Abstract
Nucleotide second messengers are universally crucial factors for the signal transduction of various organisms. In prokaryotes, cyclic nucleotide messengers are involved in the bacterial life cycle and in functions such as virulence and biofilm formation, mainly via gene regulation. Here, we show that the swimming motility of the soil bacterium Leptospira kobayashii is rapidly modulated by light stimulation. Analysis of a loss-of-photoresponsivity mutant obtained by transposon random mutagenesis identified the novel sensory gene, and its expression in Escherichia coli through codon optimization elucidated the light-dependent synthesis of cyclic adenosine monophosphate (cAMP). GFP labeling showed the localization of the photoresponsive enzyme at the cell poles where flagellar motors reside. These findings suggest a new role for cAMP in rapidly controlling the flagella-dependent motility of Leptospira and highlight the global distribution of the newly discovered photoactivated cyclase among diverse microbial species.
Similar content being viewed by others
Introduction
In all domains of life, the nucleotide messengers cAMP, cGMP, c-di-AMP, and c-di-GMP have vital roles in signaling networks, allowing organisms to recognize changes in environmental factors and regulate their physiologies and behaviors. Many cyclic nucleotide-binding proteins have been identified, e.g., the binding of cAMP or cGMP to cyclic nucleotide-gated channels triggers cation flux1. The capacitation of mammalian sperm involves phosphorylation of the associated proteins by the cAMP-dependent activation of protein kinase A, and flagellar beating is accelerated by cAMP derived from soluble adenylyl cyclase2. Additionally, the c-di-GMP level is known to be involved in the biofilm formation, motility, and virulence of bacteria3,4.
Here, we show that the motility of a species of the genus Leptospira, a member of the spirochetes, is rapidly modulated by cAMP generated upon light exposure. Motile bacteria possess varied machinery for moving in liquid or over surfaces5. A major motility form is flagella-dependent swimming: the peritrichous bacteria Escherichia coli and Salmonella enterica swim by rotating bundled flagella, and Vibrio cholerae and Pseudomonas aeruginosa are propelled by a single polar flagellum6. While these species possess flagella at the cell exterior, spirochetes such as Borrelia burgdorferi (Lyme disease pathogen) and Treponema pallidum (syphilis) have their flagella (endoflagella) within the periplasmic space7. The endoflagella of spirochetes are thought to rotate within the periplasmic space, rolling or transforming the spiral cell body to generate thrust7. We discovered that Leptospira kobayashii isolated from soil in Japan8,9 drastically alters its swimming pattern immediately after sensing light. We demonstrated the responsibility of cAMP synthesized by a novel photoresponsive adenylyl cyclase for motility control. cAMP affects gene regulation for pili synthesis in cyanobacteria10,11,12, but the soil bacteria respond to light exposure in a subsecond timeframe.
Results
Light modulates swimming of soil bacteria
The genus Leptospira possesses two endoflagella (one flagellum per cell end, Supplementary Fig. 1). Leptospira spp. swim by rotating the coiled cell body (swimming mode), and smooth swimming is frequently interrupted by rotation without migration (rotation mode)13,14. We found that L. kobayashii exclusively showed rotation in the low light condition (0.2 μmol/m2/s, “Light OFF” in Fig. 1), but smooth swimming was triggered by light exposure (26.1 μmol/m2/s, “Light ON” in Fig. 1, Movie 1, Supplementary Fig. 2). It is known that the transition between the swimming and rotation modes of Leptospira spp. is affected by viscosity15 and chemical substrates16,17, but this is the first report of light-dependent modulation of Leptospira motility. The frequency of swimming reversal decreased with light intensity, increasing the migration distance per unit time (Supplementary Fig. 3). Therefore, light-dependent motility was quantified using the swimming velocity of individual cells (i.e., migration distance per second), showing an increase in velocity up to fourfold under light (Fig. 1b, Supplementary Fig. 3). Smooth swimming reached the maximum velocity at ~ 1 s after stimulation (Fig. 1c). The light-responsivity depends on the light intensity, and the duration of unidirectional swimming increased with increasing light intensity (Supplementary Fig. 3). The response to light was observed even at ~ 1 μmol/m2/s, which is less than 1/10 of the light in a conventional experimental room illuminated by 32 W fluorescent lamps (500–600 lx) (Supplementary Fig. 3). The bacteria can respond to green and blue light but not red light (Fig. 1d). Cumulative cell fractions obtained from the velocity histograms show that green light induces more cells to swim smoothly than blue light (Fig. 1e).
Light stimulation induces unidirectional swimming of spirochetal bacteria. (a) Swimming trajectories of Leptospira kobayashii cells under low-intensity (termed as “Light OFF” in this study) and high-intensity (termed as “Light ON”) illumination. The results of 2-s tracking are shown. Colored footprints (inset, time courses in the order from red to blue) show almost zero net migration in Light OFF. Note that the bacteria retain vigorous motility in Light OFF (see the scene “Light OFF” in Movie 1). As a light source, a halogen lamp was used with a wide band-pass filter (ca. 400–700 nm). (b) Velocities of individual cells measured in Light OFF (n = 189 cells) and in Light ON (n = 150 cells). The data were obtained by three independent experiments. (c) Time courses of cell displacement. Example traces obtained from 14 cells (thin colored lines) and the averaged trace (thick black line) are shown. Neutral density filters were removed at the time point (indicated by the triangle and set as 0 s) so that the cells were suddenly exposed to stronger light. The sequential micrographs (inset) show the light-dependent initiation of directional swimming. The right panel shows the time course of velocity (gray) obtained from the mean displacement and the result of sigmoidal curve fitting (red). (d) Color dependence of the light-controlled motility. Band-pass filters with center wavelengths of 650 nm, 550 nm, and 488 nm were used for red, green, and blue illumination, respectively. The experiments were repeated three times, and ca. 100 cells were measured in total in each condition. (e) Cumulative cell fractions calculated from the histograms of Light ON shown in (d). The colors indicate those used for illumination: average values (closed circles) and standard deviations (error bars) of three independent experiments.
Identification of a photoresponsive gene
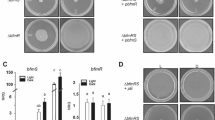
To identify the sensor gene responsible for the L. kobayashii photoresponsivity, we explored loss-of-function mutants from a kanamycin-resistant library made by transposon random mutagenesis. One of the ~ 2400 clones retained motility but lacked photoresponsivity (Fig. 2a, Movie 2), and this mutant was named Prd (photoresponsivity-deficient). The Prd mutant carried a transposon insertion in the LPTSP3_g09850 gene (Fig. 2b), but the complementation of the LPTSP3_g09850 gene was not able to restore the photoresponsivity of the mutant (Supplementary Fig. 4). Noting that the LPTSP3_g09840 gene is located immediately downstream of the LPTSP3_g09850 gene and encodes adenylate/guanylate cyclase domain-containing protein, we complemented the Prd mutant with both the LPTSP3_g09850 and LPTSP3_g09840 genes, resulting in the recovery of photoresponsivity (Fig. 2c,d). Then, the photoresponsivity of the Prd mutant was recovered to the wild-type level by complementation of the LPTSP3_g09840 gene (Fig. 2d), indicating that the LPTSP3_g09840 gene is responsible for the photoresponsivity of L. kobayashii. We termed the LPTSP3_g09840 gene lprA (leptospiral photoresponsive protein A).
Identification of a photoresponsive gene in the spirochete Leptospira. (a) Time traces of the cell displacement under light exposure. The traces of 8 cells are shown in each strain. (b) Transposon insertion in the photoresponsivity-deficient mutant Prd. The position of the transposon insertion is between 1133919 and 1133920th nucleotides of the L. kobayashii chromosome 1 (DDBJ/EMBL/GenBank accession number AP025028). (c) Immunoblotting of whole-cell lysates from the wild-type L. kobayashii, Prd, and its complemented strains using anti-LprA and anti-FlaA2 (control) antisera. (d) Effect of transposon insertion and gene complementation on photoresponsivity. Swimming velocities were measured in Light OFF (−) and Light ON (+) conditions. The average values and standard errors were determined by three independent experiments (ca. 100 cells were measured in total in each condition).
Light-dependent adenylyl cyclase activity of LprA
Photoactivated adenylyl cyclases (PACs) have been found in both eukaryotes and prokaryotes18,19,20,21,22,23,24. The alignment of LprA with known functional PACs showed that the C-terminal region of LprA contains a domain similar to the AC domain of known PACs, whereas the N-terminal putative sensor domain of LprA was distinct from the conventional PAC sensor BLUF or LOV domain (Supplementary Fig. 5). Since the light-dependent elevation of cAMP concentration was not detected in L. kobayashii cells (Supplementary Fig. 6a), we measured the enzyme activity using the E. coli overexpression system carrying the codon-optimized lprA (Fig. 3a). The overexpressed LprA showed an increase in the cAMP concentration in E. coli under light exposure (Fig. 3b). Very little cGMP was detected independently of light, and the small amount could consist of endogenous substrates (Supplementary Fig. 6b). These results suggest that LprA is a novel photoactivation-associated adenylyl cyclase. The Prd mutant cells kept rotating in the Light ON condition, but the addition of a membrane-permeable cAMP (8-bromo-cAMP) induced smooth swimming of the mutant as well as the wild-type cells exposed to light (Fig. 3c). An 8-bromo-cAMP-dependent increase in swimming velocity was also induced in the WT and Prd under the low light condition (0.2 μmol/m2/s; Supplementary Fig. 7). These results suggest that the observed modulation of flagellar rotation was induced by cAMP synthesized upon light stimulation.
Enzyme activity of LprA. (a) Immunoblotting of whole-cell lysates of L. kobayashii (1.5 × 108 cells) and E. coli expressing LprA-His (2.0 × 107 cells) with anti-LprA antiserum. Since long-time exposure was required for detecting the band of L. kobayashii, the E. coli sample was diluted at 1:10 for observing the band of the overexpressed proteins clearly. (b) Light-dependent synthesis of cAMP in E. coli carrying the codon-optimized lprA. The average values and standard deviations were determined by three independent experiments. “IPTG − ” and “Light − ” are the uninduced and unilluminated controls, respectively. (c) Effect of externally supplemented membrane-permeable cAMP analog (8-bromo-cAMP) on the Prd swimming. Video recording was started less than 1 min after the addition of 8-bromo-cAMP. The average values and standard errors were determined by three independent experiments (ca. 100 cells were measured in total in each concentration).
Bipolar localization of LprA
We examined the localization of LprA in the cell body by labeling the protein with AcGFP1 (green fluorescent protein derived from Aequorea coerulescens). AcGFP1 labeling did not affect the photoresponsivity of the bacterium (Supplementary Fig. 8a). Epifluorescence microscopy showed the localization of LprA-AcGFP1 at both ends of the cell body (Fig. 4a). Based on the fluorescence intensity of single AcGFP1 molecules, we estimated that 5.5 ± 3.8 molecules of LprA (n = 46 poles) are localized at one pole (Fig. 4b and Supplementary Fig. 8b), while those in the membrane pool diffuse along the cell body (Movie 3). Although the increase in cAMP concentration in L. kobayashii was likely to be below the detection limit (Supplementary Fig. 6), the bipolar localization of LprA could condense cAMP near the flagellar motor, resulting in a rapid response to light.
Subcellular localization of LprA. (a) Fluorescent images of LprA-AcGFP expressed in the Prd mutant (upper) and 2D intensity profiles (lower). (b) Box plot of the number of LprA-AcGFP molecules localized at one end of the cell body. The lower and upper box boundaries are 25th and 75th percentiles, respectively. The line in the middle of the box shows the median number. The lower and upper error lines indicate the smallest and largest values, respectively. 46 cell poles were analyzed.
Discussion
Organisms react to light through various sensory systems, leading to vision, signaling, and energetic activities. For example, rhodopsins, which are retinal-binding membrane proteins, play a crucial role in vision, ion pumping, and microbial taxis25,26, and cryptochromes, which are flavin-binding proteins, are involved in the growth of plants and the circadian clock of animals27. Investigating such light-driven proteins deepens our understanding of essential cellular activities. In addition, they have great potential for application as an optogenetic tool, enabling energetic or signaling modulation in live cells under arbitrary spatial and temporal conditions. We showed that L. kobayashii modulates its swimming pattern on a subsecond timescale after increasing illumination intensity and that the adenylyl cyclase discovered in this soil spirochete synthesizes cAMP upon light stimulation. The E. coli expression system experiment showed that light exposure is indispensable for activating LprA (Fig. 3b), suggesting that LprA functions as a photosensor by itself or is activated by interacting with another photosensor. Since photosensory activity needs a chromophore, the former hypothesis implies that LprA may use a common chromophore with E. coli. Although cAMP signaling has been known to be involved in pilus/flagellar biogenesis and biofilm formation in bacteria, these responses are slow and occur through gene regulation10,12,28. The current results suggest that cAMP could mediate rapid motility control, although further studies are required to determine whether cAMP acts directly on the flagellar motor.
The genus Leptospira comprises pathogenic and saprophytic species8. The species that belong to the saprophytic clade S2 containing L. kobayashii can respond to light, whereas one of the major pathogenic species, L. interrogans, swims even in low light and does not react to light despite carrying the homologous gene (Supplementary Fig. 9). Since the LprA-deficient mutant retains the ability to rotate in one position but cannot migrate by swimming (Movie 2), cAMP-dependent swimming could be important for exploring the environments of free-living species. In contrast, the pathogen motility independent of light is significant for migrating within the host body29. Interestingly, a search of the amino acid sequence database showed that many microbial species have a gene homologous to LprA in their genome (Supplementary Fig. 10). Although LprA is not involved in photoresponsive motility in all bacteria, cAMP synthesized by LprA may be used in some signal transductions. Adenylyl cyclases are categorized into six classes, e.g., class I has been found in gamma-proteobacteria, and class II is used by pathogenic bacteria, such as Bacillus anthracis and P. aeruginosa30. Since PACs that have functional similarity to LprA belong to a universal class III used by many species of eukaryotes (e.g., fungi and protozoa) and prokaryotes (e.g., eubacteria and archaea), the successful expression of functional LprA in other bacterial species is the crucial first step for optogenetic application. Understanding the molecular basis of the fast photosensory responses of LprA and cAMP signaling will be a future research topic.
Methods
Bacteria and media
Leptospira spp. were grown in enriched Ellinghausen-McCullough-Johnson-Harris (EMJH) liquid medium (BD Difco, NJ, USA) at 30 °C for 2 to 4 days (depends on the strains) until the log-phase. E. coli strains C41(DE3) and β2163 were grown in L-broth and LB medium supplemented with 0.3 mM diaminopimelate, respectively.
Screening of a photoresponsivity-deficient mutant by random transposon mutagenesis
Random insertion mutagenesis of L. kobayashii strain E30 using Himar1 transposon was conducted as described previously31,32. By conjugation with E. coli β2163 carrying pCjTKS232, about 2400 transconjugant colonies were observed on plates of EMJH agar (1% agar) containing kanamycin at a final concentration of 25 μg/ml. Each transconjugant was independently inoculated to 150 μl of liquid EMJH containing kanamycin and grown at 30 °C for 4 days. The grown bacteria were observed using a dark-field microscope (BX50, mercury lamp, 10 × objective, dry condenser; Olympus) for screening photoresponsivity-deficient mutants. The transposon insertion site was identified using the semi-random PCR technique32.
For the complementation of photoresponsive-deficient mutant, Prd, the LPTSP3_g09850 and/or lprA genes were expressed under the flgB promoter33,34. The flgB promoter region was amplified as previously described29, and the LPTSP3_g09850/lprA genes, lprA, LPTSP3_g09850 gene with FLAG tag, and lprA with FLAG tag were amplified from genomic DNA of the L. kobayashii E30, and the amplified products were cloned into the SalI-digested pCjSpLe9435 by NEBuilder HiFi DNA Assembly cloning (New England BioLabs). For the LprA with GFP joined by a flexible (GGGGS)3 linker, lprA was amplified as described above, and gfp was amplified from pAcGFP1 (Clontech) using the primer containing the (GGGGS)3 linker sequence, and the amplified products were cloned into the SalI-digested pCjSpLe94 as described above. Primers used in this study are listed in Supplementary Table 1.
Immunoblotting experiments
About 1.5 × 108 leptospiral cells suspended in SDS-PAGE sample buffer were subjected to 5–20% SDS-PAGE and Western blotting. The blot was incubated with antisera raised against the peptide fragment of LprA (NH2-LSWADRTDSIYIWK-COOH) and FlaA231 or monoclonal antibody for FLAG tag. Full-size images of immunoblotting data without cropping are shown in Supplementary Fig. 11.
Motility assay
Leptospira culture was diluted 1:20 into a fresh EMJH and was infused into a flow chamber composed of a coverslip (upper side) and a glass slide (bottom side). To examine the effect of membrane-permeable cAMP and cGMP on swimming, 8-bromo-cAMP and 8-bromo-cGMP dissolved in 10 mM Tris–HCl (pH7.0) were added to L. kobayashii cells suspended with the same buffer. Motility of leptospires was observed under a dark-field microscope (BX53, Splan 40× , NA 0.75; Olympus, Tokyo, Japan) equipped with a halogen lamp and were recorded by a supersensitive charge-coupled device (CCD) camera (WAT- 910HX, Watec Co., Yamagata, Japan) at a frame rate of 30 frames per second. Swimming trajectories and velocities of individual cells were analyzed using ImageJ software (National Institutes of Health, MD, USA) and VBA-based macros programmed in Microsoft Excel (Microsoft, WA, USA)14. Light intensity and wavelength were adjusted with neutral density (ND) filters and bandpass filters with 50-nm bandwidth (FF01-488/50-25 for blue, FF01-550/49-25 for green, FF01-650/54-25 for red; Semrock), respectively. Light intensity was measured using an illuminometer (CHE-LT1, Sanwa Supply INC.).
Observation of LprA-AcGFP1 subcellular localization
Fluorescence of LprA-AcGFP1 was observed using an inverted fluorescence microscope (IX-83, Olympus) with a 100× oil immersion objective lens (UPLSAPO100XO, NA 1.4, Olympus) and an sCMOS camera (Prime95B, Photometrics). AcGFP1 was excited by a 130 W mercury light source system (U-HGLGPS, Olympus) with a fluorescence mirror unit U-FGFP (Excitation BP 460–480; Emission BP 495–540, Olympus). Fluorescence image processing was performed with the ImageJ version 1.53 software (National Institutes of Health).
The number of LprA-AcGFP1 at the cell pole was estimated by comparing the fluorescence spot intensity at the pole with the fluorescence intensity of a single His-AcGFP1 molecule as previously reported36. His-AcGFP1 was purified from E. coli C41(DE3) cells carrying pET19b/ His-AcGFP1 using Ni–NTA agarose (Fujifilm Wako). 10 pg/ml of His-AcGFP1 solution was applied to a coverslip washed by 0.1 M KOH and observed by fluorescence microscopy. In the fluorescent images, a rectangular mask for the fluorescent spot of 30 × 30 pixels was applied to the ROI (region of interest). We defined the spot intensity as the sum of all pixel values within the rectangular mask after subtracting the total background intensity from each pixel value. The number of LprA-AcGFP1 per pole was estimated as the intensity of the cell pole divided by the average intensity of a single His-AcGFP1 molecule.
cAMP ELISA assay
A cAMP ELISA kit (ADI-900-066, Enzo Life Sciences) was used to determine intracellular cAMP levels following the manufacturer’s instructions. E. coli C41(DE3) cells carrying pET22b/lprA-His were grown in L-broth containing 100 μg/mL ampicillin with or without 1 mM IPTG at 30 °C for 5 h with shaking. The codon usages of the lprA sequence were optimized to those of E. coli for efficient protein expression. L. kobayashii cells were grown at 30 °C for 4 days in EMJH liquid medium until the late-exponential phase. Cells were photo-stimulated with white LED (1156 μmol/m2/s) for 3 min and subsequently lysed. Cell lysates were used to calculate cAMP values. The intracellular cAMP concentrations of E. coli and Leptospira cells were normalized to OD600 and OD420, respectively.
Data availability
The data supporting the findings of this study are available from the corresponding author upon request.
References
Beavo, J. A. & Brunton, L. L. Cyclic nucleotide research: Still expanding after half a century. Nat. Rev. Mol. Cell Biol. 3, 710–717 (2002).
Jansen, V. et al. Controlling fertilization and cAMP signaling in sperm by optogenetics. Elife 4, e05161 (2015).
Valentini, M. & Filloux, A. Biofilms and Cyclic di-GMP (c-di-GMP) signaling: Lessons from Pseudomonas aeruginosa and other bacteria. J. Biol. Chem. 291, 12547–12555 (2016).
Jenal, U., Reinders, A. & Lori, C. Cyclic di-GMP: Second messenger extraordinaire. Nat. Rev. Microbiol. 15, 271–284 (2017).
Miyata, M. et al. Tree of motility: A proposed history of motility systems in the tree of life. Genes Cells 25, 6–21 (2020).
Nakamura, S. & Minamino, T. Flagella-driven motility of bacteria. Biomolecules 9, 279 (2019).
Nakamura, S. Spirochete flagella and motility. Biomolecules 10, 550 (2020).
Vincent, A. T. et al. Revisiting the taxonomy and evolution of pathogenicity of the genus Leptospira through the prism of genomics. PLOS Neglec. Trop. Dis. 13, e0007270 (2019).
Nakao, R., Masuzawa, T., Nakamura, S. & Koizumi, N. Complete genome sequence of Leptospira kobayashii strain E30, isolated from soil in Japan. Microbiol. Resour. Announc. https://doi.org/10.1128/MRA.00907-21 (2021).
Yoshimura, H., Yoshihara, S., Okamoto, S., Ikeuchi, M. & Ohmori, M. A cAMP receptor protein, SYCRP1, is responsible for the cell motility of Synechocystis sp. PCC 6803. Plant Cell Physiol. 43, 460–463 (2002).
Bhaya, D. Light matters: Phototaxis and signal transduction in unicellular cyanobacteria. Mol. Microbiol. 53, 745–754 (2004).
Terauchi, K. & Ohmori, M. Blue light stimulates cyanobacterial motility via a cAMP signal transduction system. Mol. Microbiol. 52, 303–309 (2004).
Goldstein, S. F. & Charon, N. W. Multiple-exposure photographic analysis of a motile spirochete. Proc. Natl. Acad. Sci. U.S.A. 87, 4895–4899 (1990).
Nakamura, S., Leshansky, A., Magariyama, Y., Namba, K. & Kudo, S. Direct measurement of helical cell motion of the spirochete Leptospira. Biophys. J. 106, 47–54 (2014).
Takabe, K. et al. Viscosity-dependent variations in the cell shape and swimming manner of Leptospira. Microbiology 163, 153–160 (2017).
Islam, M. S., Takabe, K., Kudo, S. & Nakamura, S. Analysis of the chemotactic behaviour of Leptospira using microscopic agar-drop assay. FEMS Microbiol. Lett. 356, 39–44 (2014).
Affroze, S., Islam, Md. S., Takabe, K., Kudo, S. & Nakamura, S. Characterization of leptospiral chemoreceptors using a microscopic agar drop assay. Curr. Microbiol. 73, 202–205 (2016).
Iseki, M. et al. A blue-light-activated adenylyl cyclase mediates photoavoidance in Euglena gracilis. Nature 415, 1047–1051 (2002).
Ohki, M. et al. Structural insight into photoactivation of an adenylate cyclase from a photosynthetic cyanobacterium. PNAS 113, 6659–6664 (2016).
Stierl, M. et al. Light modulation of cellular cAMP by a small bacterial photoactivated adenylyl cyclase, bPAC, of the soil bacterium Beggiatoa. J. Biol. Chem. 286, 1181–1188 (2011).
Penzkofer, A., Stierl, M., Hegemann, P. & Kateriya, S. Photo-dynamics of the BLUF domain containing soluble adenylate cyclase (nPAC) from the amoeboflagellate Naegleria gruberi NEG-M strain. Chem. Phys. 387, 25–38 (2011).
Penzkofer, A., Tanwar, M., Veetil, S. K. & Kateriya, S. Photo-dynamics of photoactivated adenylyl cyclase TpPAC from the spirochete bacterium Turneriella parva strain HT. J. Photochem. Photobiol. B 153, 90–102 (2015).
Penzkofer, A., Tanwar, M., Veetil, S. K. & Kateriya, S. Photo-dynamics of photoactivated adenylyl cyclase LiPAC from the spirochete bacterium Leptonema illini strain 3055T. Trends Appl. Spectrosc. 11, 39–62 (2014).
Blain-Hartung, M. et al. Cyanobacteriochrome-based photoswitchable adenylyl cyclases (cPACs) for broad spectrum light regulation of cAMP levels in cells. J. Biol. Chem. 293, 8473–8483 (2018).
Inoue, K., Tsukamoto, T. & Sudo, Y. Molecular and evolutionary aspects of microbial sensory rhodopsins. Biochimica et Biophysica Acta (BBA) Bioenergetics 1837, 562–577 (2014).
Kandori, H. Biophysics of rhodopsins and optogenetics. Biophys. Rev. 12, 355–361 (2020).
Chaves, I. et al. The cryptochromes: Blue light photoreceptors in plants and animals. Annu. Rev. Plant Biol. 62, 335–364 (2011).
Liu, C., Sun, D., Zhu, J., Liu, J. & Liu, W. The Regulation of bacterial biofilm formation by cAMP-CRP. Front Microbiol. 11, 802 (2020).
Xu, J., Koizumi, N. & Nakamura, S. Crawling motility on the host tissue surfaces is associated with the pathogenicity of the zoonotic spirochete Leptospira. Front. Microbiol. 11, 1886 (2020).
Linder, J. U. Class III adenylyl cyclases: Molecular mechanisms of catalysis and regulation. Cell. Mol. Life Sci. 63, 1736 (2006).
Sasaki, Y. et al. Leptospiral flagellar sheath protein FcpA interacts with FlaA2 and FlaB1 in Leptospira biflexa. PLoS ONE 13, e0194923 (2018).
Slamti, L. & Picardeau, M. Construction of a library of random mutants in the spirochete Leptospira biflexa using a mariner transposon. Methods Mol. Biol. 859, 169–176 (2012).
Bono, J. L. et al. Efficient targeted mutagenesis in Borrelia burgdorferi. J. Bacteriol. 182, 2445–2452 (2000).
Bauby, H., Saint Girons, I. & Picardeau, M. Construction and complementation of the first auxotrophic mutant in the spirochaete Leptospira meyeri. Microbiology 149, 689–693 (2003).
Picardeau, M. Conjugative transfer between Escherichia coli and Leptospira spp. as a new genetic tool. Appl. Environ. Microbiol. 74, 319–322 (2008).
Morimoto, Y. V. et al. Assembly and stoichiometry of FliF and FlhA in Salmonella flagellar basal body. Mol. Microbiol. 91, 1214–1226 (2014).
Acknowledgements
We thank Dr. E. Isogai (Tohoku University) and Dr. H. Yoneyama (Tohoku University) for the experiment reagents and the insightful discussion. This work was supported by the JSPS KAKENHI Grant Number 18J10834 to J.X., 19K07571 to N.K., 21K06099 to Y.V.M., 21H05532 to Y.V.M., 15H01307 to S.N., 20H05524 to S.N., and 21H02727 to S.N. This work was also supported by JST PRESTO Grant Number JPMJPR204B to Y.V.M.
Author information
Authors and Affiliations
Contributions
J.X., N.K., Y.V.M., R.O., T.M., and S.N. planned the project; J.X., N.K. Y.V.M. R.O., T.M., and S.N. carried out the experiments; Y.V.M. and S.N. set up the optical system and programs for data analysis; J.X., Y.V.M., N.K., R.O., and S.N. analyzed the data; J.X., N.K., Y.V.M., and S.N. wrote the paper.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Xu, J., Koizumi, N., Morimoto, Y.V. et al. Light dependent synthesis of a nucleotide second messenger controls the motility of a spirochete bacterium. Sci Rep 12, 6825 (2022). https://doi.org/10.1038/s41598-022-10556-7
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-022-10556-7
This article is cited by
-
Genomic insights into the c-di-GMP signaling and biofilm development in the saprophytic spirochete Leptospira biflexa
Archives of Microbiology (2023)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.