Abstract
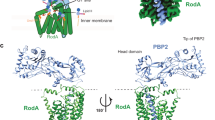
The shape, elongation, division and sporulation (SEDS) proteins are a highly conserved family of transmembrane glycosyltransferases that work in concert with class B penicillin-binding proteins (bPBPs) to build the bacterial peptidoglycan cell wall1,2,3,4,5,6. How these proteins coordinate polymerization of new glycan strands with their crosslinking to the existing peptidoglycan meshwork is unclear. Here, we report the crystal structure of the prototypical SEDS protein RodA from Thermus thermophilus in complex with its cognate bPBP at 3.3 Å resolution. The structure reveals a 1:1 stoichiometric complex with two extensive interaction interfaces between the proteins: one in the membrane plane and the other at the extracytoplasmic surface. When in complex with a bPBP, RodA shows an approximately 10 Å shift of transmembrane helix 7 that exposes a large membrane-accessible cavity. Negative-stain electron microscopy reveals that the complex can adopt a variety of different conformations. These data define the bPBP pedestal domain as the key allosteric activator of RodA both in vitro and in vivo, explaining how a SEDS–bPBP complex can coordinate its dual enzymatic activities of peptidoglycan polymerization and crosslinking to build the cell wall.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 digital issues and online access to articles
$119.00 per year
only $9.92 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout



Similar content being viewed by others
Data availability
Structure factors and refined atomic coordinates for the WT RodA–PBP2 complex and RodA(D255A)–PBP2 variant complex have been deposited in the RCSB PDB under accession codes 6PL5 and 6PL6, respectively. All other data that support the findings of this study are available from the corresponding author upon request. The source data for Figs. 2b and 2d are provided with the paper.
References
Meeske, A. J. et al. SEDS proteins are a widespread family of bacterial cell wall polymerases. Nature 537, 634–638 (2016).
Emami, K. et al. RodA as the missing glycosyltransferase in Bacillus subtilis and antibiotic discovery for the peptidoglycan polymerase pathway. Nat. Microbiol. 2, 16253 (2017).
Cho, H. et al. Bacterial cell wall biogenesis is mediated by SEDS and PBP polymerase families functioning semi-autonomously. Nat. Microbiol. 1, 16172 (2016).
Rohs, P. D. A. et al. A central role for PBP2 in the activation of peptidoglycan polymerization by the bacterial cell elongation machinery. PLoS Genet. 14, e1007726 (2018).
Taguchi, A. et al. FtsW is a peptidoglycan polymerase that is functional only in complex with its cognate penicillin-binding protein. Nat. Microbiol. 4, 587–594 (2019).
Reichmann, N. T. et al. SEDS–bPBP pairs direct lateral and septal peptidoglycan synthesis in Staphylococcus aureus. Nat. Microbiol. 4, 1368–1377 (2019).
Silhavy, T. J., Kahne, D. & Walker, S. The bacterial cell envelope. Cold Spring Harb. Perspect. Biol. 2, a000414 (2010).
Scheffers, D.-J. & Tol, M. B. LipidII: just another brick in the wall? PLoS Pathog. 11, e1005213 (2015).
Otten, C., Brilli, M., Vollmer, W., Viollier, P. H. & Salje, J. Peptidoglycan in obligate intracellular bacteria. Mol. Microbiol. 107, 142–163 (2018).
Ruiz, N. Lipid flippases for bacterial peptidoglycan biosynthesis. Lipid Insights 8, 21–31 (2016).
Sjodt, M. et al. Structure of the peptidoglycan polymerase RodA resolved by evolutionary coupling analysis. Nature 556, 118–121 (2018).
Caffrey, M. A comprehensive review of the lipid cubic phase or in meso method for crystallizing membrane and soluble proteins and complexes. Acta Crystallogr. F Struct. Biol. Commun. 71, 3–18 (2015).
Levy, N. et al. Structural basis for E. coli penicillin binding protein (PBP) 2 inhibition, a platform for drug design. J. Med. Chem. 62, 4742–4754 (2019).
Contreras-Martel, C. et al. Molecular architecture of the PBP2–MreC core bacterial cell wall synthesis complex. Nat. Commun. 8, 776 (2017).
Mravic, M. et al. Packing of apolar side chains enables accurate design of highly stable membrane proteins. Science 363, 1418–1423 (2019).
Ovchinnikov, S., Kamisetty, H. & Baker, D. Robust and accurate prediction of residue–residue interactions across protein interfaces using evolutionary information. eLife 3, e02030 (2014).
Hopf, T. A. et al. Sequence co-evolution gives 3D contacts and structures of protein complexes. eLife 3, e03430 (2014).
Cho, H., Uehara, T. & Bernhardt, T. G. Beta-lactam antibiotics induce a lethal malfunctioning of the bacterial cell wall synthesis machinery. Cell 159, 1300–1311 (2014).
Otero, L. H. et al. How allosteric control of Staphylococcus aureus penicillin binding protein 2a enables methicillin resistance and physiological function. Proc. Natl Acad. Sci. USA 110, 16808–16813 (2013).
Fay, A., Meyer, P. & Dworkin, J. Interactions between late-acting proteins required for peptidoglycan synthesis during sporulation. J. Mol. Biol. 399, 547–561 (2010).
Fraipont, C. et al. The integral membrane FtsW protein and peptidoglycan synthase PBP3 form a subcomplex in Escherichia coli. Microbiology 157, 251–259 (2011).
Welsh, M. A. et al. Identification of a functionally unique family of penicillin-binding proteins. J. Am. Chem. Soc. 139, 17727–17730 (2017).
Caffrey, M. & Cherezov, V. Crystallizing membrane proteins using lipidic mesophases. Nat. Protoc. 4, 706–731 (2009).
Kabsch, W. XDS. Acta Crystallogr. D Biol. Crystallogr. 66, 125–132 (2010).
McCoy, A. J. et al. Phaser crystallographic software. J. Appl. Crystallogr. 40, 658–674 (2007).
Fedarovich, A., Nicholas, R. A. & Davies, C. Unusual conformation of the SxN motif in the crystal structure of penicillin-binding protein A from Mycobacterium tuberculosis. J. Mol. Biol. 398, 54–65 (2010).
Emsley, P. & Cowtan, K. Coot: model-building tools for molecular graphics. Acta Crystallogr. D Biol. Crystallogr. 60, 2126–2132 (2004).
Afonine, P. V. et al. Towards automated crystallographic structure refinement with phenix.refine. Acta Crystallogr. D Biol. Crystallogr. 68, 352–367 (2012).
Strong, M. et al. Toward the structural genomics of complexes: crystal structure of a PE/PPE protein complex from Mycobacterium tuberculosis. Proc. Natl Acad. Sci. USA 103, 8060–8065 (2006).
Chen, V. B. et al. MolProbity: all-atom structure validation for macromolecular crystallography. Acta Crystallogr. D Biol. Crystallogr. 66, 12–21 (2010).
Morin, A. et al. Collaboration gets the most out of software. eLife 2, e01456 (2013).
Ashkenazy, H. et al. ConSurf 2016: an improved methodology to estimate and visualize evolutionary conservation in macromolecules. Nucleic Acids Res. 44, W344–W350 (2016).
Scheres, S. H. W. RELION: implementation of a Bayesian approach to cryo-EM structure determination. J. Struct. Biol. 180, 519–530 (2012).
Tang, G. et al. EMAN2: an extensible image processing suite for electron microscopy. J. Struct. Biol. 157, 38–46 (2007).
Qiao, Y. et al. Detection of lipid-linked peptidoglycan precursors by exploiting an unexpected transpeptidase reaction. J. Am. Chem. Soc. 136, 14678–14681 (2014).
Bendezú, F. O. & de Boer, P. A. J. Conditional lethality, division defects, membrane involution, and endocytosis in Mre and Mrd shape mutants of Escherichia coli. J. Bacteriol. 190, 1792–1811 (2008).
Smale, S. T. β-galactosidase assay. Cold Spring Harb. Protoc. 2010, pdb.prot5423 (2010).
Ducret, A., Quardokus, E. M. & Brun, Y. V. MicrobeJ, a tool for high throughput bacterial cell detection and quantitative analysis. Nat. Microbiol. 1, 16077 (2016).
Acknowledgements
Financial support for the work was provided by National Institutes of Health grant nos. U19AI109764 (A.C.K., D.Z.R., T.G.B., S.W. and D.K.), R01GM106303 (D.S.M.) and 5F31GM128233-02 (S.C.E), the Howard Hughes Medical Institute (T.G.B.) and a Canadian Institutes of Health Research doctoral research award to P.D.A.R. We thank the Advanced Photon Source GM/CA beamline staff for the excellent facilities and technical assistance during X-ray data collection and M. Welsh for the generous gift of purified lipid II substrate.
Author information
Authors and Affiliations
Contributions
M.S. performed the large-scale purification and crystallization of the RodA–PBP2 complexes, the enzymatic assays and negative-stain electron microscopy and data processing. S.C.E. collected the electron microscopy images and provided input on data processing. M.S.A.G. assisted with revisions and performed small-scale protein purification. A.T., D.K. and S.W. provided additional input regarding the enzyme assays. M.S., S.Z. and A.C.K. solved and refined the structure. K.B. and A.G.G., with supervision by D.S.M., performed additional validation of the RodA–PBP2 structure using evolutionary couplings. P.D.A.R. assessed the PBP2 mutant phenotypes with supervision from T.G.B. and D.Z.R. A.C.K. performed the overall project supervision with input from T.G.B. and D.Z.R. M.S. and A.C.K. wrote the manuscript with input from the other authors.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data
Extended Data Fig. 1 RodA–PBP2 complex interface within the membrane plane.
The transmembrane helix of PBP2 is shown as blue ribbons and transparent molecular surfaces to highlight its interaction with a, RodA transmembrane helix 8 and b, RodA transmembrane helix 9.
Extended Data Fig. 2 Evolutionary covariation analysis of RodA–PBP2 interface.
Graphical representation of 19 evolutionary couplings between RodA-PBP2.
Extended Data Fig. 3 RodA–PBP2 complex interface II.
PBP2 is shown as ribbons colored blue and RodA is shown as molecular surfaces and colored green. The insets show two views of interface II. Side chains of PBP2 residues at the RodA interface are shown as sticks with hydrophobic residues also shown as transparent molecular surface to highlight surface complementarity. Two PBP2 residues that disrupted RodA function when substituted with arginine are labeled.
Extended Data Fig. 4 Shift of TM7 of RodA in complex with PBP2 results in a large membrane-accessible cavity.
Surface view of RodA:PBP2 complex (left) and RodA (right) representing electrostatic potential (top panel) and a cross section (bottom panel). The large surface-exposed cavity is outlined in black dotted lines.
Extended Data Fig. 5 PBP2 mutants co-purify with RodA.
PBP2 variants were co-expressed with FLAG-tagged RodA and purified using anti-FLAG affinity resin. SDS-PAGE gel showing the elution from the anti-FLAG affinity resin demonstrates that PBP2 remains associated with RodA throughout the purification. Results are derived from one experiment.
Extended Data Fig. 6 Structural comparison of extracytoplasmic domains of PBP2 from T. thermophilus and H. pylori.
The catalytic serine in each transpeptidase in shown as a red sphere.
Extended Data Fig. 7 E. coli PBP2 variants co-purify with E. coli RodA.
PBP2 variants were co-expressed with FLAG-tagged RodA and purified using anti-FLAG affinity resin. SDS-PAGE gel showing the elution from the anti-FLAG affinity resin demonstrates that PBP2 remains associated with RodA throughout the purification. Low (L) and high (H) correspond to approximately 2 µg and 4 µg as measured by A280. Results are derived from one experiment.
Extended Data Fig. 8 RodA–PBP2 complex crystal lattice.
The TtRodA:PBP2 complex adopts type 1 lipid cubic phase crystal packing. RodA and PBP2 are shown in green and blue, respectively.
Extended Data Fig. 9 EM analysis of TtRodA–PBP2 complex.
a, Representative EM micrograph of negatively stained RodA:PBP2 complex solubilized in DDM detergent micelles. Scale bar denotes 50 nm. b, Representative two-dimensional class averages from a total of 32,152 particles. Scale bar represents 10 nm. Results are derived from one experiment.
Extended Data Fig. 10 Electron density map.
2Fo−Fc electron density map contoured at 1.0 σ within a 2.3 Å radius of atoms shown for a, the entire T. thermophilus RodA:PBP2 complex b, the transmembrane helix of PBP2 and c, transmembrane helices 5–7 and extracellular loop 4 of RodA.
Supplementary information
Supplementary Information
Supplementary Tables 1 and 2, and Figs. 1–3.
Source data
Source Data Fig. 2
Unprocessed western blots.
Rights and permissions
About this article
Cite this article
Sjodt, M., Rohs, P.D.A., Gilman, M.S.A. et al. Structural coordination of polymerization and crosslinking by a SEDS–bPBP peptidoglycan synthase complex. Nat Microbiol 5, 813–820 (2020). https://doi.org/10.1038/s41564-020-0687-z
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41564-020-0687-z