Abstract
Phosphatidylinositol 4,5-bisphosphate (PI(4,5)P2) is a low-abundance membrane lipid essential for plasma membrane function1,2. In plants, mutations in phosphatidylinositol 4-phosphate (PI4P) 5-kinases (PIP5K) suggest that PI(4,5)P2 production is involved in development, immunity and reproduction3,4,5. However, phospholipid synthesis is highly intricate6. It is thus likely that steady-state depletion of PI(4,5)P2 triggers confounding indirect effects. Furthermore, inducible tools available in plants allow PI(4,5)P2 to increase7,8,9 but not decrease, and no PIP5K inhibitors are available. Here, we introduce iDePP (inducible depletion of PI(4,5)P2 in plants), a system for the inducible and tunable depletion of PI(4,5)P2 in plants in less than three hours. Using this strategy, we confirm that PI(4,5)P2 is critical for various aspects of plant development, including root growth, root-hair elongation and organ initiation. We show that PI(4,5)P2 is required to recruit various endocytic proteins, including AP2-µ, to the plasma membrane, and thus to regulate clathrin-mediated endocytosis. Finally, we find that inducible PI(4,5)P2 perturbation impacts the dynamics of the actin cytoskeleton as well as microtubule anisotropy. Together, we propose that iDePP is a simple and efficient genetic tool to test the importance of PI(4,5)P2 in given cellular or developmental responses, and also to evaluate the importance of this lipid in protein localization.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 digital issues and online access to articles
$119.00 per year
only $9.92 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
Data availability
The Arabidopsis lines and plasmids generated in this study are available from the corresponding authors upon request and the transgenic lines are also available at NASC as NASC stocks N2110785–N2110797. Source data are provided with this paper.
References
Bigay, J. & Antonny, B. Curvature, lipid packing, and electrostatics of membrane organelles: defining cellular territories in determining specificity. Dev. Cell 23, 886–895 (2012).
Balla, T. Phosphoinositides: tiny lipids with giant impact on cell regulation. Physiol. Rev. 93, 1019–1137 (2013).
Noack, L. C. & Jaillais, Y. Precision targeting by phosphoinositides: how PIs direct endomembrane trafficking in plants. Curr. Opin. Plant Biol. 40, 22–33 (2017).
Noack, L. C. & Jaillais, Y. Functions of anionic lipids in plants. Annu. Rev. Plant Biol. 71, 71–102 (2020).
Heilmann, I. Phosphoinositide signaling in plant development. Development 143, 2044–2055 (2016).
Colin, L. A. & Jaillais, Y. Phospholipids across scales: lipid patterns and plant development. Curr. Opin. Plant Biol. 53, 1–9 (2019).
Gujas, B. et al. Perturbing phosphoinositide homeostasis oppositely affects vascular differentiation in Arabidopsis thaliana roots. Development 144, 3578–3589 (2017).
Ischebeck, T., Stenzel, I. & Heilmann, I. Type B phosphatidylinositol-4-phosphate 5-kinases mediate Arabidopsis and Nicotiana tabacum pollen tube growth by regulating apical pectin secretion. Plant Cell 20, 3312–3330 (2008).
Barbosa, I. C. et al. Phospholipid composition and a polybasic motif determine D6 PROTEIN KINASE polar association with the plasma membrane and tropic responses. Development 143, 4687–4700 (2016).
Simon, M. L. et al. A multi-colour/multi-affinity marker set to visualize phosphoinositide dynamics in Arabidopsis. Plant J. 77, 322–337 (2014).
Simon, M. L. et al. A PtdIns(4)P-driven electrostatic field controls cell membrane identity and signalling in plants. Nat. Plants 2, 16089 (2016).
Platre, M. P. et al. A combinatorial lipid code shapes the electrostatic landscape of plant endomembranes. Dev. Cell 45, 465–480 (2018).
Zhao, Y. et al. Phosphoinositides regulate clathrin-dependent endocytosis at the tip of pollen tubes in Arabidopsis and tobacco. Plant Cell 22, 4031–4044 (2010).
Dejonghe, W. et al. Disruption of endocytosis through chemical inhibition of clathrin heavy chain function. Nat. Chem. Biol. https://doi.org/10.1038/s41589-019-0262-1 (2019).
Yperman, K. et al. Molecular architecture of the endocytic TPLATE complex. Sci. Adv. 7, eabe7999 (2021).
Mei, Y., Jia, W. J., Chu, Y. J. & Xue, H. W. Arabidopsis phosphatidylinositol monophosphate 5-kinase 2 is involved in root gravitropism through regulation of polar auxin transport by affecting the cycling of PIN proteins. Cell Res. 22, 581–597 (2012).
Ischebeck, T. et al. Phosphatidylinositol 4,5-bisphosphate influences PIN polarization by controlling clathrin-mediated membrane trafficking in Arabidopsis. Plant Cell 25, 4894–4911 (2013).
He, K. et al. Dynamics of phosphoinositide conversion in clathrin-mediated endocytic traffic. Nature 552, 410–414 (2017).
Gadeyne, A. et al. The TPLATE adaptor complex drives clathrin-mediated endocytosis in plants. Cell 156, 691–704 (2014).
Narasimhan, M. et al. Evolutionarily unique mechanistic framework of clathrin-mediated endocytosis in plants. eLife https://doi.org/10.7554/eLife.52067 (2020).
Raucher, D. et al. Phosphatidylinositol 4,5-bisphosphate functions as a second messenger that regulates cytoskeleton-plasma membrane adhesion. Cell 100, 221–228 (2000).
Zhang, Q. et al. Phosphatidic acid regulates microtubule organization by interacting with MAP65-1 in response to salt stress in Arabidopsis. Plant Cell 24, 4555–4576 (2012).
Munnik, T. & Nielsen, E. Green light for polyphosphoinositide signals in plants. Curr. Opin. Plant Biol. 14, 489–497 (2011).
Gungabissoon, R. A., Jiang, C.-J., Drøbak, B. K., Maciver, S. K. & Hussey, P. J. Interaction of maize actin-depolymerising factor with actin and phosphoinositides and its inhibition of plant phospholipase C. Plant J. 16, 689–696 (1998).
Guo, S., Stolz, L. E., Lemrow, S. M. & York, J. D. SAC1-like domains of yeast SAC1, INP52, and INP53 and of human synaptojanin encode polyphosphoinositide phosphatases. J. Biol. Chem. 274, 12990–12995 (1999).
Huang, S., Blanchoin, L., Kovar, D. R. & Staiger, C. J. Arabidopsis capping protein (AtCP) is a heterodimer that regulates assembly at the barbed ends of actin filaments. J. Biol. Chem. 278, 44832–44842 (2003).
Staiger, C. J., Gibbon, B. C., Kovar, D. R. & Zonia, L. E. Profilin and actin-depolymerizing factor: modulators of actin organization in plants. Trends Plant Sci. 2, 275–281 (1997).
Shibasaki, Y. et al. Massive actin polymerization induced by phosphatidylinositol-4-phosphate 5-kinase in vivo. J. Biol. Chem. 272, 7578–7581 (1997).
Vaughn, L. et al. in The Plant Cytoskeleton. Advances in Plant Biology Vol. 2 (ed. Liu, B.) 307–326 (Springer, 2011).
Balla, T. Imaging and manipulating phosphoinositides in living cells. J. Physiol. 582, 927–937 (2007).
Lorenzo-Orts, L., Couto, D. & Hothorn, M. Identity and functions of inorganic and inositol polyphosphates in plants. New Phytol. 225, 637–652 (2020).
Marques-Bueno, M. M. et al. A versatile Multisite Gateway-compatible promoter and transgenic line collection for cell type-specific functional genomics in Arabidopsis. Plant J. 85, 320–333 (2016).
Saile, S. C. et al. Coiled-coil and RPW8-type immune receptors function at the plasma membrane in a phospholipid dependent manner. Preprint at bioRxiv https://doi.org/10.1101/2020.11.18.388520 (2020).
Johnson, C. M., Chichili, G. R. & Rodgers, W. Compartmentalization of phosphatidylinositol 4,5-bisphosphate signaling evidenced using targeted phosphatases. J. Biol. Chem. 283, 29920–29928 (2008).
Perera, I. Y., Love, J., Heilmann, I., Thompson, W. F. & Boss, W. F. Up-regulation of phosphoinositide metabolism in tobacco cells constitutively expressing the human type I inositol polyphosphate 5-phosphatase. Plant Physiol. 129, 1795–1806 (2002).
Christie, J. M. et al. Plant UVR8 photoreceptor senses UV-B by tryptophan-mediated disruption of cross-dimer salt bridges. Science 335, 1492–1496 (2012).
Jaillais, Y. et al. Tyrosine phosphorylation controls brassinosteroid receptor activation by triggering membrane release of its kinase inhibitor. Genes Dev. 25, 232–237 (2011).
Karimi, M., Depicker, A. & Hilson, P. Recombinational cloning with plant gateway vectors. Plant Physiol. 145, 1144–1154 (2007).
Konopka, C. A., Backues, S. K. & Bednarek, S. Y. Dynamics of Arabidopsis dynamin-related protein 1C and a clathrin light chain at the plasma membrane. Plant Cell 20, 1363–1380 (2008).
Yamaoka, S. et al. Identification and dynamics of Arabidopsis adaptor protein-2 complex and its involvement in floral organ development. Plant Cell 25, 2958–2969 (2013).
Nagel, M.-K. et al. Arabidopsis SH3P2 is an ubiquitin-binding protein that functions together with ESCRT-I and the deubiquitylating enzyme AMSH3. Proc.Natl Acad. Sci. USA 114, E7197–E7204 (2017).
Grandjean, O. et al. In vivo analysis of cell division, cell growth, and differentiation at the shoot apical meristem in Arabidopsis. Plant Cell 16, 74–87 (2004).
Stanislas, T., Hamant, O. & Traas, J. In-vivo analysis of morphogenesis in plants. Methods Cell Biol. 139, 203–223 (2017).
Doumane, M., Lionnet, C., Bayle, V., Jaillais, Y. & Caillaud, M.-C. Automated tracking of root for confocal time-lapse imaging of cellular processes. Bio Protoc. https://doi.org/10.21769/BioProtoc.2245 (2017).
Zarza, X. et al. Lipid kinases PIP5K7 and PIP5K9 are required for polyamine-triggered K(+) efflux in Arabidopsis roots. Plant J. 104, 416–432 (2020).
Munnik, T. & Zarza, X. in Plant Lipid Signaling Protocols Vol. 1009 (eds Munnik, T. & Heilmann, I.) 3–15 (Humana Press, 2013).
Wickham, H. ggplot2: Elegant Graphics for Data Analysis (Springer, 2016).
Bates, D., Mächler, M., Bolker, B. & Walker, S. Fitting linear mixed-effects models using lme4. Preprint at https://arxiv.org/abs/1406.5823 (2014).
Fox, J., & Weisberg, S. (2011). Multivariate Linear Models in R. An R Companion to Applied Regression. (SAGE, 2011).
Hothorn, T., Bretz, F., Westfall, P. & Heiberger, R. M. Multcomp: simultaneous inference for general linear hypotheses v.1.0-3 (2008).
Lenth, R. V. Least-squares means: the R package lsmeans. J. Stat. Softw. 69, 1–33 (2016).
Acknowledgements
We are grateful to the SiCE group (RDP), in particular T. Gaude and V. Bayle (RDP, Lyon), and to Y. Boutté (Laboratoire de Biogenèse Membranaire), D. Van Damme (Vlaams Instituut voor Biotechnologie (VIB)), T. Stanislas (ZMBP), F. Besnard and N. Doll (RDP), for comments and discussion. We also thank former interns L. Courgeon (ENS de Lyon) and A. Bauer (ENS de Lyon), who performed preliminary experiments during the early steps of this project and helped with plant labour, and K. Grünwald. We thank J. Berger, P. Bolland and A. Lacroix from our plant facility. We acknowledge the contribution of Structure Fédérative de Recherche Biosciences (UMS3444/CNRS, US8/Inserm, ENS de Lyon, Université Claude Bernard Lyon 1) facilities: C. Lionet, E. Chatre and J. Brocard at the LBI-PLATIM-MICROSCOPY for assistance with imaging. We are also grateful to E. Russinova (VIB) for kindly providing us with ES9-17; S. Bednarek for sharing markers and for discussion and M. Uyttewaal (INRAE) for the MBD–GFP marker. We thank E. Isono (University of Konstanz) for sharing SH3P2–GFP’s transgenic line with us. We also thank R. Boisseau (OBEE department, University of Montana) for help with the statistical analysis. This work was supported by the European Research Council (no. 3363360-APPL under FP/2007–2013, Y.J.; EU-FET 828753, T.M.), the Netherlands Organisation for Scientific Research (NWO 867.15.020; 711.017.005; T.M.), Austrian Academy of Science through the Gregor Mendel Institute (Y.B.), French National Research Agency ANR (caLIPSO; ANR-18-CE13-0025-02; Y.J.), ANR JC/JC JUNIOR INVESTIGATOR GRANT (INTERPLAY; ANR-16-CE13-0021; M.-C.C., A.F.) and a SEED FUND ENS LYON-2016 (M.-C.C.).
Author information
Authors and Affiliations
Contributions
M.D. performed the experiments, analysed the data (including statistics) and wrote the paper; A.L. performed the experiments and analysed the data (including statistics); L.C. performed the experiments and analysed the data; A.F. performed the experiments; F.D.S. performed the biochemical quantification of the PIP2 and analysed the data; J.B. performed the biochemistry experiment; O.H. supervised the work and helped with writing the paper; Y.B. supervised the biochemistry work and analysed the data; T.M. supervised the biochemical quantification of PIP2 work and analysed the data; Y.J. supervised the work and wrote the paper; and M.-C.C. performed the experiments and analysed the data, supervised the work and wrote the paper.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Peer review information Nature Plants thanks Takashi Aoyama, Bo Liu and the other, anonymous, reviewer(s) for their contribution to the peer review of this work.
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data
Extended Data Fig. 1 Drosophila dOCRL displays a PI(4,5)P2 phosphatase activity in vitro.
a, Alignment of (i) Homo sapiens OCRL (HsOCRL or OCRL1) truncation (HsOCRL234-539) successfully used in optogenetic systems in mammalian cells, and (ii) of Drosophila melanogaster dOCRL. We were concerned that this domain, that underwent natural selection in an endotherm organism with set-point temperature of 37 °C, would be less active in Arabidopsis thaliana, a plant of temperate climate which we usually grow at 21 °C. Therefore, we decided to use the phosphatase domain of dOCRL, Drosophila melanogaster homolog, as Drosophila are ectotherm and live at ~ 20 °C. Alignment of OCRL1234-539 and dOCRL allowed us to identify dOCRL168-509 as the homolog region of OCRL1 generally used in optogenetic systems. b, Coomassie blue staining monitoring purification of recombinant dOCRL phosphatase domain (His-SUMO-dOCRL168-509-His) from Escherichia coli. We first assessed dOCRL activity, which has not been reported so far, even though in vivo data suggest a conserved PI(4,5)P2 5-phosphatase activity. We ordered a synthetic gene corresponding to dOCRL168-509 and codon optimized for expression in Arabidopsis. We cloned a recombinant dOCRL protein (His-SUMO-dOCRL168-509-His) tagged with two 6-histidine tags and a SUMO tag, making it suitable for expression in Escherichia coli and purification. We purified His-SUMO-dOCRL168-509-His using a cobalt resin-based affinity chromatography and protein concentration IN: input, lysate of bacteria induced for the expression of His-SUMO- dOCRL168-509-His; FL: flow through; W: wash; E1 to E3: elution fraction 1 to 3; C1 and C2: concentrated fractions from pulled E1, E2 and E3. C1 and C2 were obtained separately on different days. His-SUMO-dOCRL168-509-His expected molecular weight is 54.8 kD; 3 Replicates (c) Malachite green phosphatase assay on His-SUMO-dOCRL169-509-His using short chain water-soluble phosphoinositides. We subjected the purified protein fractions C1 and C2 to Malachite Green Phosphatase assays, that detects phosphate released by dephosphorylations, in presence of short chain (diC:8) water-soluble phosphoinositides. Note that we only assessed dOCRL168-509 activity toward phosphoinositides reported in plant cells. Replicate 1 correspond to C1 fraction. Replicate 2 to C2 fractions. Mock: no phosphoinositide added.
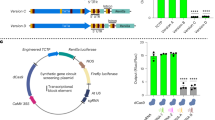
Extended Data Fig. 2 Rational behind the design of the iDePP system.
a, Schematic drawing depicting constructs used to genetically-induced PI(4,5)P2 pool modification at the PM. After dex treatment, MAP-mCh-dOCRL is anchored at the plasma membrane and dephosphorylates PI(4,5)P2, causing the PI(4,5)P2 biosensor to be released into the cytosplasm. b, Controls used in this study includes 3xmCh and a catalytic dead version of dOCRL. After dex treatment, both MAP-3xmCh and MAP-mCh-dOCRLdead are anchored at the PM, but do not affect the pool of PI(4,5)P2. The PI(4,5)P2 biosensor is found at the PM. To build the iDePP system, we had to engineer a synthetic enzyme, which consists of the isolated catalytic domain of a 5-phosphatase (which if expressed on its own would be soluble in the cytosol and likely inactive) and then target this module to the plasma membrane, for specific depletion of PI(4,5)P2). Details can be found in the Methods section.
Extended Data Fig. 3 Successful inducible expression and subcellular targeting of MAP-mCH-dOCRL and negative controls.
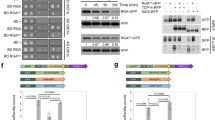
a, Inducible system used in this study. We performed site-directed D468G mutation in dOCRL catalytic domain, similarly to D523G mutation already described in human OCRL1 that abolishes its phosphatase activity. We could therefore use MAP-mCH-dOCRL168-509D468G (hereafter named MAP-mCH-dOCRLdead) as negative control, together with a MAP-3xmCH recombinant protein. UBQ10 promoter is responsible for mild/strong and ubiquitous expression of GVG (GAL4 VP16 GR) synthetic gene. GR domain binds dexamethasone (dex) and subsequently induces GVG homodimerization and nuclear import. There, GAL4 domain binds UAS DNA elements and VP16 strongly activates the expression of downstream gene: MAP-mCH-dOCRL168-509, or MAP-mCH-dOCRL168-509D468G (MAP-mCH-dOCRLdead), or MAP-3xmCH. MAP is myristoylation and palmytoylation sequence, responsible for plasma membrane targeting. The mCH corresponds to monomeric CHERRY fluorescent protein. dOCRLD468G168-509 is an inactive phosphatase domain (later on called dOCRLdead). 3xmCH correspond to three fused mCHERRY. b, We addressed the timing of expression and the localization of each construct in Arabidopsis lines stably transformed with UBQ10pro:GVG:MAP-mCH-dOCRL, or UBQ10pro:GVG:MAP-mCH-dOCRLdead, or UBQ10pro:GVG:MAP-3xmCH. In root meristematic epidermal cells, without dex treatment, none of the recombinant-proteins were detected using confocal microscopy. A 16 h treatment with 0.5 µM dex led to the detection of mCH fluorescence, indicating that induction of the genetic construct had occurred. However, we observed a mosaic induction and a significant number of roots had no cells expressing the fluorescent reporter to detectable levels. To overcome these issues, we optimized the treatment to a 16 h induction with 5 µM dex. Using this set up, we robustly observed red fluorescence using confocal microscopy where MAP-mCH-dOCRL, MAP-mCH-dOCRLdead and MAP-3xmCH. Each of the synthetic protein was efficiently targeted to membranes, including the plasma membrane, where PI(4,5)P2 accumulates, and intracellular compartments. Therefore, a 16 h 5 µM dex treatment is sufficient for an effective expression of dOCRL in Arabidopsis stable transgenic lines. For all three inducible constructs we monitored mCH fluorescence without dex treatment or after a 16h-treatment with either 0.5 µM or 5 µM dex. Signal intensity is color-coded (green fire blue scale); 2 Replicates. Scale bars, 20 µm.
Extended Data Fig. 4 Biochemical quantification of the PIP2, PIP and PA after iDePP induction.
The 32P-PIP levels in iDePP lines and controls ± dex of 3 independent replicates. a-c, Seedlings were labelled with 32Pi and incubated ± dex O/N (16–20 hrs). Each sample contained the lipid extract of three seedlings, of which 1/5th was analyzed by TLC and quantified by phosphoimaging, of which 32P-PIP2 (a), PIP (b) and PA (c) were calculated as percentage of total 32P-lipids. d-f, Time-course analysis of 32P-PIP2 (d), PIP (e) and PA (f) levels in iDePP seedlings ± dex in MAP-mCH-dOCRL line. Seedlings of MAP-mCH-dOCRL line were labelled for 20 hrs with 32Pi and co-incubated with or without dex for the times indicated (0–20 hrs). Each sample contained the lipid extract of three seedlings of which 1/5th was analyzed by TLC and quantified by phosphoimaging. First, the mean of control on all time points was calculated. The graphs represent the calculated mean of the sample reported to the mean of the control. In the plots, middle horizontal bars represent the median, while the bottom and top of each box represent the 25th and 75th percentiles, respectively. At most, the whiskers extend to 1.5 times the interquartile range, excluding data beyond. For range of value under 1,5 IQR, whiskers represent the range of maximum and minimum values. All statistical tests were two-sided.
Extended Data Fig. 5 Graphic representation of the fluorescent intensity observed for the different biosensors, used for the quantification of the dissociation index.
Orange lines represent the means. In the plots, middle horizontal bars represent the median, while the bottom and top of each box represent the 25th and 75th percentiles, respectively. At most, the whiskers extend to 1.5 times the interquartile range, excluding data beyond. For range of value under 1,5 IQR, whiskers represent the range of maximum and minimum values.
Extended Data Fig. 6 iDePP enables efficient depletion of PI(4,5)P2 in various tissues and organs.
a-c, Subcellular localization of mCIT-2xPHPLC and either MAP-mCH-dOCRL (a), or MAP-3xmCH; N = 12 (b), MAP-mCH-dOCRLdead; N = 12 (c), in cotyledon epidermal cells; N = 6. Fluorescence intensity is color-coded (green fire blue scale), cytoplasmic strands are indicated by the arrows. Scale bars, 10 µm. d-g, Induction of the iDePP system in the shoot apical meristem of Arabidopsis; N = 15 (Scale bar, 20 μm in (a-b), 10 μm in (c-d). Asterisks represent the position of the nucleus in the cell. (h) and (i) Subcellular localization of MAP-mCH-dOCRL (left panel) and mCIT-2xPHPLC (right panel) in a naked shoot meristem from an NPA-treated seedling, as viewed from the top; N = 4. In the absence of dex induction, mCIT-2xPHPLC was localized at the plasma membrane throughout the meristem, including both the central and peripheral zones (h, right panel). By contrast, upon induction of MAP-mCH-dOCRL expression, mCIT-2xPHPLC was no longer sharply localizing at the cell edge (i, right panel). Instead, mCIT-2xPHPLC accumulated in the cytosol, while still being excluded from the round central nuclei (i, right panel). Note that after dex induction, MAP-mCH-dOCRL was strongly expressed in the peripheral zone, and was only weekly expressed in the central zone (i, left panel). However, although its expression was weak in the central zone it appeared to be sufficient to impact mCIT-2xPHPLC localization. Scale bar, 20 μm. (j) and (k) Effect of PI(4,5)P2 depletion on organ initiation in NPA-treated meristems; N = 8. Altogether, these observations suggest that PI(4,5)P2 depletion seems to affect organ initiation in NPA-treated meristems. Scale bars, 700 µm.
Extended Data Fig. 7 Quantification of the growth phenotype obtained using the iDePP system.
Quantification over time (at 3, 5- and 7-days post germination) of the root length (cm) in transgenic lines MAP-mCH-dOCRL (a) and MAP-mCH-dOCRLdead (b) grown on MS ½ plate with 5 µM of dex. Details for the statistical analysis can be found in the Methods. In the plots, middle horizontal bars represent the median, while the bottom and top of each box represent the 25th and 75th percentiles, respectively. At most, the whiskers extend to 1.5 times the interquartile range, excluding data beyond. For range of value under 1,5 IQR, whiskers represent the range of maximum and minimum values.
Extended Data Fig. 8 Effects on endocytosis of the depletion of PI(4,5)P2 using the iDePP system.
a,b, Representative confocal images of Arabidopsis root cells coexpressing mCIT-P4MSidM with either MAP-mCH-dOCRL (a) or MAP-3xmCH (b). c,d, Confocal imaging showing the effect of the expression of MP-mCH-dOCRL on CLC2-GFP localization. e, Quantification of the effect of MAP-mCH-dOCRL expression on CLC2-GFP localization. The dissociation index is the ratio of (i) plasma membrane to cytosol fluorescence ratio without dex treatment, (ii) plasma membrane to cytosol fluorescence ratio after dex treatment. Statistical analysis with LMER (Type II Wald χ2 test) and posthoc tests; 3 replicates (f-g) Confocal imaging showing the effect of the expression of MAP-mCH-dOCRL on SH3P2-sGFP line (h) Quantification of the effect of MAP-mCH-dOCRL expression on SH3P2-sGFP localization. Per root, cells showing a delocalization of SH3P2-sGFP were manually counted for untreated and dex treated plants using FIJI ‘multi-point’ tool and compared by a generalized linear model, Poisson family; 3 replicates. In the plots, middle horizontal bars represent the median, while the bottom and top of each box represent the 25th and 75th percentiles, respectively. At most, the whiskers extend to 1.5 times the interquartile range, excluding data beyond. For range of value under 1,5 IQR, whiskers represent the range of maximum and minimum values. All statistical tests were two-sided. Scale bar, 10 μm.
Extended Data Fig. 9 Effects on the cytoskeleton of the depletion of PI(4,5)P2 using the iDePP system.
a, F-actin cytoskeleton labelled by LifeAct-YFPv in cells expressing MAP-mCH-dOCRL or MAP-3xmCH 16h after 5 µM dex; 4 replicates. b, Cortical microtubules labelled by TUA6-GFP, in non-treated cells (no dex) or treated (5 µM dex after 16 hours) to induce MAP-mCH-dOCRL or MAP-3xmCH expression; 3 replicates. All pictures are z-projections; no dex: no treatment; dex: dexamethasone treatment. Scale bars, 10 µm. c, Example of the images used for the quantification of the anisotropy using FibrilTool macro for imageJ. In the plots, middle horizontal bars represent the median, while the bottom and top of each box represent the 25th and 75th percentiles, respectively. At most, the whiskers extend to 1.5 times the interquartile range, excluding data beyond. For range of value under 1,5 IQR, whiskers represent the range of maximum and minimum values. All statistical tests were two-sided.
Supplementary information
Supplementary Video 1
Time-course analysis during dex induction, monitoring mCIT-2xPHPLC PI(4,5)P2 biosensor subcellular localization at the root tip in epidermal cells every five minutes.
Supplementary Video 2
Time-course analysis with dex induction of MAP-mCH-dOCRL (right panel), monitoring mCIT-2xPHPLC PI(4,5)P2 biosensor subcellular localization (left panel) at the root tip in epidermal cells every five minutes.
Supplementary Video 3
Time-course analysis without dex induction of MAP-mCH-dOCRL (right panel), monitoring mCIT-2xPHPLC PI(4,5)P2 biosensor subcellular localization (left panel) at the root tip in epidermal cells every five minutes.
Supplementary Video 4
Time-lapse analysis of the root-hair growth upon induction of PI(4,5)P2 depletion using iDePP. Time-course analysis with dex induction (5 µM) of MAP-mCH-dOCRLdead (left roots) and MAP-mCH-dOCRL (right roots), monitoring root growth and root-hair formation for seven hours (images taken every second).
Supplementary Video 5
Time-lapse analysis of the actin dynamics upon induction of PI(4,5)P2 depletion using iDePP. Time-course analysis over two minutes, monitoring Lifeact-YFPv subcellular localization in root cells expressing MAP-mCH-dOCRL (right) or not expressing MAP-mCH-dOCRL (left). Pictures were taken every three seconds.
Source data
Source Data Fig. 1
Unprocessed blot.
Source Data Fig. 2
Unprocessed blot.
Source Data Extended Data Fig. 1
Unprocessed blot.
Rights and permissions
About this article
Cite this article
Doumane, M., Lebecq, A., Colin, L. et al. Inducible depletion of PI(4,5)P2 by the synthetic iDePP system in Arabidopsis. Nat. Plants 7, 587–597 (2021). https://doi.org/10.1038/s41477-021-00907-z
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41477-021-00907-z
This article is cited by
-
Disarming PI(4,5)P2 in the plasma membrane
Nature Plants (2021)