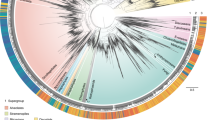
Abstract
Over millennia, ecological and evolutionary mechanisms have shaped macroecological patterns across the tree of life. Research describing these patterns at both regional and global scales has traditionally focused on the study of metazoan species. Consequently, there is a limited understanding of cross-phylum biogeographic structuring and an escalating need to understand the macroecology of both microscopic and macroscopic organisms. Here we used environmental DNA (eDNA) metabarcoding to explore the biodiversity of marine metazoans, protists and bacteria along an extensive and highly heterogeneous coastline. Our results showed remarkably consistent biogeographic structure across the kingdoms of life despite billions of years of evolution. Analyses investigating the drivers of these patterns for each taxonomic kingdom found that environmental conditions (such as temperature) and, to a lesser extent, anthropogenic stressors (such as fishing pressure and pollution) explained some of the observed variation. Additionally, metazoans displayed biogeographic patterns that suggested regional biotic homogenization. Against the backdrop of global pervasive anthropogenic environmental change, our work highlights the importance of considering multiple domains of life to understand the maintenance and drivers of biodiversity patterns across broad taxonomic, ecological and geographical scales.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 digital issues and online access to articles
$119.00 per year
only $9.92 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout



Similar content being viewed by others
Data availability
The raw Illumina sequencing data are available from the European Nucleotide Archive under study accession number PRJEB38452; sample-specific accessions are provided in Supplementary Data 1. The associated metadata, R scripts and intermediate files are available at https://doi.org/10.5281/zenodo.4564075.
Code availability
All code used in the current study can be found at https://doi.org/10.5281/zenodo.4564075.
References
Spalding, M. D. et al. Marine ecoregions of the world: a bioregionalization of coastal and shelf areas. BioScience 57, 573–583 (2007).
Awad, A. A., Griffiths, C. L. & Turpie, J. K. Distribution of South African marine benthic invertebrates applied to the selection of priority conservation areas. Divers. Distrib. 8, 129–145 (2002).
Pecl, G. T. et al. Biodiversity redistribution under climate change: impacts on ecosystems and human well-being. Science https://doi.org/10.1126/science.aai9214 (2017).
Sunday, J. M., Bates, A. E. & Dulvy, N. K. Thermal tolerance and the global redistribution of animals. Nat. Clim. Change 2, 686–690 (2012).
Wallace, A. R. The Geographical Distribution of Animals, with a Study of the Relations of Living and Extinct Faunas as Elucidating the Past Changes of the Earth’s Surface (Macmillan, 1876).
Holt, B. G. et al. An update of Wallace’s zoogeographic regions of the world. Science 339, 74–78 (2013).
Ficetola, G. F., Mazel, F. & Thuiller, W. Global determinants of zoogeographical boundaries. Nat. Ecol. Evol. 1, 89 (2017).
Kocsis, A. T., Reddin, C. J. & Kiessling, W. The stability of coastal benthic biogeography over the last 10 million years. Glob. Ecol. Biogeogr. 27, 1106–1120 (2018).
Zaffosa, A., Finnegan, S. & Peters, S. E. Plate tectonic regulation of global marine animal diversity. Proc. Natl Acad. Sci. USA 114, 5653–5658 (2017).
Costello, M. J. et al. Marine biogeographic realms and species endemicity. Nat. Commun. https://doi.org/10.1038/s41467-017-01121-2 (2017).
Beck, J. et al. What’s on the horizon for macroecology? Ecography 35, 673–683 (2012).
Sunagawa, S. et al. Ocean plankton: structure and function of the global ocean microbiome. Science 348, 1261359 (2015).
Shade, A. et al. Macroecology to unite all life, large and small. Trends Ecol. Evol. 33, 731–744 (2018).
Djurhuus, A. et al. Environmental DNA reveals seasonal shifts and potential interactions in a marine community. Nat. Commun. 11, 254 (2020).
Richter, D. J. et al. Genomic evidence for global ocean plankton biogeography shaped by large-scale current systems. Preprint at bioRxiv https://doi.org/10.1101/867739 (2019).
Naeem, S., Duffy, J. E. & Zavaleta, E. The functions of biological diversity in an age of extinction. Science 336, 1401–1406 (2012).
Tilman, D., Isbell, F. & Cowles, J. M. Biodiversity and ecosystem functioning. Annu. Rev. Ecol. Evol. Syst. 45, 471–493 (2014).
Finderup Nielsen, T., Sand-Jensen, K., Dornelas, M. & Bruun, H. H. More is less: net gain in species richness, but biotic homogenization over 140 years. Ecol. Lett. 22, 1650–1657 (2019).
Blowes, S. A. et al. The geography of biodiversity change in marine and terrestrial assemblages. Science 366, 339–345 (2019).
Dornelas, M. et al. Assemblage time series reveal biodiversity change but not systematic loss. Science 344, 296–299 (2014).
Olden, J. D. & Rooney, T. P. On defining and quantifying biotic homogenization. Glob. Ecol. Biogeogr. 15, 113–120 (2006).
Stuart-Smith, R. D. et al. Integrating abundance and functional traits reveals new global hotspots of fish diversity. Nature 501, 539–542 (2013).
Mouillot, D. et al. Rare species support vulnerable functions in high-diversity ecosystems. PLoS Biol. 11, e1001569 (2013).
Bernardo-Madrid, R. et al. Human activity is altering the world’s zoogeographical regions. Ecol. Lett. 22, 1297–1305 (2019).
Capinha, C., Essl, F., Seebens, H., Moser, D. & Pereira, H. M. The dispersal of alien species redefines biogeography in the Anthropocene. Science 348, 1248–1251 (2015).
Azam, F. & Malfatti, F. Microbial structuring of marine ecosystems. Nat. Rev. Microbiol. 5, 782–791 (2007).
Deiner, K. et al. Environmental DNA metabarcoding: transforming how we survey animal and plant communities. Mol. Ecol. 26, 5872–5895 (2017).
Emanuel, B. P., Bustamante, R. H., Branch, G. M., Eekhout, S. & Odendaal, F. J. A zoogeographic and functional approach to the selection of marine reserves on the west coast of South Africa. South Afr. J. Mar. Sci. 12, 341–354 (1992).
Griffiths, C. L., Robinson, T. B., Lange, L. & Mead, A. Marine biodiversity in South Africa: an evaluation of current states of knowledge. PLoS ONE 5, e12008 (2010).
Griffiths, C. L. et al. Impacts of human activities on marine animal life in the Benguela: a historical overview. Oceanogr. Mar. Biol. Annu. Rev. 42, 303–392 (2004).
Kaluza, P., Kolzsch, A., Gastner, M. T. & Blasius, B. The complex network of global cargo ship movements. J. R. Soc. Interface 7, 1093–1103 (2010).
Rapacciuolo, G., Beman, J. M., Schiebelhut, L. M. & Dawson, M. N. Microbes and macro-invertebrates show parallel β-diversity but contrasting α-diversity patterns in a marine natural experiment. Proc. R. Soc. B 286, 20190999 (2019).
Astorga, A. et al. Distance decay of similarity in freshwater communities: do macro- and microorganisms follow the same rules? Glob. Ecol. Biogeogr. 21, 365–375 (2012).
Wang, J. et al. Patterns of elevational beta diversity in micro- and macroorganisms. Glob. Ecol. Biogeogr. 21, 743–750 (2012).
Tittensor, D. P. et al. Global patterns and predictors of marine biodiversity across taxa. Nature 466, 1098–U1107 (2010).
Herlemann, D. P. et al. Transitions in bacterial communities along the 2000 km salinity gradient of the Baltic Sea. ISME J. 5, 1571–1579 (2011).
Broman, E. et al. Salinity drives meiofaunal community structure dynamics across the Baltic ecosystem. Mol. Ecol. 28, 3813–3829 (2019).
Shochat, E., Warren, P. S., Faeth, S. H., McIntyre, N. E. & Hope, D. From patterns to emerging processes in mechanistic urban ecology. Trends Ecol. Evol. 21, 186–191 (2006).
Halpern, B. S. et al. Recent pace of change in human impact on the world’s ocean. Sci. Rep. 9, 11609 (2019).
Kelly, R. P. et al. Genetic signatures of ecological diversity along an urbanization gradient. PeerJ 4, e2444 (2016).
Blouin, D., Pellerin, S. & Poulin, M. Increase in non-native species richness leads to biotic homogenization in vacant lots of a highly urbanized landscape. Urban Ecosyst. 22, 879–892 (2019).
Holman, L. E. et al. Detection of introduced and resident marine species using environmental DNA metabarcoding of sediment and water. Sci. Rep. 9, 11559 (2019).
Lima-Mendez, G. et al. Determinants of community structure in the global plankton interactome. Science 348, 1262073 (2015).
Baas-Becking, L. G. M. Geobiologie; of inleiding tot de milieukunde (WP Van Stockum & Zoon NV, 1934).
Hanson, C. A., Fuhrman, J. A., Horner-Devine, M. C. & Martiny, J. B. Beyond biogeographic patterns: processes shaping the microbial landscape. Nat. Rev. Microbiol. 10, 497–506 (2012).
Farjalla, V. F. et al. Ecological determinism increases with organism size. Ecology 93, 1752–1759 (2012).
Wu, W. X. et al. Contrasting the relative importance of species sorting and dispersal limitation in shaping marine bacterial versus protist communities. ISME J. 12, 485–494 (2018).
Hellweger, F. L., van Sebille, E. & Fredrick, N. D. Biogeographic patterns in ocean microbes emerge in a neutral agent-based model. Science 345, 1346–1349 (2014).
Balint, M. et al. Environmental DNA time series in ecology. Trends Ecol. Evol. 33, 945–957 (2018).
He, K. S. et al. Will remote sensing shape the next generation of species distribution models? Remote Sens. Ecol. Conserv. 1, 4–18 (2015).
Rius, M. et al. Range expansions across ecoregions: interactions of climate change, physiology and genetic diversity. Glob. Ecol. Biogeogr. 23, 76–88 (2014).
Spens, J. et al. Comparison of capture and storage methods for aqueous macrobial eDNA using an optimized extraction protocol: advantage of enclosed filter. Methods Ecol. Evol. 8, 635–645 (2017).
Leray, M. et al. A new versatile primer set targeting a short fragment of the mitochondrial COI region for metabarcoding metazoan diversity: application for characterizing coral reef fish gut contents. Front. Zool. 10, 34 (2013).
Zhan, A. et al. High sensitivity of 454 pyrosequencing for detection of rare species in aquatic communities. Methods Ecol. Evol. 4, 558–565 (2013).
Takahashi, S., Tomita, J., Nishioka, K., Hisada, T. & Nishijima, M. Development of a prokaryotic universal primer for simultaneous analysis of Bacteria and Archaea using next-generation sequencing. PLoS ONE 9, e105592 (2014).
Martin, M. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet J. 17, 10–12 (2011).
Callahan, B. J. et al. DADA2: high-resolution sample inference from Illumina amplicon data. Nat. Methods 13, 581–583 (2016).
R Core Team R: A Language and Environment for Statistical Computing v.3.6.1 (R Foundation for Statistical Computing, 2019).
Frøslev, T. G. et al. Algorithm for post-clustering curation of DNA amplicon data yields reliable biodiversity estimates. Nat. Commun. 8, 1188 (2017).
Wang, Q., Garrity, G. M., Tiedje, J. M. & Cole, J. R. Naive Bayesian classifier for rapid assignment of rRNA sequences into the new bacterial taxonomy. Appl. Environ. Microbiol. 73, 5261–5267 (2007).
Porter, T. M. & Hajibabaei, M. Automated high throughput animal CO1 metabarcode classification. Sci. Rep. 8, 4226 (2018).
Quast, C. et al. The SILVA ribosomal RNA gene database project: improved data processing and web-based tools. Nucleic Acids Res. 41, D590–D596 (2013).
Edgar, R. C. Updating the 97% identity threshold for 16S ribosomal RNA OTUs. Bioinformatics 34, 2371–2375 (2018).
Burki, F., Roger, A. J., Brown, M. W. & Simpson, A. G. The new tree of eukaryotes. Trends Ecol. Evol. 35, 43–55 (2020).
GHRSST Level 4 G1SST Global Foundation Sea Surface Temperature Analysis (JPL_OurOceanProject, 2010); https://doi.org/10.5067/GHG1S-4FP01
Zweng, M. M. et al. World Ocean Atlas 2018, Volume 2: Salinity NOAA Atlas NESDIS 82 (ed. Mishinov, A.) (NESDIS/US Department of Commerce, NOAA, 2019).
Ocean Colour Climate Change Initiative Dataset Version 4.2 (European Space Agency, 2020).
Anderson, M. J. in Wiley Stats Ref: Statistics Reference Online (eds Balakrishnan, N. et al.) 1–15 (John Wiley & Sons, 2014).
Oksanen, J. et al. Vegan: Community ecology package. R package version 2.5–6 (2011).
Kreft, H. & Jetz, W. A framework for delineating biogeographical regions based on species distributions. J. Biogeogr. 37, 2029–2053 (2010).
Salazar, G. EcolUtils: Utilities for community ecology analysis. R package version 0.1 (2018).
Anderson, M. J. Distance-based tests for homogeneity of multivariate dispersions. Biometrics 62, 245–253 (2006).
Crabot, J., Clappe, S., Dray, S. & Datry, T. Testing the Mantel statistic with a spatially-constrained permutation procedure. Methods Ecol. Evol. 10, 532–540 (2019).
McArdle, B. H. & Anderson, M. J. Fitting multivariate models to community data: a comment on distance-based redundancy analysis. Ecology 82, 290–297 (2001).
Acknowledgements
L.E.H. acknowledges the assistance of M. Czachur and T. Grevesse during field surveys, S. von der Heyden for lab consumables and assistance with fieldwork logistics, and J. Hudson and I. Haigh for assistance with remote sensing data. L.E.H. acknowledges S. Parker-Nance and the Elwandle Node of the South African Environmental Observation Network for assistance and in-country logistics. We acknowledge all marina owners and operators for field site access. We acknowledge the Environmental Sequencing Facility at the National Oceanography Centre, Southampton, for advice and sequencing assistance. We thank the IRIDIS High Performance Computing Facility and associated support services at the University of Southampton. L.E.H. was supported by the Natural Environmental Research Council (grant no. NE/L002531/1). The UK Research and Innovation Newton Fund (grant no. ES/N013913/1) supported L.E.H.’s research stay in South Africa.
Author information
Authors and Affiliations
Contributions
L.E.H. and M.R. designed the study. L.E.H. collected the samples, generated and analysed the data, prepared all figures and wrote the first draft of the paper. M.d.B., S.C., G.C., J.R. and M.R. supervised and advised the research. All authors substantially contributed to further manuscript drafts and provided final approval for publication.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Peer review information Nature Ecology & Evolution thanks Simon Jarman, Ryan Kelly and the other, anonymous, reviewer(s) for their contribution to the peer review of this work. Peer reviewer reports are available.
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data
Extended Data Fig. 1 Taxonomic identity of sequenced reads.
Bar charts indicating the proportion of reads assigned per phyla (metazoans/bacteria) or supergroup (protists) from environmental DNA metabarcoding of seawater collected from sites across South Africa. The three rows correspond with data from metazoans (top), protists (middle) and bacteria (bottom). Site name abbreviations as in Supplementary Table 9.
Extended Data Fig. 2 Beta diversity patterns of additional data subsets.
Observed patterns of β-diversity from environmental DNA metabarcoding of: a metazoans from the 18S dataset and b protists from the COI dataset; based on Jaccard dissimilarities between amplicon sequence variants along the coast of South Africa. The first column of plots shows non-metric multidimensional scaling (nMDS) ordinations. Coloured hulls show the spread of the data and lines indicate the spread around the centroid grouped by coast with the east, south and west coasts denoted by orange, green and blue respectively. Site name abbreviations as in Supplementary Table 9. Natural sites are denoted with triangles and artificial sites with filled circles. The second column of plots shows the same nMDS ordinations as the first column including the output of a generalised additive model with a 2D smoothed function for each of the significant environmental / impact variables overlaid; temperature – mean sea surface temperature (°C); impact – human marine impact score (unitless measurement, see details in main text) against the two nMDS axes. The Venn diagram charts indicate the percentage total of variance in community dissimilarity explained by each significant variable, derived using variance partitioning of a distance-based redundancy analysis.
Supplementary information
Supplementary Information
Supplementary Tables 1–9 and Notes 1–5.
Supplementary Data 1
Sample identifiers and raw sequence accession numbers.
Rights and permissions
About this article
Cite this article
Holman, L.E., de Bruyn, M., Creer, S. et al. Animals, protists and bacteria share marine biogeographic patterns. Nat Ecol Evol 5, 738–746 (2021). https://doi.org/10.1038/s41559-021-01439-7
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41559-021-01439-7
This article is cited by
-
eDNA metabarcoding warms up a hotspot of marine biodiversity: revealing underrepresented taxa in visual surveys and historical records from the Gulf of California
Marine Biodiversity (2024)
-
Alfred Russel Wallace’s legacy: an interdisciplinary conception of evolution in space and time
npj Biodiversity (2023)
-
Different Responses of Bacteria and Microeukaryote to Assembly Processes and Co-occurrence Pattern in the Coastal Upwelling
Microbial Ecology (2023)
-
Applying convolutional neural networks to speed up environmental DNA annotation in a highly diverse ecosystem
Scientific Reports (2022)