Abstract
Orthosomycin antibiotics inhibit protein synthesis by binding to the large ribosomal subunit in the tRNA accommodation corridor, which is traversed by incoming aminoacyl-tRNAs. Structural and biochemical studies suggested that orthosomycins block accommodation of any aminoacyl-tRNAs in the ribosomal A-site. However, the mode of action of orthosomycins in vivo remained unknown. Here, by carrying out genome-wide analysis of antibiotic action in bacterial cells, we discovered that orthosomycins primarily inhibit the ribosomes engaged in translation of specific amino acid sequences. Our results reveal that the predominant sites of orthosomycin-induced translation arrest are defined by the nature of the incoming aminoacyl-tRNA and likely by the identity of the two C-terminal amino acid residues of the nascent protein. We show that nature exploits this antibiotic-sensing mechanism for directing programmed ribosome stalling within the regulatory open reading frame, which may control expression of an orthosomycin-resistance gene in a variety of bacterial species.

This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout






Similar content being viewed by others
Data availability
Ribosome profiling (Ribo-seq) data have been deposited to the Gene Expression Omnibus (GEO) database under accession number GSE193270. Source data are provided with this paper.
Code availability
The scripts for Ribo-seq analysis can be found at the following link: https://github.com/mmaiensc/RiboSeq. The metagene analysis custom scripts are available at https://github.com/adamhockenberry/ribo-t-sequencing. The eUIgene.py Python script is available at https://github.com/GCA-VH-lab/eUIgene.
References
McCranie, E. K. & Bachmann, B. O. Bioactive oligosaccharide natural products. Nat. Prod. Rep. 31, 1026–1042 (2014).
McNicholas, P. M. et al. Evernimicin binds exclusively to the 50S ribosomal subunit and inhibits translation in cell-free systems derived from both Gram-positive and Gram-negative bacteria. Antimicrob. Agents Chemother. 44, 1121–1126 (2000).
Lin, J., Zhou, D., Steitz, T. A., Polikanov, Y. S. & Gagnon, M. G. Ribosome-targeting antibiotics: modes of action, mechanisms of resistance, and implications for drug design. Annu. Rev. Biochem. 87, 451–478 (2018).
Adrian, P. V. et al. Evernimicin (SCH27899) inhibits a novel ribosome target site: analysis of 23S ribosomal DNA mutants. Antimicrob. Agents Chemother. 44, 3101–3106 (2000).
Belova, L., Tenson, T., Xiong, L., McNicholas, P. M. & Mankin, A. S. A novel site of antibiotic action in the ribosome: interaction of evernimicin with the large ribosomal subunit. Proc. Natl Acad. Sci. USA 98, 3726–3731 (2001).
Krupkin, M. et al. Avilamycin and evernimicin induce structural changes in rProteins uL16 and CTC that enhance the inhibition of A-site tRNA binding. Proc. Natl Acad. Sci. USA 113, E6796–E6805 (2016).
Arenz, S. et al. Structures of the orthosomycin antibiotics avilamycin and evernimicin in complex with the bacterial 70S ribosome. Proc. Natl Acad. Sci. USA 113, 7527–7532 (2016).
Mikolajka, A. et al. Differential effects of thiopeptide and orthosomycin antibiotics on translational GTPases. Chem. Biol. 18, 589–600 (2011).
Wolf, H. Avilamycin, an inhibitor of the 30 S ribosomal subunits function. FEBS Lett. 36, 181–186 (1973).
Gibbs, M. R. et al. Conserved GTPase LepA (elongation factor 4) functions in biogenesis of the 30S subunit of the 70S ribosome. Proc. Natl Acad. Sci. USA 114, 980–985 (2017).
Choi, E. & Hwang, J. The GTPase BipA expressed at low temperature in Escherichia coli assists ribosome assembly and has chaperone-like activity. J. Biol. Chem. 293, 18404–18419 (2018).
Morse, J. C. et al. Elongation factor-Tu can repetitively engage aminoacyl-tRNA within the ribosome during the proofreading stage of tRNA selection. Proc. Natl Acad. Sci. USA 117, 3610–3620 (2020).
Orelle, C. et al. Tools for characterizing bacterial protein synthesis inhibitors. Antimicrob. Agents Chemother. 57, 5994–6004 (2013).
Ingolia, N. T., Ghaemmaghami, S., Newman, J. R. & Weissman, J. S. Genome-wide analysis in vivo of translation with nucleotide resolution using ribosome profiling. Science 324, 218–223 (2009).
Oh, E. et al. Selective ribosome profiling reveals the cotranslational chaperone action of trigger factor in vivo. Cell 147, 1295–1308 (2011).
Davis, A. R., Gohara, D. W. & Yap, M. N. Sequence selectivity of macrolide-induced translational attenuation. Proc. Natl Acad. Sci. USA 111, 15379–15384 (2014).
Kannan, K. et al. The general mode of translation inhibition by macrolide antibiotics. Proc. Natl Acad. Sci. USA 111, 15958–15963 (2014).
Vazquez-Laslop, N. & Mankin, A. S. Context-specific action of ribosomal antibiotics. Annu. Rev. Microbiol. 72, 185–207 (2018).
Eggert, U. S. et al. Genetic basis for activity differences between vancomycin and glycolipid derivatives of vancomycin. Science 294, 361–364 (2001).
Meydan, S. et al. Retapamulin-assisted ribosome profiling reveals the alternative bacterial proteome. Mol. Cell 74, 481–493 (2019).
Weaver, J., Mohammad, F., Buskirk, A. R. & Storz, G. Identifying small proteins by ribosome profiling with stalled initiation complexes. mBio 10, 02819-18 (2019).
Schuller, A. P., Wu, C. C., Dever, T. E., Buskirk, A. R. & Green, R. eIF5A functions globally in translation elongation and termination. Mol. Cell 66, 194–205 (2017).
Sothiselvam, S. et al. Binding of macrolide antibiotics leads to ribosomal selection against specific substrates based on their charge and size. Cell Rep. 16, 1789–1799 (2016).
Svetlov, M. S. et al. Context-specific action of macrolide antibiotics on the eukaryotic ribosome. Nat. Commun. 12, 2803 (2021).
Weisblum, B. Insights into erythromycin action from studies of its activity as inducer of resistance. Antimicrob. Agents Chemother. 39, 797–805 (1995).
Mann, P. A. et al. EmtA, a rRNA methyltransferase conferring high-level evernimicin resistance. Mol. Microbiol. 41, 1349–1356 (2001).
Bailey, M., Chettiath, T. & Mankin, A. S. Induction of ermC expression by ‘non-inducing’ antibiotics. Antimicrob. Agents Chemother. 52, 866–874 (2008).
Cantara, W. A. et al. Modifications modulate anticodon loop dynamics and codon recognition of E. coli tRNA(Arg1,2). J. Mol. Biol. 416, 579–597 (2012).
Curran, J. F. Decoding with the A:I wobble pair is inefficient. Nucleic Acids Res. 23, 683–688 (1995).
Muramatsu, T. et al. Codon and amino-acid specificities of a transfer RNA are both converted by a single post-transcriptional modification. Nature 336, 179–181 (1988).
Soma, A. et al. An RNA-modifying enzyme that governs both the codon and amino acid specificities of isoleucine tRNA. Mol. Cell 12, 689–698 (2003).
Voorhees, R. M. et al. The structural basis for specific decoding of AUA by isoleucine tRNA on the ribosome. Nat. Struct. Molec. Biol. 20, 641–643 (2013).
Bjork, G. R. et al. A primordial tRNA modification required for the evolution of life? EMBO J. 20, 231–239 (2001).
Masuda, I. et al. Loss of N1-methylation of G37 in tRNA induces ribosome stalling and reprograms gene expression. eLife 10, 70619 (2021).
Muto, H. & Ito, K. Peptidyl-prolyl-tRNA at the ribosomal P-site reacts poorly with puromycin. Biochem. Biophys. Res. Commun. 366, 1043–1047 (2008).
Wohlgemuth, I., Brenner, S., Beringer, M. & Rodnina, M. V. Modulation of the rate of peptidyl transfer on the ribosome by the nature of substrates. J. Biol. Chem. 283, 32229–32235 (2008).
Ude, S. et al. Translation elongation factor EF-P alleviates ribosome stalling at polyproline stretches. Science 339, 82–85 (2013).
Doerfel, L. K. et al. EF-P is essential for rapid synthesis of proteins containing consecutive proline residues. Science 339, 85–88 (2013).
Woolstenhulme, C. J. et al. Nascent peptides that block protein synthesis in bacteria. Proc. Natl Acad. Sci. USA 110, E878–E887 (2013).
Rajkovic, A. et al. Translation control of swarming proficiency in Bacillus subtilis by 5-amino-pentanolylated elongation factor P. J. Biol. Chem. 291, 10976–10985 (2016).
Beckert, B. et al. Structural and mechanistic basis for translation inhibition by macrolide and ketolide antibiotics. Nat. Commun. 12, 4466 (2021).
Pelechano, V. & Alepuz, P. eIF5A facilitates translation termination globally and promotes the elongation of many non polyproline-specific tripeptide sequences. Nucleic Acids Res. 45, 7326–7338 (2017).
Chadani, Y. et al. Intrinsic ribosome destabilization underlies translation and provides an organism with a strategy of environmental sensing. Mol. Cell 68, 528–539 (2017).
Chadani, Y. et al. Nascent polypeptide within the exit tunnel stabilizes the ribosome to counteract risky translation. EMBO J. 40, e108299 (2021).
Delsol, A. A. et al. Effect of the growth promoter avilamycin on emergence and persistence of antimicrobial resistance in enteric bacteria in the pig. J. Appl. Microbiol. 98, 564–571 (2005).
Gupta, P., Sothiselvam, S., Vazquez-Laslop, N. & Mankin, A. S. Deregulation of translation due to post-transcriptional modification of rRNA explains why erm genes are inducible. Nat. Commun. 4, 1984 (2013).
Gupta, P. et al. Nascent peptide assists the ribosome in recognizing chemically distinct small molecules. Nat. Chem. Biol. 12, 153–158 (2016).
Marks, J. et al. Context-specific inhibition of translation by ribosomal antibiotics targeting the peptidyl transferase center. Proc. Natl Acad. Sci. USA 113, 12150–12155 (2016).
Orelle, C. et al. Identifying the targets of aminoacyl-tRNA synthetase inhibitors by primer extension inhibition. Nucleic Acids Res. 41, e144 (2013).
Becker, A. H., Oh, E., Weissman, J. S., Kramer, G. & Bukau, B. Selective ribosome profiling as a tool for studying the interaction of chaperones and targeting factors with nascent polypeptide chains and ribosomes. Nat. Prot. 8, 2212–2239 (2013).
McGlincy, N. J. & Ingolia, N. T. Transcriptome-wide measurement of translation by ribosome profiling. Methods 126, 112–129 (2017).
Mangano, K. et al. Genome-wide effects of the antimicrobial peptide apidaecin on translation termination in bacteria. eLife 9, 62655 2020).
Aleksashin, N. A. et al. Assembly and functionality of the ribosome with tethered subunits. Nat. Commun. 10, 930 (2019).
Saha, C. K., Sanches Pires, R., Brolin, H., Delannoy, M. & Atkinson, G. C. FlaGs and webFlaGs: discovering novel biology through the analysis of gene neighbourhood conservation. Bioinformatics 37, 1312–1314 (2021).
Katoh, K. & Standley, D. M. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol. Biol. Evol. 30, 772–780 (2013).
Capella-Gutierrez, S., Silla-Martinez, J. M. & Gabaldon, T. trimAl: a tool for automated alignment trimming in large-scale phylogenetic analyses. Bioinformatics 25, 1972–1973 (2009).
Miller, M. A., Pfeiffer, W., Schwartz, T. The CIPRES science gateway: a community resource for phylogenetic analyses. In Proc. 2011 TeraGrid Conference: Extreme Digital Discovery (ACM, 2011).
Minh, B. Q. et al. IQ-TREE 2: new models and efficient methods for phylogenetic inference in the genomic era. Mol. Biol. Evol. 37, 1530–1534 (2020).
Larsson, A. AliView: a fast and lightweight alignment viewer and editor for large datasets. Bioinformatics 30, 3276–3278 (2014).
Krishnamoorthy, G. et al. Breaking the permeability barrier of Escherichia coli by controlled hyperporination of the outer membrane. Antimicrob. Agents Chemother. 60, 7372–7381 (2016).
Acknowledgements
We thank A. Innis and E. Leroy (University of Bordeaux) for structural insights, S. Meydan (University of Illinois at Chicago) for advice with some experiments, M. Svetlov for help with the figures and M. Ibba (Ohio State University) for sharing the B. subtilis strains. We are grateful to Y.-M. Hou (Thomas Jefferson University) for insights into the role of tRNA modification in decoding and to P. Mann and T. Black (Merck) for the advice on antibiotic-sensitive bacterial strains and information about preclinical development of evernimicin (Ziracin). This work was supported by the NIH grant R35-GM127134 to A.S.M. K.M. was supported by a NIH Training Grant (5T32AT007533). G.C.A. was supported by grants from the Knut and Alice Wallenberg Foundation (KAW 2020.0037), Umeå University Medical Faculty (Biotechnology grant to G.C.A), Kempestiftelserna (SMK-1858.3 to G.C.A), Carl Tryggers Stiftelse för Vetenskaplig Forskning (CTS19:24), and the Swedish Research Council (2019-01085). C.K.S. acknowledges support from Stiftelsen J.C. Kempes Stipendiefond.
Author information
Authors and Affiliations
Contributions
K.M., J.M., N.V.-L. and A.S.M. designed research. D. K. carried out Ribo-seq experiments, emtA induction studies and some toeprinting experiments. J.M. performed the initial analysis of the Ribo-seq data. K.M. carried out comprehensive analysis of the Ribo-seq data, performed the majority of the toeprinting experiments and analyzed inducibility of the emtA gene. C.K.S. and G.C.A. analyzed conservation of the EmtA methyltransferase, phylogenetic distribution of the emtA genes and conservation of the emtAL ORF. K.M., J.M., G.C.A., N.V.-L. and A.S.M. analyzed data. K.M, N.V.-L. and A.S.M. wrote the manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Nature Chemical Biology thanks the anonymous reviewers for their contribution to the peer review of this work.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data
Extended Data Fig. 1 Action of EVN is gene specific and does not involve an obligatory translation initiation arrest.
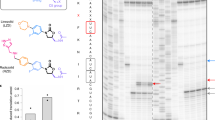
a, In vitro toeprinting experiments to analyze EVN-dependent translation arrest. Selection of the model genes was based on the results of Ribo-seq analysis (hupA) or those tested in previous reports (hns and ermBL)7,13. Open arrows indicate toeprint bands representing ribosomes stalled at the start codons by the action of initiation inhibitor oncocin (Onc)21. Black arrows point to the bands representing EVN-stalled ribosomes. Shown is a representative gel from at least two independent experiments. b, The nucleotide sequences of the relevant portions of the mRNA templates used in the toeprinting experiments shown in (a) and amino acid sequences of the encoded proteins. The open and filled arrows are as in (a). c, Ribosome profiling does not show any pronounced EVN-induced ribosome stalling at the hns start codon in E. coli cells. The first strong drug-induced translation arrest (green arrow) occurs at the codon 5 of hns.
Extended Data Fig. 2 EVN preserves the polysome profiles.
Sucrose gradient analysis of polysomes prepared from untreated E. coli cells (no drug) or treated with 0.5 µg/mL (1x MIC) or with 50 µg/mL (100x MIC) EVN during 30 min or 4 min, respectively.
Extended Data Fig. 3 Conditions and reproducibility of the Ribo-seq experiments to elucidate the mode of action of EVN.
a, Residual global protein synthesis (evaluated as incorporation of [35S]-Met into the TCA-insoluble protein fraction) of E. coli cells exposed to 50 µg/mL EVN for 4 min. Incorporation of [35S]-Met in a sample of the bacterial culture taken right before the addition of EVN was set as 100%. Open and closed circles represent the results of two independent experiments. b-d, Reproducibility of the two independent Ribo-seq experiments as judged by: (b) the EVN stall scores of all the considered codon positions (n = 118,222, Pearson’s r = 0.72); (c) of the mean stall score for codons at the ribosomal A site (n = 61, Pearson’s r = 0.96); (d) of the mean score for considered tripeptide motifs (n = 4039, Pearson’s r = 0.83) described in Figs. 2 and 3.
Extended Data Fig. 4 The action of EVN in bacterial cells is context specific.
Comparison of ribosome occupancy profiles of individual genes in untreated cells (no drug) with those treated with EVN.
Extended Data Fig. 5 An extended sequence motif is required for EVN-dependent ribosome stalling.
a,b, Ribosome occupancy profiles of individual genes where, in the presence of EVN, ribosomes translate successfully or not CGA codons (a) or PEP sequences (belonging to the identified PXP EVN arrest motif, see Fig. 3a, b) (b).
Extended Data Fig. 6 EVN-mediated ribosome stalling at a tripeptide PSP exemplifying the PXP motif.
a, Modifications of the gltX template to facilitate toeprinting analysis of EVN-mediated stalling at the PSP arrest sequence. The thin arrows indicate the mutations introduced in the wild-type sequence of the first 14 codons of the gltX gene encoding the EVN arrest motif Pro9-Ser10-Pro11. Gly13 codon was mutagenized to Trp13 in order to trap translating ribosomes at the preceding Thr12 (open arrow) when Trp-tRNA is depleted by the addition of indolmycin, an inhibitor of tryptophanyl-tRNA synthetase. In addition, the Lys4 codon was mutagenized to Asn4 codon to disrupt the +X + motif Lys2-Ile3-Lys4 of GltX, where mild EVN-mediated stalling occurs during in vitro translation (grey arrow). The red and open arrows indicate toeprint band representing EVN-bound ribosomes stalled at Ser10 codon and the ribosomes trapped at the Thr12 codon due to the depletion of Trp-tRNA, respectively. b, Toeprinting analysis of EVN-mediated ribosome stalling at the original PXP motif Pro9-Ser10-Pro11 (X = Ser) of gltX or mutant templates where the Ser10 codon was mutated to encode different amino acids (Met, Leu, Ile, or Ala). c, Synonymous mutations of the P-site Ser10 codon have little effect on stalling, whereas replacement of the Ser10codon with the Ala codon has a strong effect. Representative gels from two independent experiments are shown in panels a, b and c.
Extended Data Fig. 7 EmtA homologs are found in diverse bacterial species.
Alignment of representative EmtA sequences identified in sequenced bacterial genomes (see Extended Data Fig. 8 for the complete phylogenetic tree).
Extended Data Fig. 8 Maximum likelihood phylogeny of emtA homologs.
Branch lengths are proportional to the number of substitutions. The tree is annotated with circles showing the presence (red) or absence (blue) of the emtAL uORF in 5’UTR of the emtA genes, where the 5’UTR is confidently alignable. The sequences of the encoded leader peptides are indicated. The asterisk indicates peptides from two annotated neighboring ORFs in Paenactinomyces guangxiensis, corresponding to the N and C terminal regions of EmtA. The interruption of the emtA ORF suggests pseudogenation caused by a frameshift.
Extended Data Fig. 9 Distal segments of the nascent peptide in the ribosomal exit tunnel contribute to EVN-dependent translation arrest within the emtAL ORF.
a, The VFL motif within the EmtAL peptide affords only partial translation arrest. Top: gel electrophoretic analysis of accumulation of MMMAVF-tRNA and MMMAVFL-tRNA during in vitro translation of the truncated emtAL template at increasing concentrations of EVN. Bottom: quantification of the gel (from two independent experiments) showing the fraction of incomplete translation product (MMMAVF-tRNA). b, Toeprinting analysis of EVN-mediated ribosome stalling in templates encoding the E. faecium emtAL ORF where the identities of codons 2–4 were simultaneously modified by introducing single-nucleotide compensatory frame-shifting mutations (indicated by black arrows and red letters). Representative gels from two independent experiments are shown in panels a and b.
Extended Data Fig. 10 Leader ORF emtAL in the 5’ UTR of the E. faecium emtA gene.
Possible secondary structure of the 5’UTR of the E. faecium emtA gene. The emtAL ORF and its Shine-Dalgarno sequence are highlighted in green. The emtA ORF and its Shine-Dalgarno sequence are shown in red.
Supplementary information
Supplementary Table 1
DNA oligonucleotides used in the study.
Supplementary Data
Ranking of tripeptides by the EVN stalling score.
Source data
Source Data Fig. 1
Uncropped toeprinting gels.
Source Data Fig. 2
Uncropped toeprinting gel.
Source Data Fig. 3
Uncropped toeprinting gels.
Source Data Fig. 4
Uncropped toeprinting and protein gels.
Source Data Extended Data Fig. 1
Uncropped toeprinting gel.
Source Data Extended Data Fig. 6
Uncropped toeprinting gels.
Source Data Extended Data Fig. 9
Uncropped protein and toeprinting gels.
Rights and permissions
Springer Nature or its licensor holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Mangano, K., Marks, J., Klepacki, D. et al. Context-based sensing of orthosomycin antibiotics by the translating ribosome. Nat Chem Biol 18, 1277–1286 (2022). https://doi.org/10.1038/s41589-022-01138-9
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41589-022-01138-9
This article is cited by
-
Motif-ation matters
Nature Chemical Biology (2023)