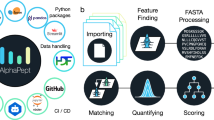
Abstract
Shotgun proteomics aims to identify and quantify the thousands of proteins in complex mixtures such as cell and tissue lysates and biological fluids. This approach uses liquid chromatography coupled with tandem mass spectrometry and typically generates hundreds of thousands of mass spectra that require specialized computational environments for data analysis. PatternLab for proteomics is a unified computational environment for analyzing shotgun proteomic data. PatternLab V (PLV) is the most comprehensive and crucial update so far, the result of intensive interaction with the proteomics community over several years. All PLV modules have been optimized and its graphical user interface has been completely updated for improved user experience. Major improvements were made to all aspects of the software, ranging from boosting the number of protein identifications to faster extraction of ion chromatograms. PLV provides modules for preparing sequence databases, protein identification, statistical filtering and in-depth result browsing for both labeled and label-free quantitation. The PepExplorer module can even pinpoint de novo sequenced peptides not already present in the database. PLV is of broad applicability and therefore suitable for challenging experimental setups, such as time-course experiments and data handling from unsequenced organisms. PLV interfaces with widely adopted software and community initiatives, e.g., Comet, Skyline, PEAKS and PRIDE. It is freely available at http://www.patternlabforproteomics.org.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout













Similar content being viewed by others
Code availability
The software used in this protocol can be found at http://patternlabforproteomics.org
References
Washburn, M. P., Wolters, D. & Yates, J. R. III Large-scale analysis of the yeast proteome by multidimensional protein identification technology. Nat. Biotechnol. 19, 242–247 (2001).
Eng, J. K., McCormack, A. L. & Yates, J. R. An approach to correlate tandem mass spectral data of peptides with amino acid sequences in a protein database. J. Am. Soc. Mass Spectrom. 5, 976–989 (1994).
Zhang, B., Chambers, M. C. & Tabb, D. L. Proteomic parsimony through bipartite graph analysis improves accuracy and transparency. J. Proteome Res. 6, 3549–3557 (2007).
Elias, J. E. & Gygi, S. P. Target–decoy search strategy for increased confidence in large-scale protein identifications by mass spectrometry. Nat. Methods 4, 207–214 (2007).
Yates, J. R. III et al. Toward objective evaluation of proteomic algorithms. Nat. Methods 9, 455–456 (2012).
Barboza, R. et al. Can the false-discovery rate be misleading? Proteomics 11, 4105–4108 (2011).
Carvalho, P. C. et al. Search engine processor: filtering and organizing peptide spectrum matches. Proteomics 12, 944–949 (2012).
Moosa, J. M., Guan, S., Moran, M. F. & Ma, B. Repeat-preserving decoy database for false discovery rate estimation in peptide identification. J. Proteome Res. 19, 1029–1036 (2020).
Ma, B. et al. PEAKS: powerful software for peptide de novo sequencing by tandem mass spectrometry. Rapid Commun. Mass Spectrom. 17, 2337–2342 (2003).
Perkins, D. N., Pappin, D. J., Creasy, D. M. & Cottrell, J. S. Probability-based protein identification by searching sequence databases using mass spectrometry data. Electrophoresis 20, 3551–3567 (1999).
Keller, A., Eng, J., Zhang, N., Li, X. & Aebersold, R. A uniform proteomics MS/MS analysis platform utilizing open XML file formats. Mol. Syst. Biol. 1, 0017 (2005).
Kohlbacher, O. et al. TOPP—the OpenMS proteomics pipeline. Bioinformatics 23, e191–e197 (2007).
McDonald, W. H. et al. MS1, MS2, and SQT-three unified, compact, and easily parsed file formats for the storage of shotgun proteomic spectra and identifications. Rapid Commun. Mass Spectrom. 18, 2162–2168 (2004).
Xu, T. et al. ProLuCID: An improved SEQUEST-like algorithm with enhanced sensitivity and specificity. J. Proteom. 129, 16–24 (2015).
Carvalho, P. C., Fischer, J. S. G., Chen, E. I., Yates, J. R. & Barbosa, V. C. PatternLab for proteomics: a tool for differential shotgun proteomics. BMC Bioinform. 9, 316 (2008).
Carvalho, P. C., Hewel, J., Barbosa, V. C. & Yates, J. R. III Identifying differences in protein expression levels by spectral counting and feature selection. Genet. Mol. Res. 7, 342–356 (2008).
Liu, H., Sadygov, R. G. & Yates, J. R. III A model for random sampling and estimation of relative protein abundance in shotgun proteomics. Anal. Chem. 76, 4193–4201 (2004).
Carvalho, P. C., Yates Iii, J. R. & Barbosa, V. C. Analyzing shotgun proteomic data with PatternLab for proteomics. Curr. Protoc. Bioinform. Chapter 13, Unit 13.13.1–15 (2010).
Zhang, S.-R. et al. The Null-Test for peptide identification algorithm in Shotgun proteomics. J. Proteom. 163, 118–125 (2017).
Carvalho, P. C., Fischer, J. S. G., Xu, T., Yates, J. R., III & Barbosa, V. C. PatternLab: from mass spectra to label-free differential shotgun proteomics. Curr. Protoc. Bioinform. Chapter 13, Unit13.19 (2012).
Carvalho, P. C., Yates, J. R. III & Barbosa, V. C. Improving the TFold test for differential shotgun proteomics. Bioinformatics 28, 1652–1654 (2012).
Carvalho, P. C. et al. Analyzing marginal cases in differential shotgun proteomics. Bioinformatics 27, 275–276 (2011).
de Saldanha da Gama Fischer, J. et al. Chemo-resistant protein expression pattern of glioblastoma cells (A172) to perillyl alcohol. J. Proteome Res. 10, 153–160 (2011).
Leprevost, F. V. et al. PepExplorer: a similarity-driven tool for analyzing de novo sequencing results. Mol. Cell Proteom. https://doi.org/10.1074/mcp.M113.037002 (2014).
Fischer, J. et al. A scoring model for phosphopeptide site localization and its impact on the question of whether to use MSA. J. Proteom. https://doi.org/10.1016/j.jprot.2015.01.008 (2015).
Eng, J. K. et al. A deeper look into Comet–implementation and features. J. Am. Soc. Mass Spectrom. 26, 1865–1874 (2015).
Carvalho, P. C. et al. Integrated analysis of shotgun proteomic data with PatternLab for proteomics 4.0. Nat. Protoc. 11, 102–117 (2015).
Santos, M. D. M. et al. Mixed-data acquisition: next-generation quantitative proteomics data acquisition. J. Proteom. 222, 103803 (2020).
MacLean, B. et al. Skyline: an open source document editor for creating and analyzing targeted proteomics experiments. Bioinformatics 26, 966–968 (2010).
Gatchalian, J. et al. A non-canonical BRD9-containing BAF chromatin remodeling complex regulates naive pluripotency in mouse embryonic stem cells. Nat. Commun. 9, 5139 (2018).
Prieto, D. et al. S100-A9 protein in exosomes from chronic lymphocytic leukemia cells promotes NF-κB activity during disease progression. Blood 130, 777–788 (2017).
Sogues, A. et al. Essential dynamic interdependence of FtsZ and SepF for Z-ring and septum formation in Corynebacterium glutamicum. Nat. Commun. 11, 1641 (2020).
Horstmann, J. A. et al. Methylation of Salmonella typhimurium flagella promotes bacterial adhesion and host cell invasion. Nat. Commun. 11, 2013 (2020).
Camillo-Andrade, A. C. et al. Proteomics reveals that quinoa bioester promotes replenishing effects in epidermal tissue. Sci. Rep. 10, 19392 (2020).
Richards, A. L. et al. One-hour proteome analysis in yeast. Nat. Protoc. 10, 701–714 (2015).
UniProt Consortium. Update on activities at the Universal Protein Resource (UniProt) in 2013. Nucleic Acids Res. 41, D43–D47 (2013).
Zahn-Zabal, M. et al. The neXtProt knowledgebase in 2020: data, tools and usability improvements. Nucleic Acids Res. 48, D328–D334 (2020).
Li, H. et al. Evaluating the effect of database inflation in proteogenomic search on sensitive and reliable peptide identification. BMC Genomics 17, 1031 (2016).
Ma, B. Novor: real-time peptide de novo sequencing software. J. Am. Soc. Mass Spectrom. 26, 1885–1894 (2015).
Thompson, A. et al. Tandem mass tags: a novel quantification strategy for comparative analysis of complex protein mixtures by MS/MS. Anal. Chem. 75, 1895–1904 (2003).
Ong, S.-E. et al. Stable isotope labeling by amino acids in cell culture, SILAC, as a simple and accurate approach to expression proteomics. Mol. Cell Proteom. 1, 376–386 (2002).
Santos, M. D. M. et al. A quantitation module for isotope-labeled peptides integrated into PatternLab for proteomics. J. Proteom. 202, 103371 (2019).
Vizcaíno, J. A. et al. The mzIdentML data standard version 1.2, supporting advances in proteome informatics. Mol. Cell Proteom. 16, 1275–1285 (2017).
Perez-Riverol, Y. et al. The PRIDE database and related tools and resources in 2019: improving support for quantification data. Nucleic Acids Res. 47, D442–D450 (2019).
Martens, L. et al. mzML—a community standard for mass spectrometry data. Mol. Cell Proteom. 10, R110.000133–R110.000133 (2011).
Eng, J. K., Searle, B. C., Clauser, K. R. & Tabb, D. L. A face in the crowd: recognizing peptides through database search. Mol. Cell Proteom. 10, R111.009522 (2011).
Eng, J. K. & Deutsch, E. W. Extending Comet for global amino acid variant and post‐translational modification analysis using the PSI extended FASTA format. Proteomics 20, 1900362 (2020).
Wippel, H. H. et al. Comparing intestinal versus diffuse gastric cancer using a PEFF-oriented proteomic pipeline. J. Proteom. https://doi.org/10.1016/j.jprot.2017.10.005 (2017).
Punta, M. et al. The Pfam protein families database. Nucleic Acids Res. 40, D290–D301 (2012).
Pandurangan, A. P., Stahlhacke, J., Oates, M. E., Smithers, B. & Gough, J. The SUPERFAMILY 2.0 database: a significant proteome update and a new webserver. Nucleic Acids Res. 47, D490–D494 (2019).
Zybailov, B. et al. Statistical analysis of membrane proteome expression changes in Saccharomyces cerevisiae. J. Proteome Res. 5, 2339–2347 (2006).
Brunoro, G. V. F. et al. Reevaluating the Trypanosoma cruzi proteomic map: the shotgun description of bloodstream trypomastigotes. J. Proteom. 115, 58–65 (2015).
Benjamini, Yoav & Hochberg, Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J. R. Stat. Soc. 57, 289–300 (1995).
Kurt, L. U. et al. RawVegetable—a data assessment tool for proteomics and cross-linking mass spectrometry experiments. J. Proteom. 225, 103864 (2020).
Bonilauri, B. et al. Proteogenomic analysis reveals proteins involved in the first step of adipogenesis in human adipose-derived stem cells. Stem Cells Int. 2021, 1–14 (2021).
Leprevost, F. et al. On best practices in the development of bioinformatics software. Front. Genet. 5, 199 (2014).
Shalit, T., Elinger, D., Savidor, A., Gabashvili, A. & Levin, Y. MS1-based label-free proteomics using a quadrupole orbitrap mass spectrometer. J. Proteome Res. 14, 1979–1986 (2015).
Keshishian, H. et al. Quantitative, multiplexed workflow for deep analysis of human blood plasma and biomarker discovery by mass spectrometry. Nat. Protoc. 12, 1683–1701 (2017).
Acknowledgements
We thank W. Nagib, from Fiocruz, for creating the new PatternLab logo and entrance screen and J. Eng, from the University of Washington, for all the support and adaptations in the Comet search engine. We thank Conselho Nacional de Desenvolvimento Científico e Tecnológico (CNPq), Fundação de Amparo à Pesquisa do Estado do Rio de Janeiro (FAPERJ), Fiocruz, and Coordenação de Aperfeiçoamento de Pessoal de Nível Superior (CAPES) for financial support. R.H.V. (grant 304523/2019-4), V.C.B. (grant 300987/2019-6) and P.C.C. (grant 308930/2020-7) are CNPq research fellows. J.R.Y. acknowledges NIH P41 GM103533.
Author information
Authors and Affiliations
Contributions
P.C.C., J.R.Y. and V.C.B. have participated since the initial version of PatternLab, published in 2008. M.D.M.S., D.B.L., M.A.C., L.U.K., L.C.M. and P.C.C. served as developers, implementing the many features that enabled the transition from PL4 to PLV. J.S.G.F., P.F.d.A., A.G.C.N.F., R.H.V., M.O.T., G.V.F.B., T.A.C.B.S., R.M.S., A.C.C.-A., M.B., F.C.G. and R.D. are all experts in proteomics and worked closely with the computational team in developing new features, improving user experience and performing in-depth testing.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Nature Protocols thanks Annalisa Santucci, Yafeng Zhu and the other, anonymous, reviewer(s) for their contribution to the peer review of this work.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Related links
Key references using this protocol
Gatchalian, J. et al. Nat. Commun. 9, 5139 (2018): https://doi.org/10.1038/s41467-018-07528-9
Prieto, D. et al. Blood 130, 777–788 (2017): https://doi.org/10.1182/blood-2017-02-769851
Sogues, A. et al. Nat. Commun. 11, 1641 (2020): https://doi.org/10.1038/s41467-020-15490-8
Horstmann, J. A. et al. Nat. Commun. 11, 2013 (2020): https://doi.org/10.1038/s41467-020-15738-3
Key data used in this protocol
Camillo-Andrade, A. C. et al. Sci. Rep. 10, 19392 (2020): https://doi.org/10.1038/s41598-020-76325-6
Shalit, T. et al. Proteome Res. 14, 1979–1986 (2015): https://doi.org/10.1021/pr501045t
This protocol is an update to Nat. Protoc. 11, 102–117 (2015): https://doi.org/10.1038/nprot.2015.133
Rights and permissions
About this article
Cite this article
Santos, M.D.M., Lima, D.B., Fischer, J.S.G. et al. Simple, efficient and thorough shotgun proteomic analysis with PatternLab V. Nat Protoc 17, 1553–1578 (2022). https://doi.org/10.1038/s41596-022-00690-x
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41596-022-00690-x
This article is cited by
-
Intra-Individual Paired Mass Spectrometry Dataset for Decoding Solar-Induced Proteomic Changes in Facial Skin
Scientific Data (2024)
-
Eukaryotic-like gephyrin and cognate membrane receptor coordinate corynebacterial cell division and polar elongation
Nature Microbiology (2023)
-
Genomic–proteomic analysis of a novel Bacillus thuringiensis strain: toxicity against two lepidopteran pests, abundance of Cry1Ac5 toxin, and presence of InhA1 virulence factor
Archives of Microbiology (2023)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.