, ibex and mouflon are abundant prey items in Yazd along with very few gazelles67,68. In terms of livestock presence, very large numbers of small livestock (sheep and goats) are found in the range of the Northern Subpopulation, while Southern Subpopulations are only hosting few herds69; about 1,500 dromedary camel graze freely in Naybandan, about the same number in Touran and about 400 in Miandasht but few are found in Yazd areas (Table 3).
Sample collection
Scat samples available for this study had two origins: (1) between May to September 2017, LK, EH, HA and TGH performed line transects across all protected areas to collect samples (N = 355). Surveys targeted trails, riverbeds, and shrub communities in the vicinity of previously known cheetah marking sites or other potential marking sites, such as large trees and big rocks. Given that the identification of large carnivores' scats in the field is prone to mistakes, we aimed for broad sampling, excluding only those prominent white scats from striped hyaenas (Hyaena hyaena)70, to include all potential cheetah scats; and (2) between 2014 and 2017, Conservation of Asiatic cheetah project (CACP) project members collected samples (N = 21) from Touran BR. In total, 376 putatively cheetah samples were available from Touran (181), Miandasht (81), Naybandan (40) and Yazd (74). Given the very low humidity of these areas, all collected samples were completely dry, thus they were stored in individual Ziploc bags, and then stored at room temperature until being transferred to the laboratory, where they were stored at − 20 °C until extraction. The fieldwork in the protected areas, collection of the samples and transferring them to the laboratory, were carried out under the permission from CACP. The geographic location of all samples was recorded with a Global Positioning System device on site in the WGS84 datum. Additionally, 15 tissue and hair samples of wild and domestic sheep and goats were collected to supplement the sequence database retrieved from NCBI (see “Laboratory analysis” section and Supplementary Table 3).
Laboratory analysis
The first step for the molecular analysis was the identification of the depositor of each scat sample (Fig. 2). The E.Z.N.A Tissue DNA Kit (Omega Bio-tek, USA) was used to extract DNA from each scat sample following the manufacturer’s instructions, with the following modifications: manufacturer's TL buffer was replaced with Gordon’s lysis buffer (0.1 M Tris–HCl, 0.1 M EDTA, 0.01 M NaCl, 1% N–lauroyl sarcosine, pH 7.5–871) and half of an inhibitEX® tablet (QIAGEN, GmbH, Hilden) was added after the first lysis step and before adding the OB protease to remove potential PCR inhibitors. Samples were extracted in batches of 24, each batch including a negative extraction control. Following extraction, DNA was amplified using the 16S_mam primers72 (Supplementary Table 4), modified to contain Illumina adaptors to allow the posterior addition of sample-specific barcodes. The PCRs were carried out in a final volume of 10 μl, comprising 5 μl of Qiagen Multiplex PCR Master Mix (Qiagen, Germany), 0.3 μl of each 10 pM primer and 2 μl of DNA extract. Cycling conditions were: initial denaturation at 95 °C for 15 min; 40 cycles of denaturation at 95 °C for 30 s, annealing at 52 °C for 30 s and extension at 72 °C for 30 s; a final extension at 72 °C for 10 min. Amplification success was checked by visually inspecting 2 μl of each PCR product on a 2% agarose gel stained with GelRed (Biotium, USA). Successfully amplified samples were then prepared for Illumina sequencing and sequences were blasted against the NCBI nt database to identify the predator species (see “Library preparation” and “Bioinformatics pipeline” sections).
The second step was the identification of food items in each scat sample (Fig. 2). From the samples that were confirmed to be of cheetah origin, DNA was extracted again following the protocol of Frantz et al. (2003)73 after applying the GuSCN /silica method74. As this method uses bigger portions of the scat, it allowed for DNA with better quality and quantity and minimising subsampling bias that can occur when smaller portions of scats are sampled75,76. Samples were extracted in batches of 32, with each batch including a negative extraction control. DNA was amplified using 12S primers77 (Supplementary Table 4; herein referred to as 12SV5.1), modified to contain Illumina adaptors. PCRs included a newly developed cheetah blocking primer for this primer set (Supplementary Table 4). PCR composition and conditions were the same as in the previous step but using an annealing temperature of 47 °C. Successfully amplified samples were then prepared for Illumina sequencing and sequences were blasted against the NCBI nt database to identify prey species (see the Library preparation and Bioinformatics pipeline sections). In both steps 1 and 2, to minimise the potential for contamination, extractions and manipulation of scat samples were carried out in a positive-pressure laboratory (CIBIO-InBio, Vairão Campus, Portugal), physically separated from other laboratory rooms that handle already extracted DNA material and PCR products, following strict protocols including disposable laboratory wear, UV sterilization of all equipment before entering the room, and cleaning laboratory surfaces with bleach between extraction batches. Instruments were sterilised by ethanol flaming twice between each sample. Additionally, each PCR plate included at least one PCR negative control to monitor for potential contaminations.
The third step consisted of distinguishing domestic from wild Ovis and Capra food items (Fig. 2). The metabarcode targeted by the 12SV5.1 primer pair does not provide resolution to achieve this. Instead, the mtDNA control region (D-Loop) was chosen for this aspect of the study, as this has been used previously to distinguish subspecies and lineages of goats78. A database was created comprising over 600 sequences of the control region from Iranian sheep and goats, mainly retrieved from NCBI, supplemented with 15 sequences obtained from tissue and hair-derived DNA extracts collected from wild and domestic sheep and goats in Iran. Each database entry was checked according to the data from original paper and designated as “Ovis wild”, “Ovis domestic”, “Capra wild” or “Capra domestic” (see Supplementary Table 3). Primers were designed to amplify a 100–122 bp fragment (excluding primer binding sites) within the region amplified by the CAP-FI and CAP-RI primer pair used by Luikart et al. (2001)78. Primers included degenerate bases to facilitate amplification of multiple species of Ovis and Capra.
PCR composition and conditions were the same as in the previous steps, but with an annealing temperature of 45 °C and a PCR cycle number of 38. Successfully amplified samples were then prepared for Illumina sequencing and sequences were blasted against the custom database (see “Library preparation” and “Bioinformatics pipeline” section).
In the fourth step, we assessed the individual ID of cheetah samples, using a set of 21 microsatellites developed for Felis catus79 and tested on cheetahs80,81,82,83. DNA samples were amplified using a multiple-tubes approach so as to reduce the genotyping errors84, using four replicates. PCR products were separated by size on an ABI 3130xl DNA analyzer (Applied Biosystems, Foster City, California). Microsatellite alleles were scored using the GeneScan500 LIZ size standard and GeneMapper v4.0 (Applied Biosystems, Foster City, California), and checked manually. Multiplex 2 was used first on all samples to assess the quality and samples with more than 70% of amplification were chosen to test with remaining multiplexes. For sex identification, we used ZFx gene in Multiplex 2. For details on multiplex combination and Thermocycling parameters allele range see Supplementary Table 4.
Library preparation
Amplicons from the initial PCRs from the first, second and third steps (see above) were prepared for sequencing using the Illumina platform following Egeter et al. (2018)85, with the following minor modifications: a ratio of 0.9 X AMPure XP beads (Beckman Coulter, USA) was used for the purification step; quantification of each purified PCR product was conducted using the EPOCH plate reader (BioTek, USA). Final pools were sequenced on either the Illumina Miseq (CIBIO, Portugal) or Hiseq (GeneWiz) platforms.
Bioinformatic pipeline
Reads were demultiplexed according to the sample-specific indexes using BASESPACE (basespace.illumina.com). Sequence data were processed using the MBC pipelines package (Galhardo et al. in prep.; see Supplementary Methods for exact commands). Within the package, paired-end reads were aligned using flash286 with the settings: –max-overlap = 100 -D -m 10 -t 1. Sequences outside the expected amplicon lengths were removed. Remaining sequences were demultiplexed using vsearch87 with fastq_maxee = 1 and singletons were removed. The zero-radius Operational Taxonomic Units (OTUs) were mapped against relevant databases, using the blastn algorithm, and 100 results per query were kept. To obtain full-length alignments only, rather than the partial alignments blastn can often return, the gap and extension penalties were relaxed and only alignments with a query cover of 98% were kept (see Supplementary Methods for exact commands). Blast results to the taxa that were not present in Iran67 were removed.
OTUs were placed in taxonomic bins using the metabin program in metabinkit (v0.1.0; https://doi.org/10.5281/zenodo.3855032). To choose taxonomic thresholds we: 1) measured the within-taxon and among-taxon percentage identity variation for all taxa in the blastn results, and; 2) run metabin numerous times with varying thresholds to ensure that the correct assignments were being retrieved for each database entry (see Supplementary Methods for more details). The final percentage identity taxonomic thresholds were species = 99%, genus = 99%, family = 99% and above-family = 99%. Within metabin mammal species that do not occur in the study area67 were disabled. To remove potential crosstalk between samples, detections that were < 0.1% of the total read count for the respective taxon were removed, as were detections that were < 0.1% of the total read count for the respective sample. To further remove any potentially spurious results, all detection < 40 reads were discarded. Detailed taxa-tables, produced from 16S_mam, 12SV5.1 and Egeter-2019-Cap primer, were presented in Supplementary Table 5, 6 and 7 respectively.
Consensus genotypes over four replicates were assembled following three rules: heterozygous genotypes were accepted if the same genotype was observed in two independent PCRs; homozygous genotypes were accepted if the genotype was observed in or three independent PCRs88; only one allele was accepted (and missing data for the second allele) at a locus when observed in each of two available replicates. We discarded replicate samples from analysis as determined by identical genotypes using Gimlet v1.3.289. The same software was used to measure mean allelic dropout and false allele rates across loci, samples, and PCRs. Finally, we estimated the probability of identical genotypes being shared by chance (probability of identity, PID, and Probability of Identity among siblings PIDsibs) to assess if our markers’ resolution was sufficient to reliably differentiate among closely related individuals, including siblings.
Statistical analysis
Given that the abundance data were collected using variable protocols (see “Sampling and barcoding constraints and innovations”), we were unable to estimate prey preference according to prey abundance, and instead we used more simple and descriptive analyses. To quantify Relative Frequency of Occurrence (RFO) of food items in diet composition, each prey species detected in one scat sample was assumed to represent a single predatory event15. Given that metabarcoding methods did not rely on visual identification of prey remaining, we did not used any correction factor90. The RFO is the relation of identified prey items to the number of all prey items found in all scats following Breuer 2005 and references therein91. We excluded food items that were observed in very low number of scats (< 3) or in very low reads in comparison to other food items. In the former case we consider that they were not repeated enough times to have significant impact on results, and in the latter case we consider that they could be the result of field contaminations (e.g., from prey's scat). RFO was calculated for each of the four regions and across the entire study area. Prey population size was extracted from DoE (Department of Environment of Iran) regional office census report of each province (Unpublished data, DoE). Given that cheetahs display large home-range sizes (1136 km2)52, all of each region was considered as hunting area of cheetah. Since we observed evidence of cheetah movements between protected areas in Yazd (i.e. identification of the same individual in several areas), all of them were considered as a single region.
Data availability
The raw sequencing data has been deposited at the European Nucleotide Archive (ENA) under the accession number PRJEB39665 (https://www.ebi.ac.uk/ena/browser/view/PRJEB39665).
References
Ceballos, G. & Ehrlich, P. R. Mammal population losses and the extinction crisis. Science 296, 904–907 (2002).
Barnosky, A. D. et al. Has the Earth’s sixth mass extinction already arrived?. Nature 471, 51–57 (2011).
Ripple, W. J. et al. Status and ecological effects of the world’s largest carnivores. Science 343, 1241484 (2014).
Carbone, C. & Gittleman, J. L. A common rule for the scaling of carnivore density. Science 295, 2273–2276 (2014).
Durant, S. M. et al. The global decline of cheetah Acinonyx jubatus and what it means for conservation. Proc. Natl. Acad. Sci. 114, 528–533 (2017).
Jowkar, H. et al. Acinonyx jubatus ssp. venaticus. The IUCN Red List of Threatened Species 2008: e.T220A13035342. (2008).
Khalatbari, L., Yusefi, G. H., Martínez-Freiría, F., Jowkar, H. & Brito, J. C. Availability of prey and natural habitats are related with temporal dynamics in range and habitat suitability for Asiatic Cheetah. Hystrix 29, 145–151 (2018).
Asadi, H. The Environmental Limitations and Future of the Asiatic Cheetah in Iran. (1997).
CACP. Annual Report. (2014).
Khalatbari, L., Jowkar, H., Yusefi, G. H., Brito, J. C. & Ostrowski, S. The current status of Asiatic cheetah in Iran. Cat News 66, 10–13 (2017).
Marker, L. L. et al. Ecology of free-ranging cheetahs. in Cheetahs: Biology and Conservation (eds. Marker, L. L., Boast, L. K. & Schmidt-Kuntzel, A.) 107–119 (Elsevier, 2017). doi:https://doi.org/10.1016/B978-0-12-804088-1.00008-3
Hayward, M. W., Hofmeyr, M., O’Brian, J. & Kerley, G. I. H. Prey preferences of the cheetah (Acinonyx jubatus) (Felidae: Carnivora): morphological limitations or the need to capture rapidly consumable prey before kleptoparasites arrive?. J. Zool. 270, 615–627 (2006).
Mills, M. G. L., Broomhall, L. S. & Toit, J. T. Cheetah Acinonyx jubatus feeding ecology in the Kruger National Park and a comparison across African savanna habitats: is the cheetah only a successful hunter on open grassland plains?. Wildlife Biol. 10, 177–186 (2004).
Wachter, B., Jauernig, O. & Breitenmoser, U. Determination of prey hair in faeces of free-ranging Namibian cheetahs with a simple method. Cat News 44, 8–9 (2006).
Marker, L. L., Muntifering, J. R., Dickman, A. J., Mills, M. G. L. & Macdonald, D. W. Quantifying prey preferences of free-ranging Namibian cheetahs. South Afr. J. Wildl. Res. 33, 43–53 (2003).
Wacher, T. et al. Sahelo-Saharan Interest Group Wildlife Surveys, Part 4: Ahaggar Mountains, Algeria (March 2005). (2005).
Thuo, D. et al. An insight into the prey spectra and livestock predation by cheetahs in Kenya using faecal DNA metabarcoding. Zoology 143, 125853 (2020).
Broekhuis, F., Thuo, D. & Hayward, M. W. Feeding ecology of cheetahs in the Maasai Mara, Kenya and the potential for intra- and interspecific competition. J. Zool. 304, 65–72 (2018).
Cooper, A. B., Pettorelli, N. & Durant, S. M. Large carnivore menus: factors affecting hunting decisions by cheetahs in the Serengeti. Anim. Behav. 73, 651–659 (2007).
Mills, M. G. L. Living near the edge: A review of the ecological relationships between large carnivores in the arid Kalahari. African J. Wildl. Res. 45, 127–137 (2015).
Rostro-García, S., Kamler, J. F. & Hunter, L. T. B. To kill, stay or flee: The effects of lions and landscape factors on habitat and kill site selection of cheetahs in South Africa. PLoS ONE 10, e0117743 (2015).
Laurenson, M. K. Behavioural costs and constraints of lactation in free-living cheetahs. Anim. Behav. 50, 815–826 (1995).
Farhadinia, M. S. & Hemami, M.-R. Prey selection by the critically endangered Asiatic cheetah in central Iran. J. Nat. Hist. 44, 1239–1249 (2010).
Farhadinia, M. S. et al. Feeding ecology of the Asiatic cheetah Acinonyx jubatus venaticus in low prey habitats in northeastern Iran: Implications for effective conservation. J. Arid Environ. 87, 206–211 (2012).
Zahedian, B. & Nezami, B. Cheetah (Acinonyx jubatus venaticus) (Felidae: Carnivora) feeding ecology in Central Plateau of Iran and effects of prey poor management. J. Wildl. Biodivers. 3, 22–30 (2019).
Zamani, N. et al. Predation of montane deserts ungulates by Asiatic cheetah Acinonyx jubatus venaticus in Central Iran. Folia Zool. 66, 50–57 (2017).
Monterroso, P. et al. Factors affecting the (in)accuracy of mammalian mesocarnivore scat identification in South-western Europe. J. Zool. 289, 243–250 (2013).
Morin, D. J. et al. Bias in carnivore diet analysis resulting from misclassification of predator scats based on field identification. Wildl. Soc. Bull. 40, 669–677 (2016).
Caro, T. M. Cheetahs of the Serengeti Plains: Group Living in an Asocial Species (University of Chicago Press, 1994).
Floyd, T. J., Mech, L. D. & Jordan, P. A. Relating wolf scat content to prey consumed. J. Wildl. Manage. 42, 528–532 (1978).
Jethva, B. D. & Jhala, Y. V. Computing biomass consumption from prey occurrences in Indian wolf scats. Zoo Biol. 23, 513–520 (2004).
Pompanon, F. et al. Who is eating what: diet assessment using next generation sequencing. Mol. Ecol. 21, 1931–1950 (2012).
Taberlet, P., Coissac, E., Pompanon, F., Brochmann, C. & Willerslev, E. Towards next-generation biodiversity assessment using DNA metabarcoding. Mol. Ecol. 21, 2045–2050 (2012).
Mata, V. A. et al. How much is enough? Effects of technical and biological replication on metabarcoding dietary analysis. Mol. Ecol. 28, 165–175 (2019).
Shehzad, W. et al. Prey preference of Snow Leopard (Panthera uncia) in South Gobi Mongolia. PLoS ONE 7, e32104 (2012).
Monterroso, P. et al. Feeding ecological knowledge: the underutilised power of faecal DNA approaches for carnivore diet analysis. Mamm. Rev. 49, 97–112 (2019).
Shehzad, W. et al. Carnivore diet analysis based on next-generation sequencing: Application to the leopard cat (Prionailurus bengalensis) in Pakistan. Mol. Ecol. 21, 1951–1965 (2012).
Thuo, D. et al. Food from faeces: Evaluating the efficacy of scat DNA metabarcoding in dietary analyses. PLoS ONE 14, e0225805 (2019).
Araujo, M. S., Bolnick, D. I. & Layman, C. A. The ecological causes of individual specialisation. Ecol. Lett. 14, 948–958 (2011).
Balme, G. A., Roex, N., Rogan, M. S. & Hunter, L. T. B. Ecological opportunity drives individual dietary specialization in leopards. J. Anim. Ecol. 89, 589–600 (2020).
Bolnick, D. I. et al. The ecology of individuals: incidence and implications of individual specialization. Am. Nat. 161, 1–28 (2003).
Harrington, L. A., Harrington, A. L., Hughes, J., Stirling, D. & Macdonald, D. W. The accuracy of scat identification in distribution surveys: American mink, Neovison vison, in the northern highlands of Scotland. Eur. J. Wildl. Res. 56, 377–384 (2010).
Weiskopf, S. R., Kachel, S. M. & McCarthy, K. P. What are snow leopards really eating? Identifying bias in food-habit studies. Wildl. Soc. Bull. 40, 233–240 (2016).
Durant, S. M., Caro, T. M., Collins, D. A., Alawi, R. M. & Fitzgibbon, C. D. Migration patterns of Thomson’s gazelles and cheetahs on the Serengeti Plains. Afr. J. Ecol. 26, 257–268 (1988).
Lindsey, P. A. et al. Minimum prey and area requirements of the vulnerable cheetah Acinonyx jubatus: implications for reintroduction and management of the species in South Africa. Oryx 45, 587–599 (2011).
Farhadinia, M. S., Akbari, H., Eslami, M. & Adibi, M. A. A review of ecology and conservation status of Asiatic cheetah in Iran. Cat News Spec. Issue 18–26 (2016).
Asadi, H. Some Observation on Hunting Behaviours of the Iranian Cheetah in Captivity. (1997).
Heptner, V. G. & Sludskii, A. A. Mammals ofthe Soviet Union volume II part 2 Carnivora (hyaenas and cats). (Vysshaya Shkola Publishers, 1974).
Ziaie, H. A Field Guide to the Mammals of Iran. (Iran Wildlife Center, 2008).
Wilson, J. W. et al. Cheetahs, Acinonyx jubatus, balance turn capacity with pace when chasing prey. Biol. Lett. 9, 20130620 (2013).
Grohé, C., Lee, B. & Flynn, J. J. Recent inner ear specialization for high-speed hunting in cheetahs. Sci. Rep. 8, 2301 (2018).
Cheraghi, F. et al. Inter-dependent movements of Asiatic Cheetahs Acinonyx jubatus venaticus and a Persian Leopard Panthera pardus saxicolor in a desert environment in Iran (Mammalia: Felidae). Zool. Middle East 65, 283–292 (2019).
Ghoddousi, A., Soofi, M., Hamidi, A. K. & Lumetsberger, T. Assessing the role of livestock in big cat prey choice using spatiotemporal availability patterns. PLoS ONE 11, e0153439 (2016).
Khorozyan, I., Ghoddousi, A., Soofi, M. & Waltert, M. Big cats kill more livestock when wild prey reaches a minimum threshold. Biol. Conserv. 192, 268–275 (2015).
Zeder, M. A. Domestication and early agriculture in the Mediterranean Basin: Origins, diffusion, and impact. Proc. Natl. Acad. Sci. 105, 11597–11604 (2008).
Daberger, M. Systematic prioritization of livestock grazing rights buyout in the last viable population of Asiatic cheetah (Acinonyx jubatus venaticus) in Iran. (Humboldt University Berlin, 2021).
Wolf, C. & Ripple, W. J. Prey depletion as a threat to the world’s large carnivores. R. Soc. Open Sci. 3, 160252 (2016).
Melzheimer, J. et al. Communication hubs of an asocial cat are the source of a human—carnivore conflict and key to its solution. Proc. Natl. Acad. Sci. 117, 33325–33333 (2020).
Malakoutikhah, S., Fakheran, S., Tarkesh, M. & Senn, J. Assessing future distribution, suitability of corridors and efficiency of protected areas to conserve vulnerable ungulates under climate change. Divers. Distrib. 26, 1383–1396 (2020).
Long, R. A., Donovan, T. M., Mackay, P., Zielinski, W. J. & Buzas, J. S. Comparing scat detection dogs, cameras, and hair snares for surveying carnivores. J. Wildl. Manage. 71, 2018–2025 (2007).
Becker, M. S. et al. Using dogs to find cats: Detection dogs as a survey method for wide-ranging cheetah. J. Zool. 302, 184–192 (2017).
Johnson, W. E. & O’Brien, S. J. Phylogenetic reconstruction of the Felidae using 16S rRNA and NADH-5 mitochondrial genes. J. Mol. Evol. 44, S98–S116 (1997).
Reese, E. M., Winters, M., Booth, R. K. & Wasser, S. K. Development of a mitochondrial DNA marker that distinguishes domestic dogs from Washington state gray wolves. Conserv. Genet. Resour. 12, 497–501 (2020).
Ormerod, S. J. Applied issues with predators and predation: Editor’s introduction. J. Appl. Ecol. 39, 181–188 (2002).
Boast, L. K., Good, K. & Klein, R. Translocation of problem predators: Is it an effective way to mitigate conflict between farmers and cheetahs Acinonyx jubatus in Botswana?. Oryx 50, 537–544 (2016).
Darvish Sefat, A. A. Atlas of Protected Areas of Iran (University of Tehran, 2006).
Yusefi, G. H., Faizolahi, K., Darvish, J., Safi, K. & Brito, J. C. The species diversity, distribution, and conservation status of the terrestrial mammals of Iran. J. Mammal. 100, 55–71 (2019).
Karami, M., Ghadirian, T. & Faizolahi, K. The Atlas of the Mammals of Iran. (Iran Department of the Environment, 2016).
Abangah Consulting Engineer Company. Reconvene expanded Livestock Control Committee (LCC) in Touran and establish the LCC for Miandasht with participation of all stakeholders. (2017).
Mills, M. G. L. & Hofer, H. Hyaenas. Status Survey and Conservation Action Plan. (IUCN/SSC Hyaena Specualist Group, 1998).
Maudet, C., Luikart, G., Dubray, D., Von Hardenberg, A. & Taberlet, P. Low genotyping error rates in wild ungulate faeces sampled in winter. Mol. Ecol. Notes 4, 772–775 (2004).
Deagle, B. E., Kirkwood, R. & Jarman, S. N. Analysis of Australian fur seal diet by pyrosequencing prey DNA in faeces. Mol. Ecol. 18, 2022–2038 (2009).
Frantz, A. C. et al. Reliable microsatellite genotyping of the Eurasian badger (Meles meles) using faecal DNA. Mol. Ecol. 12, 1649–1661 (2003).
Boom, R. et al. Rapid and simple method for purification of nucleic acids. J. Clin. Microbiol. 28, 495–503 (1990).
Rosel, P. E. & Kocher, T. D. DNA-based identification of larval cod in stomach contents of predatory fishes. J. Exp. Mar. Bio. Ecol. 267, 75–88 (2002).
Deagle, B. E. et al. Molecular scatology as a tool to study diet: analysis of prey DNA in scats from captive Steller sea lions. Mol. Ecol. 14, 1831–1842 (2005).
Riaz, T. et al. ecoPrimers: inference of new DNA barcode markers from whole genome sequence analysis. Nucleic Acids Res. 39, e145 (2011).
Luikart, G. et al. Multiple maternal origins and weak phylogeographic structure in domestic goats. Proc. Natl. Acad. Sci. 98, 5927–5932 (2001).
Menotti-Raymond, M. et al. A genetic linkage map of microsatellites in the domestic cat (Felis catus). Genomics 57, 9–23 (1999).
Charruau, P. et al. Phylogeography, genetic structure and population divergence time of cheetahs in Africa and Asia: Evidence for long-term geographic isolates. Mol. Ecol. 20, 706–724 (2011).
Driscoll, C. A., Menotti-Raymond, M., Nelson, G., Goldstein, D. & O’Brien, S. J. Genomic microsatellites as evolutionary chronometers: A test in wild cats. Genome Res. 12, 414–423 (2002).
Kotze, A., Ehlers, K., Cilliers, D. C. & Grobler, J. P. The power of resolution of microsatellite markers and assignment tests to determine the geographic origin of cheetah (Acinonyx jubatus) in Southern Africa. Mamm. Biol. 73, 457–462 (2008).
Marker, L. L. et al. Molecular genetic insights on cheetah (Acinonyx jubatus) ecology and conservation in Namibia. J. Hered. 99, 2–13 (2008).
Taberlet, P. et al. Reliable genotyping of samples with very low DNA quantities using PCR. Nucleic Acids Res. 24, 3189–3194 (1996).
Egeter, B. et al. Challenges for assessing vertebrate diversity in turbid Saharan water-bodies using environmental DNA. Genome 61, 807–814 (2018).
Magoc, T. & Salzberg, S. L. FLASH: Fast length adjustment of short reads to improve genome assemblies. Bioinformatics 27, 2957–2963 (2011).
Rognes, T., Flouri, T., Nichols, B., Quince, C. & Mahé, F. VSEARCH: A versatile open source tool for metagenomics. PeerJ 4, e2584 (2016).
Godinho, R. et al. Real-time assessment of hybridization between wolves and dogs: Combining noninvasive samples with ancestry informative markers. Mol. Ecol. Resour. 15, 317–328 (2015).
Valière, N. GIMLET: A computer program for analysing individual identification data. Mol. Ecol. 2, 377–379 (2002).
Wachter, B. et al. An advanced method to assess the diet of free-ranging large carnivores based on scats. PLoS ONE 7, e38066 (2012).
Breuer, T. Diet choice of large carnivores in northern Cameroon. Afr. J. Ecol. 43, 181–190 (2005).
Wilson, M. F. J., O’Connell, B., Brown, C., Guinan, J. C. & Grehan, A. J. Multiscale terrain analysis of multibeam bathymetry data for habitat mapping on the continental slope. Mar. Geodesy 30, 2 (2007).
Fick, S. E. & Hijmans, R. J. WorldClim 2: New 1-km spatial resolution climate surfaces for global land areas. Int. J. Climatol. 37, 4302–4315 (2017).
Acknowledgements
We want to thank DoE rangers who helped us during the field work. We also acknowledge Ahmad Ajami, Bahman Janafi, Mahghol Kazari, Ladan Salamat and Gholam Hosein Yusefi for their help and participation in the field work, and Susana Lopes and Diana Castro for their support in the laboratory analyses. This work was supported by National Funds through FCT-Fundação para a Ciência e a Tecnologia (FCT) in the scope of the project UIDB/50027/2020. The authors also acknowledge research funding from the European Union's Horizon 2020 research and innovation programme under grant agreement No 668981, by AGRIGEN–NORTE-01-0145- FEDER-000007, supported by Norte Portugal Regional Operational Programme (NORTE2020), under the PORTUGAL 2020 Partnership Agreement, through the European Regional Development Fund (ERDF), and by Rufford Foundation (21607-1). L.K., and JC.B. were supported by FCT (PD/BD/132429/2017 and CEECINST/00014/2018/CP1512/CT0001, respectively).
Author information
Authors and Affiliations
Contributions
L.K., B.E. and JC.B. designed the study, L.K., H.A., E.H., T.GH. and AH.K.H. conducted the fieldwork. L.K. and B.E. designed the molecular work and L.K. performed the lab work. L.K., B.E., U.B. and JC.B. wrote the manuscript. All authors discussed and commented on the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Khalatbari, L., Egeter, B., Abolghasemi, H. et al. Assessing Asiatic cheetah’s individual diet using metabarcoding and its implication for conservation. Sci Rep 12, 11403 (2022). https://doi.org/10.1038/s41598-022-15065-1
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-022-15065-1
This article is cited by
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.