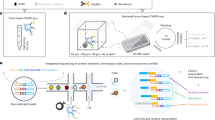
Abstract
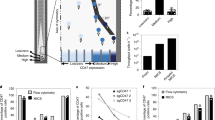
The identification of genetic regulators of cell secretions is challenging because it requires the sorting of a large number of cells according to their secretion patterns. Here we report the development and applicability of a high-throughput microfluidic method for the analysis of the secretion levels of large populations of immune cells. The method is linked with a kinome-wide loss-of-function CRISPR screen, immunomagnetically sorting the cells according to their secretion levels, and the sequencing of their genomes to identify key genetic modifiers of cell secretion. We used the method, which we validated against flow cytometry for cytokines secreted from primary mouse CD4+ (cluster of differentiation 4-positive) T cells, to discover a subgroup of highly co-expressed kinase-coding genes that regulate interferon-gamma secretion by these cells. We validated the function of the kinases identified using RNA interference, CRISPR knockouts and kinase inhibitors and confirmed the druggability of selected kinases via the administration of a kinase inhibitor in an animal model of colitis. The technique may facilitate the discovery of regulatory mechanisms for immune-cell activation and of therapeutic targets for autoimmune diseases.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 digital issues and online access to articles
$99.00 per year
only $8.25 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
Data availability
The sgRNA-sequencing data are available from figshare via the identifier https://doi.org/10.6084/m9.figshare.24151401. The raw and analysed datasets generated during the study are available for research purposes from the corresponding author on reasonable request. Source data are provided with this paper.
References
Satija, R. & Shalek, A. K. Heterogeneity in immune responses: from populations to single cells. Trends Immunol. 35, 219–229 (2014).
Shalek, A. K. et al. Single-cell RNA-seq reveals dynamic paracrine control of cellular variation. Nature 510, 363–369 (2014).
Ma, C. et al. A clinical microchip for evaluation of single immune cells reveals high functional heterogeneity in phenotypically similar T cells. Nat. Med. 17, 738–743 (2011).
Behar, M., Barken, D., Werner, S. L. & Hoffmann, A. The dynamics of signaling as a pharmacological target. Cell 155, 448–461 (2013).
Li, Y. et al. Microfluidics cell loading-dock system: ordered cellular array for dynamic lymphocyte-communication study. Adv. Biosyst. 1, e1700085 (2017).
Xue, Q. et al. Analysis of single-cell cytokine secretion reveals a role for paracrine signaling in coordinating macrophage responses to TLR4 stimulation. Sci. Signal. 8, ra59 (2015).
Lovelace, P. & Maecker, H. T. Multiparameter intracellular cytokine staining. Methods Mol. Biol. 699, 165–178 (2011).
Czerkinsky, C. C., Nilsson, L. A., Nygren, H., Ouchterlony, O. & Tarkowski, A. A solid-phase enzyme-linked immunospot (ELISPOT) assay for enumeration of specific antibody-secreting cells. J. Immunol. Methods 65, 109–121 (1983).
Ahlborg, N. & Axelsson, B. Dual- and triple-color fluorospot. Methods Mol. Biol. 792, 77–85 (2012).
Norman, M. et al. Ultrasensitive high-resolution profiling of early seroconversion in patients with COVID-19. Nat. Biomed. Eng. 4, 1180–1187 (2020).
Rissin, D. M. et al. Single-molecule enzyme-linked immunosorbent assay detects serum proteins at subfemtomolar concentrations. Nat. Biotechnol. 28, 595–599 (2010).
Love, J. C., Ronan, J. L., Grotenbreg, G. M., van der Veen, A. G. & Ploegh, H. L. A microengraving method for rapid selection of single cells producing antigen-specific antibodies. Nat. Biotechnol. 24, 703–707 (2006).
Jin, A. et al. A rapid and efficient single-cell manipulation method for screening antigen-specific antibody-secreting cells from human peripheral blood. Nat. Med. 15, 1088–1092 (2009).
Zhou, Y. et al. Evaluation of single-cell cytokine secretion and cell–cell interactions with a hierarchical loading microwell chip. Cell Rep. 31, 107574 (2020).
Lu, Y. et al. Highly multiplexed profiling of single-cell effector functions reveals deep functional heterogeneity in response to pathogenic ligands. Proc. Natl Acad. Sci. USA 112, E607–E615 (2015).
Chen, Z. et al. Multiplexed, sequential secretion analysis of the same single cells reveals distinct effector response dynamics dependent on the initial basal state. Adv. Sci. 6, 1801361 (2019).
Turcanu, V. & Williams, N. A. Cell identification and isolation on the basis of cytokine secretion: a novel tool for investigating immune responses. Nat. Med. 7, 373–376 (2001).
Eyer, K. et al. Single-cell deep phenotyping of IgG-secreting cells for high-resolution immune monitoring. Nat. Biotechnol. 35, 977–982 (2017).
Mazutis, L. et al. Single-cell analysis and sorting using droplet-based microfluidics. Nat. Protoc. 8, 870–891 (2013).
Lee, S., de Rutte, J., Dimatteo, R., Koo, D. & Di Carlo, D. Scalable fabrication and use of 3D structured microparticles spatially functionalized with biomolecules. ACS Nano 16, 38–49 (2022).
Segaliny, A. I. et al. Functional TCR T cell screening using single-cell droplet microfluidics. Lab Chip 18, 3733–3749 (2018).
Binek, A. et al. Flow cytometry has a significant impact on the cellular metabolome. J. Proteome Res. 18, 169–181 (2019).
Torres, A. J., Hill, A. S. & Love, J. C. Nanowell-based immunoassays for measuring single-cell secretion: characterization of transport and surface binding. Anal. Chem. 86, 11562–11569 (2014).
Chiou, P. Y., Ohta, A. T. & Wu, M. C. Massively parallel manipulation of single cells and microparticles using optical images. Nature 436, 370–372 (2005).
Le, K. et al. A novel mammalian cell line development platform utilizing nanofluidics and optoelectro positioning technology. Biotechnol. Prog. 34, 1438–1446 (2018).
Fu, Z. et al. Highly selective cleavage of cytokines and chemokines by the human mast cell chymase and neutrophil cathepsin G. J. Immunol. 198, 1474–1483 (2017).
Zhao, W. et al. Cell-surface sensors for real-time probing of cellular environments. Nat. Nanotechnol. 6, 524–531 (2011).
Manz, R., Assenmacher, M., Pflüger, E., Miltenyi, S. & Radbruch, A. Analysis and sorting of live cells according to secreted molecules, relocated to a cell-surface affinity matrix. Proc. Natl Acad. Sci. USA 92, 1921–1925 (1995).
Kida, A. et al. Cell surface-fluorescence immunosorbent assay for real-time detection of hybridomas with efficient antibody secretion at the single-cell level. Anal. Chem. 85, 1753–1759 (2013).
Brosterhus, H. et al. Enrichment and detection of live antigen-specific CD4(+) and CD8(+) T cells based on cytokine secretion. Eur. J. Immunol. 29, 4053–4059 (1999).
Armbrecht, L. et al. Quantification of protein secretion from circulating tumor cells in microfluidic chambers. Adv. Sci. 7, 1903237 (2020).
van Neel, T. L., Berry, S. B., Berthier, E. & Theberge, A. B. Localized cell-surface sampling of a secreted factor using cell-targeting beads. Anal. Chem. 92, 13634–13640 (2020).
Ferguson, F. M. & Gray, N. S. Kinase inhibitors: the road ahead. Nat. Rev. Drug Discov. 17, 353–377 (2018).
Poudineh, M. et al. Tracking the dynamics of circulating tumour cell phenotypes using nanoparticle-mediated magnetic ranking. Nat. Nanotechnol. 12, 274–281 (2017).
Labib, M. et al. Tracking the expression of therapeutic protein targets in rare cells by antibody-mediated nanoparticle labelling and magnetic sorting. Nat. Biomed. Eng. 5, 41–52 (2021).
Labib, M. et al. Single-cell mRNA cytometry via sequence-specific nanoparticle clustering and trapping. Nat. Chem. 10, 489–495 (2018).
Barrat, F. J., Crow, M. K. & Ivashkiv, L. B. Interferon target-gene expression and epigenomic signatures in health and disease. Nat. Immunol. 20, 1574–1583 (2019).
Langer, V. et al. IFN-γ drives inflammatory bowel disease pathogenesis through VE-cadherin-directed vascular barrier disruption. J. Clin. Invest. 129, 4691–4707 (2019).
Takashima, S. et al. T cell-derived interferon-γ programs stem cell death in immune-mediated intestinal damage. Sci. Immunol. 4, eaay8556 (2019).
Han, Q. et al. Polyfunctional responses by human T cells result from sequential release of cytokines. Proc. Natl Acad. Sci. USA 109, 1607–1612 (2012).
Imam, T., Park, S., Kaplan, M. H. & Olson, M. R. Effector T helper cell subsets in inflammatory bowel diseases. Front. Immunol. 9, 1212 (2018).
Neurath, M. F. Cytokines in inflammatory bowel disease. Nat. Rev. Immunol. 14, 329–342 (2014).
Haep, L. et al. Interferon gamma counteracts the angiogenic switch and induces vascular permeability in dextran sulfate sodium colitis in mice. Inflamm. Bowel Dis. 21, 2360–2371 (2015).
Gardet, A. et al. LRRK2 is involved in the IFN-gamma response and host response to pathogens. J. Immunol. 185, 5577–5585 (2010).
Wang, F. et al. Interferon-gamma and tumor necrosis factor-alpha synergize to induce intestinal epithelial barrier dysfunction by up-regulating myosin light chain kinase expression. Am. J. Pathol. 166, 409–419 (2005).
Colic, M. et al. Identifying chemogenetic interactions from CRISPR screens with drugZ. Genome Med. 11, 52 (2019).
Mair, B. et al. High-throughput genome-wide phenotypic screening via immunomagnetic cell sorting. Nat. Biomed. Eng. 3, 796–805 (2019).
Fan, F. et al. Pharmacological targeting of kinases MST1 and MST2 augments tissue repair and regeneration. Sci. Transl. Med. 8, 352ra108 (2016).
Elkins, J. M. et al. Comprehensive characterization of the published kinase inhibitor set. Nat. Biotechnol. 34, 95–103 (2016).
Asshoff, M. et al. Momelotinib inhibits ACVR1/ALK2, decreases hepcidin production, and ameliorates anemia of chronic disease in rodents. Blood 129, 1823–1830 (2017).
Rix, U. et al. A comprehensive target selectivity survey of the BCR-ABL kinase inhibitor INNO-406 by kinase profiling and chemical proteomics in chronic myeloid leukemia cells. Leukemia 24, 44–50 (2010).
Yamamoto, N., Honma, M. & Suzuki, H. Off-target serine/threonine kinase 10 inhibition by erlotinib enhances lymphocytic activity leading to severe skin disorders. Mol. Pharmacol. 80, 466–475 (2011).
Egger, B. et al. Characterisation of acute murine dextran sodium sulphate colitis: cytokine profile and dose dependency. Digestion 62, 240–248 (2000).
Mashimo, H., Wu, D. C., Podolsky, D. K. & Fishman, M. C. Impaired defense of intestinal mucosa in mice lacking intestinal trefoil factor. Science 274, 262–265 (1996).
Ito, R. et al. Interferon-gamma is causatively involved in experimental inflammatory bowel disease in mice. Clin. Exp. Immunol. 146, 330–338 (2006).
Chiba, H., Kojima, T., Osanai, M. & Sawada, N. The significance of interferon-gamma-triggered internalization of tight-junction proteins in inflammatory bowel disease. Sci. STKE 2006, pe1 (2006).
Biton, M. et al. T helper cell cytokines modulate intestinal stem cell renewal and differentiation. Cell 175, 1307–1320.e1322 (2018).
Parnas, O. et al. A genome-wide CRISPR screen in primary immune cells to dissect regulatory networks. Cell 162, 675–686 (2015).
Dong, M. B. et al. Systematic immunotherapy target discovery using genome-scale in vivo CRISPR screens in CD8 T cells. Cell 178, 1189–1204.e1123 (2019).
Henriksson, J. et al. Genome-wide CRISPR screens in T helper cells reveal pervasive crosstalk between activation and differentiation. Cell 176, 882–896.e818 (2019).
Schmidt, R. et al. CRISPR activation and interference screens decode stimulation responses in primary human T cells. Science 375, eabj4008 (2022).
Acknowledgements
The research reported in this publication was supported by the Canadian Institutes of Health Research (grant number FDN-148415), the Natural Sciences and Engineering Research Council of Canada (grant number 2016-06090), the Province of Ontario through the Ministry of Research, Innovation and Science (grant number RE05-009) and the National Cancer Institute of the National Institutes of Health (grant number 1R33CA204574). The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health or the other funding agencies.
Author information
Authors and Affiliations
Contributions
M.L., S.A., E.H.S. and S.O.K. conceived and designed the experiments; M.L., Z.W., Y.K., S.L. and H.Y. performed the experiments and analysed the data. A.A. performed the CRISPR KO experiments. P-Y.L. cultured and maintained the cells. M.L., Z.W., S.A., E.H.S. and S.O.K. contributed to the preparation and editing of the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Nature Biomedical Engineering thanks the anonymous reviewer(s) for their contribution to the peer review of this work.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Main Supplementary Information
Supplementary methods, table and figures.
Source data
Source data for Fig. 3
Source data.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Labib, M., Wang, Z., Kim, Y. et al. Identification of druggable regulators of cell secretion via a kinome-wide screen and high-throughput immunomagnetic cell sorting. Nat. Biomed. Eng 8, 263–277 (2024). https://doi.org/10.1038/s41551-023-01135-w
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41551-023-01135-w