Abstract
Rhabdomyosarcomas (RMS) represent a family of aggressive soft tissue sarcomas that present in both children and adults. Pathologic risk stratification for RMS has been based on histologic subtype, with poor outcomes observed in alveolar rhabdomyosarcoma (ARMS) and the adult-type pleomorphic rhabdomyosarcoma (PRMS) compared to embryonal rhabdomyosarcoma (ERMS). Genomic sequencing studies have expanded the spectrum of RMS, with several new molecularly defined entities, including fusion-driven spindle cell/sclerosing rhabdomyosarcoma (SC/SRMS) and MYOD1-mutant SC/SRMS. Comprehensive genomic analysis has previously defined the mutational and copy number spectrum for the more common ERMS and ARMS and revealed corresponding methylation signatures. Comparatively, less is known about epigenetic correlates for the rare SC/SRMS or PRMS histologic subtypes. Herein, we present exome and RNA sequencing, copy number analysis, and methylation profiling of the largest cohort of molecularly characterized RMS samples to date. In addition to ARMS and ERMS, we identify two novel methylation subtypes, one having SC/SRMS histology and defined by MYOD1 p. L122R mutations and the other matching adult-type PRMS. Selected tumors from adolescent patients grouped with the PRMS methylation class, expanding the age range of these rare tumors. Limited follow-up data suggest that pediatric tumors with MYOD1-mutations are associated with an aggressive clinical course.
Similar content being viewed by others
Introduction
Rhabdomyosarcomas (RMS) are a family of aggressive soft tissue sarcomas presenting primarily in the pediatric population and more rarely in adults1. RMS are separated into distinct histologic variants, including pleomorphic rhabdomyosarcoma (PRMS), alveolar rhabdomyosarcoma (ARMS), embryonal rhabdomyosarcoma (ERMS), and the evolving category of spindle cell/sclerosing rhabdomyosarcoma (SC/SRMS)1. The two primary subtypes of RMS encountered in the pediatric population, ERMS and ARMS, can be distinguished molecularly as nearly 85% of ARMS are defined by gene fusions between PAX3-FOXO1 or PAX7-FOXO11, whereas ERMS are characterized by disparate mutations in the RAS pathway, effectors of the PI3 Kinase pathway, or in genes that control the cell cycle2. The spindle cell/sclerosing category of RMS is characterized by disparate presentations and recurrent molecular alterations, including MYOD1-mutant SC/SRMS3, interosseous SC/SRMS with TFCP2 or NCOA2 rearrangements4, and congenital SC/SRMS with VGLL2, NCOA2, or CITED2 gene rearrangements5,6. Diagnosis of the MYOD1-mutant subtype of SC/SRMS, characterized by recurrent MYOD1 p.L122R missense mutations, has important clinical implications as it is characterized by more aggressive clinical behavior3,7. PRMS, which primarily present in adults, is characterized by a complex karyotype and an absence of recurrent molecular alterations8.
Placing rhabdomyosarcoma into a specific pathologic subtype presents a significant challenge to clinical practice. Whereas a subset of morphologically ambiguous cases can be molecularly classified using FOXO1 fusion status, fifteen percent of ARMS are fusion negative9. Similarly, SC/SRMS encompasses a variety of molecularly distinct entities that1 are exceptionally rare2, share morphologic overlap with ERMS, and3 are characterized by disparate molecular alterations for which testing is not routinely available in most pathology laboratories. In patients presenting in late adolescence or early adulthood, the distinction between pediatric ERMS with anaplasia and adult-type PRMS can be difficult, given the paucity of defining diagnostic molecular abnormalities in each group.
Several investigators have reported that RMS can be separated into at least two primary epigenetic groups based on differences in their genome-wide methylation profiles. These groups correspond closely to the embryonal and alveolar subtypes, with the latter being highly enriched for PAX3-FOXO1 and PAX7-FOXO1 fusions10,11,12. Despite these observations, complete representation of all subtypes of rhabdomyosarcoma has not been included in previous cohorts, leaving the question of how histologically-defined SC/SRMS and PRMS are epigenetically related to ERMS and ARMS.
We hypothesized that with sufficient representation, additional molecular subtypes could be identified in RMS. To test this hypothesis, we performed genome-wide methylation profiling on 154 rhabdomyosarcomas and unrelated pediatric skeletal muscle controls representing all primary histologic types of both adult and pediatric disease and correlated the findings with copy number profiling, next-generation sequencing, and clinical outcome analysis.
Materials and methods
All methods were carried out in accordance with relevant guidelines and regulations. All experimental protocols were approved by the St. Jude Children’s Research Hospital Institutional Review Board (#XPD17-163). Informed consent was not required under the Office for Human Research Protections (OHRP) guidelines regarding the disposition of deidentified human tissues for human subjects research, and was waived by the St. Jude Children’s Research Hospital Institutional Review Board.
Data generation and methylation array processing
One hundred and fifty-eight samples from St. Jude Children’s Research Hospital (SJCRH, pediatric) and Stanford Hospital and Clinics (adult) were analyzed using Illumina Methylation BeadChip (EPIC) arrays according to the manufacturer’s instructions. Data were generated from formalin-fixed paraffin-embedded (FFPE) tissue samples. DNA of 158 patients was extracted and hybridized to Illumina Infinium HumanMethylationEPIC BeadChip (850 K) arrays. All methylation data were analyzed in R (http://www.r-project.org, version 3.5.3), using multiple of packages from Bioconductor and other repositories. Specifically, array data were preprocessed using the minfi package (v.1.28.4)13. Background correction with dye-bias normalization was performed for all samples using noob (normal-exponential out-of-band) with the “single” dye method14. Filtering was performed to remove probes located on sex chromosomes, probes containing nucleotide polymorphisms (dbSNP132 Common) within five base pairs of the targeted CpG-site, or probes mapping to multiple sites on hg19 (allowing for one mismatch), and cross-reactive probes.
Survival analysis was performed after a manual chart review to extract survival status For deceased patients, the time from the initial diagnosis to death was calculated, and for live patients, the time from the initial diagnosis to the last clinic visit was calculated. Survival curves were generated using Graphpad Prism 9ing.
Unsupervised clustering and copy number variation analysis
Principal component analysis was performed using the top 10,000 variably methylated probes. The number of statistically significant principal components was determined by agDimension function in the PCDimension package (v.1.1.11)15. Dimensionality reduction using 5 statistically significant principal components (k = 5) was used for Uniform Manifold Approximation and Projection (UMAP v.0.2.6)16 with non-default parameters: theta = 0, pca = F, perplexity = 4. To identify distinct clusters in the methylation data, the density-based spatial clustering of applications with noise (DBSCAN) algorithm17 was applied to the UMAP coordinates with minPts = 4 and eps = 0.65. To evaluate the separation of DBSCAN clusters in the UMAP coordinates, silhouette analysis was performed on the Euclidian distances among the samples using the R package cluster (v.2.1.0). For hierarchical clustering, Kendall correlation was calculated as a distance measure between samples using the top 10,000 most variable probes among the SJ samples, and the unsupervised hierarchical clustering was performed on the computed distance by complete linkage agglomeration method.
Copy number variation (CNV) analysis from methylation array data was performed using the conumee package (version 1.16.0)18. Chromosomal gain or loss was determined using a 0.18 threshold. Statistically significant frequent copy number variations (CNVs) were determined using GISTIC version 2.0.2329. Copy number profiles output as segments obtained from conumee R package were used as inputs for GISTIC2. Gain and loss were categorized with CNV values greater than 0.18 or smaller than -0.18, respectively. CNVs were also divided into those that are broad (defined as exceeding half of the length of a chromosome arm) and focal (shorter than this). We considered events with False Discovery Rate q-values < 0.25 as significant at a 90% confidence level. An “arm-level peel-off” correction was enabled to assign all CNVs in the same chromosome arm of the same sample to be part of a single event when determining whether multiple significantly recurrent events exist on that chromosome arm.
Somatic mutation detection of matched tumor-normal WES samples
Paired-end sequencing reads were mapped with BWA19 to human genome GRCh38. We used an ensemble approach to call somatic mutations (SNV/indels) with multiple published tools, including Mutect220, SomaticSniper21, VarScan222, MuSE23, and Strelka224. The consensus calls by at least two callers were considered as confident mutations. The consensus call sets were further reviewed for the variant allele frequency, supporting read depth, mapping quality, and strand bias to remove additional artifacts. The variant annotation was performed by Annovar25.
Mutation detection of tumor-only FFPE WGS samples
Paired-end reads were mapped against human genome GRCh38 by BWA. Variants were called by Mutect2 using the tumor-only calling mode. The FFPE artifacts due to formaldehyde deamination of cytosines (C > T) were filtered via the GATK FilterByOrientationBias tool26. A panel of normal (PON) was constructed using 15 tonsil FFPE samples and compared with the call-sets to exclude FFPE artifacts. Multiple filtering steps were applied to exclude potential calling artifacts. The variants passing the filtering steps fulfilled the following criteria: coverage depth of the variant > 10, variant allele frequency > 0.02, alternative allele count > = 4, allele population frequency in public databases < 0.01 (gnomadAD, 1000 genomes, ExAC and Exome Sequencing Projects), mappability > 0.7, not co-localized with repeat elements and GC percentage between 0.4 and 0.6. The variant annotation was performed using Annovar.
Mutation detection of tumor-only FFPE RNA samples
The adapters in sequencing reads were trimmed with “trim_galore” (v0.4.4, https://www.bioinformatics.babraham.ac.uk/projects/trim_galore/, -q 20 –phred 33 –paired) and were mapped using STAR27. The GATK SplitNCigarReads tool was used for adjusting the cigar string of RNAseq BAMs, and the resulting BAMs were run through BQSR to calibrate base quality. The variant was called by GATK HaplotypeCaller and filtered by the VariantFiltration tool (-window 35 -cluster 3 –filter-name FS -filter "FS > 30.0" –filter-name QD -filter "QD < 2.0"). Further filtering of the variants was performed using the same criteria as described in the tumor-only FFPE calling. The variant annotation was performed using Annovar.
Fluorescent in-situ hybridization studies
Dual-color FISH was performed on 4 µm paraffin-embedded tissue sections (Abbott Molecular: FOXO1 (Centromeric) SpectrumGreen Cat# 05J48-014; FOXO1 (Telomeric) SpectrumOrange Cat# 05J48-013). Probes were co-denatured with the target cells on a slide moat at 90 °C for 12 min. The slides were incubated overnight at 37 °C on a slide moat and then washed in 4 M Urea/2xSSC at 25 °C for 1 min. Nuclei were counterstained with DAPI (200 ng/ml) (Vector Labs) for viewing on an Olympus BX51 fluorescence microscope equipped with a 100-W mercury lamp; FITC, Rhodamine, and DAPI filters; 100X PlanApo (1.40) oil objective; and a Jai CV digital camera. Images were captured and processed using the Cytovision software from Leica Biosystems (Richmond, IL).
Results
Description of Cohort
Patients were selected from the anatomic pathology archives based on sample availability for histopathologic review and methylome analysis. Histopathologic and clinical data were tabulated from the electronic medical record (Supplemental Table 1). The remaining “pediatric” cohort included samples from patients treated in a pediatric hospital setting, with a few patients representing young adults. This cohort included samples from 70 female and 76 male patients. Age ranged from 3 months to 27 years (mean 7.8 years, median 7 years). Initial histologic diagnosis included: ARMS (n = 53), ERMS (n = 46), SC/SRMS (n = 3), RMS not otherwise specified (n = 44). Cases designated as NOS were enriched by our consultation practice, and included cases for which a definitive diagnosis was not reached in the initial clinical workup. This occurred secondary to poor sample quality, inability to perform ancillary studies, and in the case of inconclusive histologic findings.
The adult pleomorphic rhabdomyosarcoma subset (n = 8) included 4 female and 4 male patients. Age ranged from 61 to 79 years (mean 69.8 years, median 68.5 years).
DNA methylation profiling of rhabdomyosarcoma
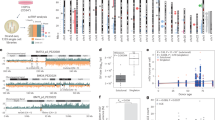
To determine if their genome-wide methylation signature could separate the types of rhabdomyosarcoma, we performed Infinium EPIC 850 K methylation array testing on a cohort of 154 rhabdomyosarcomas and four controls (skeletal muscle samples), followed by cluster analysis. Four main clusters emerged, which we designated ARMS, ERMS, SC/SRMS, and PRMS based on the most dominant histopathologic diagnosis represented in each cluster (Fig. 1). Normal control tissue grouped closely with ERMS, a finding which has been observed previously12,28.
Unsupervised Clustering Analysis of 158 samples to identify potential molecular subtypes. (A) UMAP dimension reduction of 158 samples using the 5 statistically significant principal components determined from the 10,000 most variably methylated probes, as measured by the standard deviation of the probe-level beta values across samples. The analysis yielded 4 distinct clusters, representing 4 molecular subtypes (B) Silhouette analysis showing the average silhouette widths of the 4 molecular groups identified by DBSCAN based on Euclidian distances among 158 samples on UMAP coordinates. Red—ARMS, blue—ERMS, green—PRMS, and orange—SC/SRMS, black: controls (which were clustered with ERMS subtype). (C) Hierarchical clustering of the St. Jude cohort.
Copy number analysis in subtypes of rhabdomyosarcoma
We evaluated for recurrent copy number abnormalities, focal or chromosome level, in the RMS molecular groups using the methylation array data. While few recurrent broad copy number abnormalities were detected in alveolar rhabdomyosarcoma, the ERMS group demonstrated typical changes previously reported to be associated with that subtype, including enrichment for gains of chromosomes 2, 8, 11, 12, 13, and 20 (Fig. 2A) and most of these gains were found to be statistically significant by GISTIC2.0 (Supplemental Table 2)29. Focal changes in ARMS included a gain of 12q13 and 13q14, containing MYO1A, STAT6, and FOXO1 genes, while the ERMS group had a gain of 12q15, containing both the FRS2 and MDM2 genes. The molecularly defined PRMS subtype demonstrated complex copy number changes with a frequent gain of 1p (Supplemental Table 2). Other abnormalities included loss of 13, including focal changes at the RB1 locus. Among focal changes, the PRMS group was also characterized by frequent gain of 6q24.3, a region containing FBXO30, an F-box gene known to show myocyte-specific expression (Fig. 2). The SC/SRMS group was characterized by frequent broad gain of chromosomes 11 and 22q, and loss of 10p, 13q, and 16q (Supplemental Table 2). Focal deletion in 9p21.3 encompassing the CDKN2A locus was also identified in the SC/SRMS group (Fig. 2B).
Chromosomal copy number variations among RMS molecular groups compared to reference tumors. (A) Copy number frequency plots of ARMS, ERMS, PRMS, and SC/SRMS molecular groups were constructed by conumee R package using copy number profiles of reference tumors at 0.18 threshold. (B) Copy number gains and losses in each molecular group determined by GISTIC 2.0. Green line indicates the q-value threshold to be considered statistically significant. Red: chromosomal gain, blue: chromosomal loss.
Next-generation sequencing and fluorescence in situ hybridization of rhabdomyosarcomas
We performed next-generation sequencing of 109 RMS (57 ERMS, 39 ARMS, 9 PRMS, 4 SC/SRMS) tumors with sufficient material that had undergone methylation analysis (Fig. 3). The tumors in the ARMS methylation class were characterized by a high proportion of PAX3/7-FOXO1 fusions (49/52, 94%), and FOXO1 fusions were not found outside the ARMS methylation group. In contrast, RMS samples in the ERMS group were enriched for mutations in activators of the RAS and AKT pathways, including NRAS, HRAS, KRAS, PIK3CA, and NF1. Of the two novel molecular groups, those falling in the SC/SRMS demonstrated MYDO1 L122R mutations in all samples (n = 8, 4 via DNA comprehensive sequencing, 4 manually extracted from RNAseq data). This group also showed an increased incidence of FGFR1 mutations (in 50% of cases, vs. 0% in ARMS and PRMS, 2.4% in ERMS). Mutations in ATRX and BCOR were proportionally higher within the SC/SRMS molecular group than in the other molecular groups. The two ERMS with MYOD1 alterations did not harbor the L122R variant (both showed p.R121C, considered to have uncertain significance). The PRMS molecular group was devoid of recurrent activating mutations but did have a high frequency of inactivating mutations in tumor suppressors, including TP53, RB1, NF1, and PTEN Similar molecular alterations were identified in the histologically classified pediatric ERMS cases that clustered with the PRMS methylation group, including mutations in RB1 and TP53. No cases of rhabdomyosarcoma with VGLL2, NCOA2, or CITED fusions were identified.
Mutation detection among RMS molecular groups using WGS and RNA-seq. Next generation sequencing was performed on tumors with sufficient material from the methylation cohort. Gene mutations (violet—nonsense, dark red—frameshift, green—missense, light gray—not detected) with VAF ≥ 20% are shown along with FISH results for FOXO1 fusion status (dark blue—positive, gold—negative, light gray – unknown) and PCR results (light purple—PAX3-FOXO1 fusion positive, dark purple—PAX7-FOXO1 fusion positive, light gray—unknown) in each RMS sample. The heatmap was split into ARRMS (red), ERMS (blue), PRMS (green), and SC/SRMS (orange).
Break-apart FOXO1 fluorescence in situ hybridization (FISH) was attempted in all cases with sufficient materials (n = 88) and was successful in 85 cases (97%). A positive rearrangement was identified in 49 of 52 ARMS and zero cases of ERMS (0/32).
Histologic correlates of PRMS and SC/SRMS
Clinicopathologic evaluation of the RMS samples was performed both by chart review and microscopic evaluation, emphasizing the histology of the novel molecular groups. The methylation-defined ARMS and ERMS were concordant with the corresponding histopathologic designation in most instances (51 of 53 ARMS; 96%) and (45 of 46 ERMS; 98%). The SC/SRMS with MYOD1 abnormality were designated as spindle cell RMS in a subset of initial cases (2 of 3 initially diagnosed as SC/SRMS were truly MYOD1 mutant; 67%), though most were designated as RMS not otherwise specified (NOS) clinically (n = 6). The tumors, in most instances, were dominated by a spindle-cell morphology (Fig. 4A,B), though in some examples, the characteristic hyalinized stroma was the dominant phenotype (Fig. 4C). For those SC/SRMS-MYOD1 mutant tumors that were available for immunostaining, we observed strong and diffuse immunoreactivity for MYOD1 (Fig. 4D).
Histology of tumors in SCRMS with MYOD-1 alteration methylation group. The dominant phenotype consisted of bland spindle cells (A,B) with hyalinized stroma (C). When immunohistochemistry was available for MYOD1, the tumors were characterized by strong, diffuse immunoreactivity (D). Scale bars represent 40 µm.
While the PRMS methylation group predominately consisted of cases corresponding to the adult-type PRMS histologic class, a small proportion of pediatric patients clustered in that molecular group (n = 3). These pediatric tumors in this methylation subtype demonstrated severe morphologic anaplasia, analogous to the histologic appearance of the adult PRMS tumors (see Fig. 5).
The tumors included in this study displayed enrichment for those with diagnostic ambiguity (clinically classified as RMS not otherwise specified, NOS). A molecular class was assignable in all RMS, NOS cases (n = 43, Fig. 6A) with the majority of cases assigned to the ERMS group (n = 28), and a few cases falling into ARMS (n = 6), SC/SRMS (n = 6), and PRMS groups (n = 3).
Suggested molecular re-classification of RMS and Clinical Outcome. The histologic type designated at diagnosis was compared to the molecular classification by methylation profiling and displayed as a Sankey diagram (A). Methylation profiling was able to classify a significant number of tumors that could not be classified using traditional histology. Clinical outcome data in the Kaplan–Meier plot (B) highlights poor outcome in the pleomorphic, alveolar, and spindle/sclerosing groups. Numerical data (C) displaying number at risk over a ten year period.
Clinical Correlation of RMS methylation groups
The consultative nature of the cohort limited the amount of follow-up data available (Fig. 6B-C). Of the three pediatric and adolescent cases clustered with adult type-PRMS, follow-up was available for one patient who died of disease (Table 1). Similarly, clinical follow-up was available for three patients with MYOD1-mutant SCRMS, all of whom succumbed to their disease (Table 2). The adults with PRMS patients had a high mortality rate, with five of the six patients with clinical follow-up dying of disease (overall 83%).
Discussion
Rhabdomyosarcomas represent a heterogeneous group of soft tissue sarcomas associated with poor clinical outcomes. Historically, risk stratification within RMS has primarily been based on histopathologic subtype, with ARMS tumors getting more intensive therapy30,31. Because FOXO1 fusions characterize most ARMS, these can be identified molecularly. Notably, only 94% of our ARMS molecular group were found to harbor a FOXO1 gene rearrangement by FISH. This observation correlates with the historically recognized fusion-negative ARMS group and demonstrates the limitations of relying on FISH alone for molecular stratification32.
Reliable diagnostic or prognostic biomarkers for all remaining RMS groups have not been established. ERMS are characterized by heterogeneous mutations that lead to activation of the RAS or AKT pathways but have no single recurrent mutation or fusion that can easily be tracked32. The most recent edition of the WHO Classification of Tumours, Soft Tissue and Bone Tumours now includes a histologic type encompassing the spindle cell/sclerosing subtypes of RMS, a subset of which harbor MYOD1 L122R point mutations and have particularly aggressive clinical behavior and inferior outcomes7. Other tumors in the SC/SCRMS group are reported to contain heterogeneous fusions, including those involving VGLL2 or NCOA2, and have a less aggressive clinical course.
DNA methylation profiling has emerged as a promising method for discovering molecular heterogeneity in solid tumors, including soft tissue sarcomas33,34,35,36. Additionally, by combining the technology with supervised machine learning methods, neoplastic and nonneoplastic tissues can be classified using their genome-wide DNA methylation signatures in a single clinical assay. Methylation signatures correlate with cellular developmental pathways and closely recapitulate existing histologic and molecular classification schemas. Previous studies utilizing methylation profiling to interrogate RMS reported two methylation groups corresponding to the dominant ARMS and ERMS subtypes11. A methylation class corresponding to MYDO1-mutant SC/SRMS was recently reported as part of a comprehensive sarcoma classifier37. Our cohort provides independent validation of the SC/SCRMS group with MYOD1 mutation and extends those findings to include a methylation class corresponding to the adult-type pleomorphic RMS histologic groups.
Despite the reported molecular heterogeneity of SC/SRMS, we only detect a methylation group for the SC/SRMS tumors with MYOD1 L122R missense mutations. SC/SRMS with MYOD1 mutations are well-established and associated with a dismal prognosis38. Despite enriching for RNA sequencing in tumors histologically diagnosed as SC/SRMS, we did not find a corresponding methylation group of SC/SRMS containing gene fusions. This could suggest that SC/SRMS with fusions form a subset of ERMS or that fusion-positive SC/SRMS were represented infrequently in our cohort, with insufficient representation to form a distinct cluster. One histologically-defined SC/SRMS grouped with ERMS by methylation profiling. Unfortunately, sufficient material could not be obtained from the case to evaluate for fusions by RNA sequencing. Conceptually, this classification process is also further hindered by the knowledge that subsets of ERMS will display spindled morphology, a finding that will continue to hamper morphologic classification systems moving forward.
We also identified a second novel methylation group corresponding to the histologic adult-type PRMS group. Our PRMS methylation group demonstrated no recurrent driver mutations but was characterized by genomic instability and a high proportion of mutations in tumor suppressor genes. Although histologically-defined PRMS has primarily been considered specific to the adult population, with a peak age in the 60 s and 70 s1, we identified three pediatric patients in the molecularly-defined PRMS group (aged 9, 14, and 16 years). This finding challenges the notion of adult- and pediatric RMS being completely distinct diseases and suggests that a subset of pediatric ERMS with anaplasia may be molecularly indistinguishable from adult-type PRMS. In this cohort, only 3 of 13 cases classified as ERMS with anaplasia clustered with the adult-type PRMS, suggesting morphologic features alone are insufficient to identify such cases. SC/SRMS has also been found across the entire age spectrum, from infancy to adulthood, suggesting that the rare molecular groups may have a specific predilection to cross age boundaries.
Sample size limitations are often encountered in studies exploring rare malignancies. Despite the relatively low case numbers, our unsupervised analysis supports that these represents bona fide molecular groups. The number of cases for each of the rare groups (11 for PRMS and 8 for SC/SRMS, respectively) is in line with the size of molecular groups that were used to train supervised classifiers in clinical laboratories for brain tumors (19 of 91 subclasses have between 8 and 11 examples)39 and sarcomas (34 of 65 subclasses are limited to 8–11 examples)37.
Clinical outcome data were limited in our cohort, and specifically for the new novel methylation groups. The single pediatric case in the PRMS methylation subtype with follow-up experienced rapid clinical decline and died of disease 43 days following diagnosis. The tumors in the SC/RMS methylation class also were associated with poor outcomes in our cohort. Additional characterization of the clinical correlations and outcome data will be required to characterize these novel molecular groups further.
Data availability
The methylation datasets generated during the current study are available in the GEO datasets repository. Accession number GSE167059.
References
WHO Classification of Tumours Editorial Board. Soft tissue and bone tumours. Vol 3. 5th ed. International Agency for Research on Cancer: Lyon (France), 2020.
Shern, J. F. et al. Comprehensive genomic analysis of rhabdomyosarcoma reveals a landscape of alterations affecting a common genetic axis in fusion-positive and fusion-negative tumors. Cancer Discov 4, 216–231 (2014).
Agaram, N. P. et al. MYOD1-mutant spindle cell and sclerosing rhabdomyosarcoma: an aggressive subtype irrespective of age A reappraisal for molecular classification and risk stratification. Mod. Pathol. 32, 27–36 (2019).
Agaram, N. P. et al. Expanding the spectrum of intraosseous rhabdomyosarcoma: Correlation between 2 distinct gene fusions and phenotype. Am. J. Surg. Pathol. 43, 695–702 (2019).
Alaggio, R. et al. A molecular study of pediatric spindle and sclerosing rhabdomyosarcoma: Identification of novel and recurrent VGLL2-related fusions in infantile cases. Am. J. Surg. Pathol. 40, 224–235 (2016).
Antonescu, C. R. et al. Frequent PLAG1 gene rearrangements in skin and soft tissue myoepithelioma with ductal differentiation. Genes Chromosomes Cancer 52, 675–682 (2013).
Rekhi, B., Upadhyay, P., Ramteke, M. P. & Dutt, A. MYOD1 (L122R) mutations are associated with spindle cell and sclerosing rhabdomyosarcomas with aggressive clinical outcomes. Mod. Pathol. 29, 1532–1540 (2016).
Skapek, S. X. et al. Rhabdomyosarcoma. Nat. Rev. Dis. Primers 5, 1 (2019).
Rudzinski, E. R. et al. Histology, fusion status, and outcome in metastatic rhabdomyosarcoma: A report from the Children’s Oncology Group. Pediatr. Blood Cancer 64, 2 (2017).
Mahoney, S. E., Yao, Z., Keyes, C. C., Tapscott, S. J. & Diede, S. J. Genome-wide DNA methylation studies suggest distinct DNA methylation patterns in pediatric embryonal and alveolar rhabdomyosarcomas. Epigenetics 7, 400–408 (2012).
Seki, M. et al. Integrated genetic and epigenetic analysis defines novel molecular subgroups in rhabdomyosarcoma. Nat. Commun. 6, 7557 (2015).
Stewart, E. et al. Identification of therapeutic targets in rhabdomyosarcoma through integrated genomic, epigenomic, and proteomic analyses. Cancer Cell 34, 411–26.e19 (2018).
Aryee, M. J. et al. Minfi: a flexible and comprehensive Bioconductor package for the analysis of Infinium DNA methylation microarrays. Bioinformatics 30, 1363–1369 (2014).
Triche, T. J. Jr., Weisenberger, D. J., Van Den Berg, D., Laird, P. W. & Siegmund, K. D. Low-level processing of Illumina Infinium DNA Methylation BeadArrays. Nucleic Acids Res. 41, e90 (2013).
Wang, M., Kornblau, S. M. & Coombes, K. R. Decomposing the apoptosis pathway into biologically interpretable principal components. Cancer Inform 17, 1176935118771082 (2018).
Becht E, McInnes L, Healy J, et al. Dimensionality reduction for visualizing single-cell data using UMAP. Nat Biotechnol 2018.
Hahsler Mea. dbscan: Fast Density-Based Clustering with R. Journal of Stastical Software 2019;91.
Hovestadt V ZM. conumee: Enhanced copy-number variation analysis using Illumina DNA methylation arrays. In.
Li, H. & Durbin, R. Fast and accurate long-read alignment with Burrows-Wheeler transform. Bioinformatics 26, 589–595 (2010).
Cibulskis, K. et al. Sensitive detection of somatic point mutations in impure and heterogeneous cancer samples. Nat. Biotechnol. 31, 213–219 (2013).
Larson, D. E. et al. SomaticSniper: identification of somatic point mutations in whole genome sequencing data. Bioinformatics 28, 311–317 (2012).
Koboldt, D. C. et al. VarScan 2: somatic mutation and copy number alteration discovery in cancer by exome sequencing. Genome Res. 22, 568–576 (2012).
Fan, Y. et al. MuSE: accounting for tumor heterogeneity using a sample-specific error model improves sensitivity and specificity in mutation calling from sequencing data. Genome Biol. 17, 178 (2016).
Kim, S. et al. Strelka2: fast and accurate calling of germline and somatic variants. Nat. Methods 15, 591–594 (2018).
Wang, K., Li, M. & Hakonarson, H. ANNOVAR: functional annotation of genetic variants from high-throughput sequencing data. Nucleic Acids Res. 38, e164 (2010).
Van der Auwera, G. A. et al. From FastQ data to high confidence variant calls: The genome analysis toolkit best practices pipeline. Curr. Protoc. Bioinformatics 43, 11 (2013).
Dobin, A. et al. STAR: ultrafast universal RNA-seq aligner. Bioinformatics 29, 15–21 (2013).
Sun, W. et al. Distinct methylation profiles characterize fusion-positive and fusion-negative rhabdomyosarcoma. Mod. Pathol. 28, 1214–1224 (2015).
Parham, D. M. & Barr, F. G. Classification of rhabdomyosarcoma and its molecular basis. Adv. Anat. Pathol. 20, 387–397 (2013).
Han, D. et al. Prognostic factors in patients with rhabdomyosarcoma using competing-risks analysis: A study of cases in the SEER database. J. Oncol. 2020, 2635486 (2020).
Chen, C., Dorado Garcia, H., Scheer, M. & Henssen, A. G. Current and Future Treatment Strategies for Rhabdomyosarcoma. Front. Oncol. 9, 2 (2019).
Arnold, M. A. et al. Histology, fusion status, and outcome in alveolar rhabdomyosarcoma with low-risk clinical features: a report from the children’s oncology group. Pediatr. Blood Cancer 63, 634–639 (2016).
Clay, M. R. et al. DNA methylation profiling reveals prognostically significant groups in pediatric adrenocortical tumors: A report from the international pediatric adrenocortical tumor registry. JCO Precis. Oncol. 2, 1–21 (2019).
Miele, E. et al. DNA methylation profiling for diagnosing undifferentiated sarcoma with capicua transcriptional receptor (CIC) alterations. Int. J. Mol. Sci. 2, 1 (2020).
Koelsche, C. et al. DNA methylation profiling distinguishes Ewing-like sarcoma with EWSR1-NFATc2 fusion from Ewing sarcoma. J. Cancer Res. Clin. Oncol. 145, 1273–1281 (2019).
Koelsche, C. et al. Array-based DNA-methylation profiling in sarcomas with small blue round cell histology provides valuable diagnostic information. Mod. Pathol. 31, 1246–1256 (2018).
Koelsche, C. et al. Sarcoma classification by DNA methylation profiling. Nat. Commun. 12, 498 (2021).
Kohsaka, S. et al. A recurrent neomorphic mutation in MYOD1 defines a clinically aggressive subset of embryonal rhabdomyosarcoma associated with PI3K-AKT pathway mutations. Nat. Genet. 46, 595–600 (2014).
Capper, D. et al. DNA methylation-based classification of central nervous system tumours. Nature 555(7697), 469–474 (2018).
Acknowledgements
This work was partially supported by the National Cancer Institute support Grant (P30 CA021765) and the American Lebanese Syrian Associated Charities (ALSAC).
Author information
Authors and Affiliations
Contributions
M.C. and B.O. performed study concept and design; M.C., B.O., A.P., E.S., G.C., C.C., Q.T., D.H., T.C., and M.D., performed development of methodology and writing, review, and revision of the paper; M.C., B.O., A.P, D.H., T.C., C.C, and Q.T. provided acquisition, analysis, and interpretation of data, and statistical analysis. All authors read and approved the final paper.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Clay, M.R., Patel, A., Tran, Q. et al. Methylation profiling reveals novel molecular classes of rhabdomyosarcoma. Sci Rep 11, 22213 (2021). https://doi.org/10.1038/s41598-021-01649-w
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-021-01649-w
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.