Abstract
Purpose
Pathogenic variants in SETD1B have been associated with a syndromic neurodevelopmental disorder including intellectual disability, language delay, and seizures. To date, clinical features have been described for 11 patients with (likely) pathogenic SETD1B sequence variants. This study aims to further delineate the spectrum of the SETD1B-related syndrome based on characterizing an expanded patient cohort.
Methods
We perform an in-depth clinical characterization of a cohort of 36 unpublished individuals with SETD1B sequence variants, describing their molecular and phenotypic spectrum. Selected variants were functionally tested using in vitro and genome-wide methylation assays.
Results
Our data present evidence for a loss-of-function mechanism of SETD1B variants, resulting in a core clinical phenotype of global developmental delay, language delay including regression, intellectual disability, autism and other behavioral issues, and variable epilepsy phenotypes. Developmental delay appeared to precede seizure onset, suggesting SETD1B dysfunction impacts physiological neurodevelopment even in the absence of epileptic activity. Males are significantly overrepresented and more severely affected, and we speculate that sex-linked traits could affect susceptibility to penetrance and the clinical spectrum of SETD1B variants.
Conclusion
Insights from this extensive cohort will facilitate the counseling regarding the molecular and phenotypic landscape of newly diagnosed patients with the SETD1B-related syndrome.
Similar content being viewed by others
INTRODUCTION
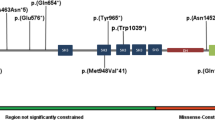
SETD1B encodes a lysine-specific histone methyltransferase that methylates histone H3 at position lysine-4 (H3K4me1, H3K4me2, H3K4me3) as part of a multisubunit complex known as COMPASS [1, 2]. The SETD1B protein consists of 1,966 amino acids and has several (presumed) functional domains (Fig. 1). The N-terminus contains an RNA recognition motif (RRM), whereas the middle region is characterized by two long disordered regions that differ from other homologs [3, 4], a conserved lysine–serine–aspartic acid (LSD) motif [5] and a coiled-coil structure. At the C-terminus, SETD1B harbors a catalytic SET domain crucial for histone methyltransferase activity, bordered proximally by the N-SET domain including a conserved WDR5-interacting (WIN) motif [6], and distally by the post-SET domain. H3K4me3 is enriched at promoter and transcription start sites whereas H3K4me1 and H3K4me2 are enriched at enhancer sites, therefore being associated with active gene transcription and euchromatin [7]. Indeed epigenetic changes have been observed in both animal models and patient material [8,9,10] at promoters and intergenic regions, confirming that SETD1B epigenetically controls gene expression and chromatin state. In addition, SETD1B is constrained for both missense and loss-of-function variants [11].
The RRM (residues 94–182), coiled-coil (CC) (residues 1173–1204), N-SET (residues 1668–1821), SET (residues 1822–1948) and post-SET (residues 1949–1966) domains in respectively magenta, orange, cyan, green and brown, the largely disordered regions (residues 320–682 and residues 1338–1640) in light gray, and the LSD (exon 5, residues 577–583) and WIN (exon 12, residues 1745–1750, within N-SET) motifs both in blue.
Consistent with this, pathogenic variants in SETD1B have been associated with a syndromic intellectual developmental disorder including seizures and language delay (IDDSELD, OMIM 619000). To date, clinical features have been described for 11 affected individuals with (likely) pathogenic SETD1B sequence variants [8, 12,13,14,15]. Individuals with microdeletions encompassing SETD1B have also been described [8, 16,17,18,19]; however, most of these deletions encompass additional genes making phenotypic comparisons challenging. In this study, we further delineate the clinical phenotype associated with SETD1B sequence variants, by describing 36 additional individuals. Comparing these new cases to the published ones provides a comprehensive molecular and clinical characterization of the SETD1B-related syndrome. In addition, using protein modeling, in vitro assays, and genome-wide methylation signatures we investigate the effects of selected variants. Together, this expands the molecular and phenotypic landscape associated with SETD1B variants.
MATERIALS AND METHODS
Cohort inclusion
After identification of three individuals with SETD1B variants at Erasmus MC Clinical Genetics, additional cases were identified using GeneMatcher [20] and the Dutch Datasharing Initiative [21] and via our network of collaborators. Individuals were included based on SETD1B variants detected in research or routine clinical diagnostics. Affected individuals were investigated by their referring physicians.
Next-generation sequencing of affected individuals
Full details are provided in the Supplementary Methods and Supplementary Fig. S1.
Variant classification
SETD1B variants were initially classified as variants of uncertain significance (VUS), likely pathogenic, or pathogenic at the performing laboratory or local referring sites. Literature and public database search identified 30 individuals with SETD1B sequence variants (Supplementary Table S1). Reclassification of SETD1B sequence variants was performed according to American College of Medical Genetics and Genomics/Association for Molecular Pathology (ACMG/AMP) Standards and Guidelines [22] (Supplementary Table S1), using reference sequence NM_001353345. For retrieval of population allele frequencies and in silico predictions Alamut® Visual 2.15 (Feb 2020) was used.
Facial gestalt and severity scoring analysis
To generate a composite facial gestalt, Face2Gene (FDNA Inc., Boston, MA, USA) research application was used (default settings). Details of severity scoring are described in Supplementary Methods.
Structural protein modeling
Sequences were retrieved from Uniprot, SWISS-MODEL [23] to produce homology models; RaptorX [3] for predicting secondary structure and disorder; ConSurf [24] for conservation analysis; and eukaryotic linear motif (ELM [25]) for short linear protein motif assessment. Models were manually inspected, and variants evaluated, using Pymol (pymol.org).
Experimental procedures
For in vitro experiments flag-tagged wild type (kindly provided by David Skalnik, Indiana University [5]) and variant SETD1B protein and HA-tagged ASH2 were overexpressed in HEK293 cells. Protein expression, isolation, western blotting, and immunocytochemistry were performed following standard procedures [26,27,28]. Genome-wide methylation profiles were obtained as described [8]. Details on experimental procedures and statistical analysis are provided in Supplementary Methods.
RESULTS
Molecular spectrum of SETD1B sequence variants
A total of 36 individuals with either heterozygous (n = 32, n = 28 confirmed de novo, n = 1 inherited from affected parent, n = 1 inherited from unaffected parent), compound heterozygous (n = 2, biallelic inheritance from unaffected parents) or homozygous (n = 2, siblings, biallelic inheritance from unaffected parents) SETD1B sequence variants were included in this cohort. Thirty-three variants were detected, of which 2 were recurrent. This includes 8 truncating (n = 6 nonsense, n = 2 frameshift), 1 extension, 1 in-frame inversion, and 23 missense variants (Fig. 1). Fourteen variants were classified as pathogenic, ten as likely pathogenic, and nine as uncertain significance. For individuals with VUS, no alternative candidate disease-causing variant was identified. In literature, 26 additional (4 recurrent) SETD1B variants have been reported including 7 truncating, 1 splicing, 1 extension, 3 in-frame insertions or deletions, and 14 missense variants (Fig. 1). Variants are distributed along the protein (Fig. 1), with the majority of (likely) pathogenic missense variants located within the SET domain region.
Clinical spectrum
The cohort consists of 24 males and 12 females, whose age at last evaluation ranged from 1 to 44 years (median 9 years, interquartile range [IQR] 6–15 years). Table 1 gives an overview of the core clinical phenotype, and Fig. 2 displays the facial appearance of individuals for whom photographs were available. The phenotype of individuals with VUS (either biallelic or heterozygous) matched that of the overall cohort (Table 2). More details can be found in Supplementary Case Reports and Supplementary Fig. 2–4.
Photographs of 16 individuals (plus one affected mother) with indicated SETD1B variants. Dysmorphic features included, among others, a slightly elongated face, high anterior hairline, thick arched or straight eyebrows, deep-set eyes, a prominent nose, and thin upper lips. Lower right corner shows facial composite images for all individuals, or only those with a likely pathogenic or pathogenic variant (note: individual 13 and mother were not included in the composite, given the image angle and glasses hindering Face2Gene program analysis). LP likely pathogenic variant, P pathogenic variant, V variant of uncertain significance.
Development and neurological findings
Most individuals were born after an uneventful pregnancy at full term, with an unremarkable neonatal period and anthropomorphic measurements in the normal range. Seven individuals (7/31, 23%) had postnatal hypoglycemia. Virtually all individuals (34/36, 94%) showed global developmental delay in early infancy. Notably, individuals 14 and 16 without documented developmental delay are the youngest individuals (respectively 2 and 1 years old). Motor development was delayed in 32 individuals (32/36, 89%), with independent ambulation acquired between 1.0 and 4.5 years of age (median 1.6, IQR 1.3–2.5, one individual is nonambulatory). Motor performance remained an issue, with clumsiness, coordination difficulties, and poor fine motor movements reported. Hypotonia was documented for 16 individuals (16/35, 46%), often manifesting in neonatal or childhood period. Language development was delayed in the majority of individuals (33/36, 92%), with first words acquired between 0.5 and 3.0 years (median 2.0, IQR 1.1–2.1). Five individuals were nonverbal at time of data collection (15%, respectively 2.5, 3, 3.5, 11, and 19 years old), and at least five additional individuals (15%) speak far fewer words than appropriate for their age. Regression of previously acquired skills was reported in nine individuals, especially with regard to language, without an obvious link to epileptic activity. At the last investigation, intellectual disability was present in 28 individuals (28/32, 88%), ranging from mild (n = 9), to moderate (n = 8) and severe (n = 4) (not specified n = 7). Formal IQ testing was performed in 11 individuals with an average score of 60 (IQR 48–67) (mild). Autistic features were observed in 24 individuals (24/36, 67%); other behavioral issues included hyperactivity (13/34, 38%), sleep disturbance (10/32, 31%), anxiety (11/35, 31%), anger or aggressive behavior (11/35, 31%, including self-mutilation for individuals 4 and 10), and obsessive compulsive behavior (individual 7, 26, 30). Epilepsy developed in 28 individuals (28/36, 78%) with a median age of seizure onset of 3 years (IQR 1.0–5.3). Eight individuals remained seizure-free up to an age of 16 years (range 2–16, median 6.0, IQR 4.5–7.3 years). At their onset, the majority of classifiable seizures were generalized (n = 19) and minority focal (n = 5), and included motor (n = 9) or nonmotor (n = 13) involvement, with variable development into seizure types over time (Table 1). Seizure frequency varied (sporadic to very frequent) and was at least daily in the majority of patients. Fever-sensitive seizures were reported in three individuals. Whereas seizures were (partially) controlled using various antiepileptic drugs in eighteen individuals, seizures responded poorly or remained intractable in seven individuals. Brain MRI (Supplementary Fig. S2) was performed in 33 individuals and was often unremarkable (23/31, 74%). Abnormal MRI findings included nonspecific minor subcortical white matter hyperintensities (individual 1); cystic encephalomalacia with ventriculomegaly (individual 4); reduced white matter volume and thin corpus callosum (individual 10); bilateral abnormal signals at frontal, temporal, and occipital lobes (individual 16); extensive irregular gyral pattern with reduced sulcation (individual 19); slightly delayed myelination and small heterotopic gray matter (individual 21); periventricular leukomalacia (individual 30, possible due to an underlying hypoplastic left heart disease); and mild diffuse cerebral volume loss with ex vacuo enlargement of lateral and third ventricles (individual 32).
Additional findings
Ophthalmological findings included strabismus (n = 5), amblyopia (n = 2), myopia (n = 2), astigmatism (n = 3), and cortical vision impairment (n = 1). Eight individuals showed gastrointestinal symptoms, including reflux, constipation, and feeding problems. Ten individuals had dermatological symptoms (eczema, rough or dry skin, café au lait spots, hypo- or hyperpigmentation). A number of individuals displayed skeletal abnormalities (scoliosis [n = 5], kyphosis [n = 2], joint hypermobility [n = 4]). (Recurrent) respiratory and urinary tract infections were reported in six individuals. No malignancies were identified. (Truncal) overweight or obesity was present in 17 individuals (Supplementary Table S2).
Facial appearance
Facial appearance varied from no discernible (5 individuals) to mild dysmorphic features (31 individuals, 86%) (Fig. 2). Dysmorphisms included prominent rounded nasal tip/bulbous nose (n = 15), high anterior hairline (n = 11), (uplifted) large earlobes (n = 10), overfolded superior helices (n = 6), low-set ears (n = 5), thin upper lip (n = 9), pointed/prominent chin (n = 6), deep-set eyes (n = 5), synophrys (n = 4), full cheeks (n = 4), elongated/narrow face (n = 5) and/or bitemporal narrowing (n = 4), and frontal bossing (n = 4). Also, tapering fingers (n = 5), brachydactyly (n = 3), small hands (n = 5), and nail hypoplasia (n = 4) were reported (Supplementary Fig. S3).
Structural modelling of variants
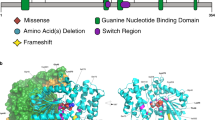
The eight truncating variants (p.[His8fs], p.[Phe95*], p.[Tyr96*], p.[Glu412fs], p.[Arg1329*], p.[Arg1524*], p.[Gln1666*], p.[Ala1730*]) are likely to be targeted for nonsense-mediated decay, but if not would result in removal of the SET region eliminating catalytic activity. Variants p.(His10Gln) and p.(Glu94Asp) are located in a disordered region preceding the RRM (Figs. 1 and 3a) and could affect the specificity of the potential interactions mediated by RRM’s N-terminus [28]. The nucleotide inversion leading to p.(Asn113_Asp121delins9) and substitution p.(Met170Thr) are located in the canonical β1α1β2β3α2β4 RRM region, whereas p.(Gly195Val) is located at the C-terminal loop of α3 (Fig. 3a). Residues 113–121 are located in the α1 helix known to participate in protein–protein interactions in RRM proteins [28]. Furthermore, the RRM domain interacts with COMPASS component WDR82 [5]. Thus, substitution of this 9-residue stretch could severely compromise the RRM fold and its interactions. p.(Met170Thr) and p.(Gly195Val) could affect substrate recognition of RRM because both residues are involved in RNA binding [29]. p.(Thr281Ile) and p.(Thr318Met) are located downstream of the RRM, in a disordered serine, threonine and proline-rich region containing numerous predicted phosphorylation sites [25]. Hence, p.(Thr281Ile) and p.(Thr318Met) might affect the phosphorylation landscape of this region. Substitutions p.(Arg429Trp), p.(Pro545Arg), p.(Pro698Ser), p.(Pro793arg), p.(Arg927His), p.(Arg982Gln), p.(Ala1010Val), p.(Ala1129Val), p.(Pro1328Ser), and p.(Arg1424Gln) are all located in the middle, largely disordered region of SETD1B. The middle portions of Setd1 proteins are divergent [1], suggesting they may have a role in differential genomic targeting of COMPASS through interaction with different targeting proteins. This role might be affected by the mostly nonconservative nature of these substitutions. p.(Ala1129Val), however, is predicted to introduce a noncanonical 5’ splice donor site at nucleotide position c.3384, which would result in a truncated protein p.(Ala1129fs) with eliminated SET catalytic domains. p.(Arg1748Cys) is located in the WIN motif (Fig. 3a) and expected to significantly decrease interaction between SETD1B and WDR5, which is essential for COMPASS assembly and SETD1B participation in H3K4 methylation [6]. Substitutions p.(Arg1792Trp), p.(Arg1825Pro), and p.(Lys1827Arg) are located at the interface with the nucleosome (Fig. 3a) and therefore likely affect interaction with histones and complex stability. Variants p.(Ala1901Val), p.(Ala1901Glu), p.(Tyr1941fs), and p.(Glu1948Lys) are located in the catalytic SET domain (Fig. 3a). Ala1901 is situated in a loop that is part of the S-Adenosyl methionine (SAM) substrate-binding pocket, but is facing away toward an opposing β-strand that is part of the structural core of the SET domain. The substitution of alanine by the larger and negatively charged glutamic acid would create a large stress on the core of the SET domain and potentially disrupt the structural frame maintaining the SAM substrate-binding site and interactions with the adjacent subunits of the complex, whereas alanine to valine substitution introduces a small physicochemical difference which is likely to create some disturbance. p.(Tyr1941fs) would extend the protein, altering the SET domain and post-SET region that are involved in catalysis and cofactor binding, thus likely rendering SETD1B inactive (Fig. 3a). This C-terminal segment is highly conserved [24]. It covers a substantial portion of the binding pocket for histone H3 and the SAM substrate (including the SAM-binding Tyr1943), and three cysteine residues that together with Arg1962 coordinate a zinc atom. Glu1948 is located in a loop adjacent to the histone H3 binding site and, when superimposed to the yeast COMPASS EM structure (PDB:6ven), it is found to be close to the DNA binding surface between Set1 and Bre2 (homolog of ASH2) (Fig. 3a). The replacement of the glutamic acid by a lysine changes the charge of that side chain and could affect interactions of this region.
(a) Homology models of SETD1B domains: RRM domain (top left), based on the crystal structure of the RRM of human SETD1A (PDB ID 3S8S, identity = 66%, QMEAN = 0.25). The segment of Asn113 to Asp121 is colored in blue. This region is known to support different protein–protein interactions in other RRM proteins. Met170 and Gly195 are shown as blue sticks. Homology model of the N-SET and catalytic SET domains of SETD1B (gray cartoon), based on the EM structure of the yeast COMPASS in a complex with a ubiquitinated nucleosome (PDB ID 6ven, identity= 40.21%, QMEAN = −5.10) (center superimposed to the template PDB, and zoom-in panels). The region containing Arg1748 was observed more accurately in the X-ray structure of the WDR5:SETD1B Win motif peptide binary complex (PDB ID 4es0 [6], top right): Arg1748 (blue sticks) is inserted into the pocket of WDR5 (yellow) and interacts with the backbone oxygen atoms of Ser91, Phe133 and Cys261 (hydrogen bonds shown as yellow dashed lines). Arg1792 (blue sticks) and the substitution by Trp (dark gray sticks) interacting with surrounding residues in the adjacent alpha helix (e.g., Glu1796, gray sticks) or with the SWD1 subunit (RBBP5 in humans, shown in violet). The insets with Arg1825 and Arg1827 show the proximity of these residues (dark blue sticks) to histone H2A (light blue cartoon). The SET domain containing Ala1901Val, Ala1901Glu, Tyr1941fs and Glu1948Lys was modeled more accurately based on the crystal structure of the yeast COMPASS catalytic module (PDB ID 6chg [40], identity=62%, QMEAN = −1.78). Ala1901 and Glu1948 are presented as blue sticks in the center figure and right insets. The Ala1901Val and Ala1901Glu substitutions (dark gray sticks) could compromise the stability of the adjacent SAM (olive sticks) binding site and the interaction with the SWD1 subunit (RBBP5 in humans, violet cartoon), which in turn contacts ubiquitin (red cartoon). Tyr1941fs alters a segment of SET and Post-SET regions involved in catalysis and cofactor binding (blue cartoon in center figure and right inset): SAM (olive sticks) and histone H3 (green sticks) binding pocket, the key Tyr1943 residue (yellow sticks), three Cys and one Arg (yellow sticks) coordinating a zinc atom (shown as a sphere). The Glu1948Lys substitution (blue/dark gray sticks in center figure and right inset) could disturb potential interactions between the flexible loops and the adjacent subunit (Bre2, homologous to human ASH2, is shown in teal cartoon). (b) Overexpression of wild-type and variant SETD1B protein in HEK293 cells 48 hours post-transfection assessed by western blot. CC cell control, lysate of mock transfected HEK293 cells (one-way analysis of variance [ANOVA] p = 0.09). (c) Nuclear localization of SETD1B variants in HEK293 cells. Upper panel—SETD1B detected by anti-Flag antibody; lower panel—overlay of nuclear staining (DAPI, cyan) and SETD1B (red); scale bar 20 μm. Images representative of 2 independent experiments are shown. (d) Colocalization of SETD1B and ASH2 in HEK293 cells. Left to right: nuclear staining (DAPI), ASH2 (anti-HA tag), SETD1B (anti-Flag tag), merge of ASH2 (green) and SETD1B (red); scale bar 20μm. Pearson’s r value (range: −1, negative correlation, 1, max correlation) calculated with coloc2 plugin (ImageJ), Z-stacks of min. 12 nuclei were used for the analysis. t-test **p = 0.005. (e) Thermal shift analysis of the SET domain. Left: Tm of GST-SETD1B proteins and GST control. Right: change in Tm of the proteins in presence of SAM substrate. Two independent protein preparations were used for the assay performed in triplicates. One-way ANOVA multiple comparison test *p < 0.05, ***p < 0.0001. (f,g) Analysis of methylation profiles. (f) Hierarchical clustering (rows represent methylation probes, columns–samples). (g) MDS plot (control samples in blue, proband samples in red, SETD1B cases from the database in pink). Sample numbers correspond to case numbers: individual 3 p.([His10Gln];[Arg927His]) (3.1 and 3.2 are the parents of individual 3), individual 4 p.([Glu94Asp];[Pro1328Ser]), individual 5 p.(Phe95*), individual 7 p.(Asn113_Asp121delins9), individual 18 p.(Arg982Gln), individual 19 p.(Ala1010Val), individual 20 p.(Ala1129Val), individual 31 p.(Ala1901Glu), and individual 33 p.(Glu1948Lys).
Functional evaluation of selected SETD1B variants
Based on the structural modeling, seven variants in different regions of SETD1B were selected for in vitro studies: p.(His10Gln) and p.(Glu94Asp) N-terminal of RRM; p.(Asn113_Asp121delins9) in RRM; p.(Thr318Met) C-terminal of RRM; and p.(Ala1901Val), p.(Ala1901Glu), and p.(Glu1948Lys) in the catalytic SET domain.
First, stability of SETD1B in cells was evaluated by western blotting of wild-type and variant SETD1B overexpressed in HEK293 cells (Fig. 3b, Supplementary Fig. S4A). No significant differences in protein levels were observed, suggesting that the evaluated variants do not affect protein stability. Genomic targeting of SETD1B might depend on the central region and the catalytic domain, whereas RRM could reinforce chromatin binding [1, 30], resulting in distribution in the nucleus and not in the nucleoli. Therefore, SETD1B nuclear distribution of wild type and variant was assessed by immunofluorescence of transiently transfected HEK293 cells. Overexpressed FLAG-SETD1B was detected in the cytoplasm and nucleus. Nuclear localization patterns of SETD1B remained similar between wild type and variants, except for p.(Asn113_Asp121delins9), which failed to localize to the nucleus (Fig. 3c). Exclusion from the nucleus correlates with an inability to bind chromatin, resulting in loss of function of this variant. As suggested by structural modeling, Glu1948 could be involved in interaction with COMPASS subunit ASH2. Co-transfection and immunostaining were performed to evaluate colocalization (Fig. 3d). Both overexpressed SETD1B and ASH2 were detected in the nucleus and cytoplasm of transfected HEK293 cells, with a higher colocalization correlation for wild type compared to p.(Glu1948Lys) (Pearson’s correlation value of 0.5 and 0.3 respectively). To evaluate the effect of p.(Ala1901Val), p.(Ala1901Glu), and p.(Glu1948Lys), protein stability and ligand binding were evaluated using thermal shift analysis of the catalytic domain (Fig. 3e). After GST-tagged SETD1B SET domain expression of wild type and variants, melting temperature (Tm) was compared (Fig. 3e, left panel; Supplementary Fig. S4C, D). The Tm of p.(Glu1948Lys) was 1.2 °C higher compared to wild type, which indicates that this substitution increases stability of the SET domain, which can result in disturbance of interactions within COMPASS, perhaps at the interface between SETD1B, the nucleosome, and the ASH2 subunit, as suggested by colocalization analysis of this variant with ASH2 subunit (Fig. 3d). Substitutions p.(Ala1901Val), p.(Ala1901Glu) resulted in a 0.3 °C negative and positive shift of Tm respectively, suggesting that these substitutions have minor effects on thermal stability and thus on conformation of the SET domain. However, since these substitutions are predicted to influence interactions between SETD1B and the SAM substrate, the effect on Tm in presence of SAM was evaluated (Fig. 3e, right panel). Generally, substrate-binding stabilizes proteins resulting in an increased Tm, and indeed a mean Tm increase of 0.3 °C was observed for wild type. The Tm changes of the control GST-protein remained < 0.1 °C, suggesting no contribution of GST tag to the SAM interactions. The increase of 0.17 °C Tm for both p.(Ala1901Val) and p.(Ala1901Glu) indicates no significant effect on SAM interaction.
A specific DNA methylation profile (episignature) for individuals with heterozygous loss-of-function pathogenic SETD1B variants has been described [8]. We performed episignature analysis for nine individuals (individuals 3, 4, 5, 7, 18, 19, 20, 31, 33), and the parents of individual 3 (Fig. 3f–g, Supplementary Fig. S4F). Individuals 5 (p.[Phe95*]), 7 (p.[Asn113_Asp121delins9]), 20 (p.[Ala1129Val]), 31 (p.[Ala1901Glu], and 33 (p.[Glu1948Lys]) showed the previously established SETD1B episignature; individual 18 (p.[Arg982Gln]) showed an inconclusive result, whereas individuals 3 (p.[His10Gln];[Arg927His], nor his parents 3.1 and 3.2), 4 (p.[Glu94Asp]);([Pro1328Ser]), and 19 (p.[Ala1010Val]) did not show the episignature associated with heterozygous loss-of-function SETD1B variants.
Taken together, through structural modeling and functional analyses we provide evidence for reduced function and therefore pathogenicity of p.(Phe95*), p.(Asn113_Asp121delins9), p.(Ala1129Val), p.(Ala1901Glu), and p.(Glu1948Lys), whereas functional consequences and clinical significance remains uncertain for p.(Thr318Met), p.(Arg982Gln), p.(Ala1010Val), p.(Ala1901Val), p.([His10Gln]);([Arg927His]), and p.([Glu94Asp]);([Pro1328Ser]).
DISCUSSION
We report on the molecular and phenotypic spectrum of 36 individuals with sequence variants in SETD1B, representing the largest cohort reported to date. Previous work suggested a possible gain-of-function effect of pathogenic variants in SETD1B [14]; however, further reports [8, 12, 13, 15,16,17,18,19], including this work, point toward a loss-of-function mechanism. Clinical features of our cohort compared to previously reported individuals with a (likely) pathogenic SETD1B variant [8, 12,13,14,15] are provided in Table 2.
The emerging phenotype of SETD1B-associated disorder consists of global developmental delay, language delay including regression, intellectual disability, autism, and epilepsy. Other often observed neurobehavioral issues include hyperactivity, anxiety, anger, or aggressive behavior, and sleep disturbance. Importantly, in most cases, developmental delay predates seizure onset, and eight individuals (up to 16 years old) are seizure-free. This indicates that SETD1B dysfunction severely impacts physiological neurodevelopment even in the absence of epilepsy, suggesting the condition is a developmental encephalopathy, with or without epilepsy. Previously alterations of SETD1B were mainly associated with myoclonic absences [13] and predominantly refractory epilepsy. Although myoclonic absence seizures were often observed in our cohort—confirming this association—other seizure types were regularly encountered at onset, including focal or generalized tonic–clonic seizures. Epilepsy was well or partially controlled in most cases, with 7/26 (27%) remaining refractory to treatment. Brain imaging was unremarkable in most cases and observed abnormalities were without a consistent phenotype. Our cohort identifies a number of mild but consistent dysmorphisms in 30 individuals, including a prominent rounded nasal tip and bulbous nose, high anterior hairline, a thin upper lip, mild ear dysmorphisms, deep-set eyes, and mild hand abnormalities including tapering fingers, brachydactyly, small hands, and nail hypoplasia. Finally, previous work reported potential susceptibility to malignancy in SETD1B-related disorder [12]. Malignancies were not identified in our cohort, although this remains important for follow-up given the relatively young age of the cohort.
To identify possible genotype–phenotype correlation, a severity score was calculated for each individual in our cohort based on clinical features (Supplementary Methods). No association could be identified between the clinical severity score and the effect or location of the corresponding SETD1B variant (Supplementary Fig. S5). Intriguingly, there is a significant overrepresentation of males in both our cohort and in literature, with a total of 36 males and 16 females with SETD1B sequence variants reported (binominal test two-tailed p = 0.008) (Supplementary Methods). The reason for this remains unclear. Incidence of hypotonia and seizures did not differ between males and females in our cohort (hypotonia respectively 12/24, 50% and 4/11, 36%; seizures respectively 19/24, 79% and 9/12, 75%), and seizure onset was similar (respectively range and median years 0–12, 3 and 0–11, 2). Behavioral issues were seen more often in males than females (autistic behavior respectively 19/24, 79% and 5/12, 42%; hyperactivity respectively 10/23, 43% and 3/11, 27%; anxiety respectively 9/23, 39% and 2/12, 17%; aggression respectively 9/23, 39% and 2/12, 17%; sleep disturbance respectively 8/20, 40% and 2/12, 17%), although differences were not significant between both sexes. The clinical severity score is significantly lower in females compared to males, especially when considering behavioral features as a group (Supplementary Fig. S5). It is thus possible that females present with a milder phenotype that may not prompt medical evaluation. However, ascertainment bias for the neurodevelopmental phenotype could also contribute to the male predominance. Nevertheless, it is tempting to speculate that sex-linked traits could affect susceptibility to clinical penetrance and spectrum of SETD1B variants, as female-protective effects have been proposed for other neurodevelopmental disorders [31, 32].
We report four males from three families with biallelic variants in SETD1B, in which variants were inherited from unaffected parents. The two consanguineous siblings (individuals 11 and 12) share, besides the homozygous VUS in SETD1B, also homozygous VUS in NBAS (associated with immune defects) and NOS1 (associated with achalasia). If disease causing, these variants could explain parts of the phenotypes of these individuals, but not their neurological findings. For both individuals, as well as for the other two individuals with biallelic SETD1B VUS, no alternative candidate variants were identified. Pathogenicity of the biallelic variants could not be experimentally proven by in vitro assays for p.(His10Gln) p.(Glu94Asp) and p.(Thr318Met), nor did p.([His10Gln]);([Arg927His]) and p.([Glu94Asp]);([Pro1328Ser]) show the episignature previously associated with heterozygous SETD1B loss-of-function variants. However, this does not exclude the involvement of these variants in yet unknown SETD1B functions. Given that the phenotype of these individuals is similar to the heterozygous individuals (Table 2), and complete absence of SETD1B is lethal in several species [10, 33, 34], we speculate that the combined action of both alleles in biallelic cases results in a phenotype similar to that observed in heterozygous cases by reducing the remaining SETD1B activity below a required threshold. A small subset of genes that typically harbor de novo variants has already been associated with recessive inheritance [35]. Further investigations remain necessary to establish causality of these variants, and the possibility of recessive inheritance of the SETD1B-related disorder.
SETD1B adds to a growing list of chromatin modifying genes implicated in neurodevelopmental disorders. SETD1B is one of the six H3K4 methyltransferases present in mammals, and remarkably loss of function of each is associated with human disease (KMT2A: Wiedemann–Steiner syndrome [OMIM 605130]; KMT2B: early-onset dystonia [OMIM 617284]; KMT2C: Kleefstra syndrome type 2 [OMIM 617768]; KMT2D: Kabuki syndrome [OMIM 147920]), with the latest additions to this list being SETD1A and SETD1B (also known as KMT2F and KMT2G, respectively). SETD1B is paralogous to SETD1A (derived from the orthologue Set1) and both associate with the same noncatalytic COMPASS components. SETD1A and SETD1B, however, show nonoverlapping localization within the nucleus and thus likely make nonredundant contributions to epigenetic control of chromatin structure and gene regulation [1]. This might explain why both SETD1A and SETD1B knockout mice are embryonically lethal, albeit at different developmental stages [33]. Also, in adult mice, SETD1B knockout is lethal and provokes severe defects in hematopoiesis [34]. Heterozygous pathogenic variants in SETD1A have been described in individuals with developmental delay, intellectual disability, subtle facial dysmorphisms, and behavioral and psychiatric problems [36] (OMIM 619056). Interestingly, despite the anticipated nonredundant contributions of SETD1A and SETD1B to epigenetic control, the clinical phenotype of both related disorders shares many similarities [36]. These include global developmental delay with motor and language delay, intellectual disability, and behavioral abnormalities. SETD1A variants have also been found in schizophrenia cohorts [36] and mouse models support SETD1A involvement in schizophrenia [37]. One likely pathogenic SETD1B variant without clinical information was identified in a schizophrenia cohort [38], but psychosis was not reported in our SETD1B cohort. Given the relatively young age of the cohort, this will be an important point for follow-up. Noticeable differences between both syndromes are the incidence of epilepsy, which is more common for SETD1B (20% in SETD1A [36], 78% in this cohort), and the absence of a male predominance for SETD1A (9 males of 19 cases [36, 39]).
Germline mutants of Set1, the orthologue of SETD1A and SETD1B in Drosophila melanogaster, are embryonically lethal [10], whereas postmitotic neuronal knockdown shows that Set1 is required for memory in flies, suggesting a role in postdevelopment neuronal function [36]. In Caenorhabditis elegans, the SETD1A/SETD1B orthologue Set-2 is important for transcription of neuronal genes, axon guidance, and neuronal functions [9], further underscoring the importance of both SETD1A and SETD1B for neural function. Interestingly, whereas we found multiple missense variants in the functional domain of SETD1B (RRM, N-SET, SET), in SETD1A only one missense variant is reported within a functional domain (post-SET). Finally, of the 23 missense variants found in SETD1B, 17 are in regions that are homologous in SETD1A. Of note, p.(Arg982Gln) in the disordered region is at a homologous position in SETD1A previously described in a patient with early-onset epilepsy (NM_014712.2(SETD1A):c.2737C>T, p.(Arg913cys]) [39]. It will be interesting to decipher the downstream epigenetic alterations causative for the resulting overlaps and differences in phenotype between both syndromes.
Data availability
The data that support the findings of this study are available from the corresponding author, with the exception of primary patient sequencing data, as they are derived from patient samples with unique variants that are impossible to guarantee anonymity for. Our institutional guidelines do not allow sharing these raw exome or genome sequencing data, as this is not part of the patient consent procedure.
References
Lee JH, Tate CM, You JS, Skalnik DG. Identification and characterization of the human Set1B histone H3–Lys4 methyltransferase complex. J Biol Chem. 2007;282:13419–28.
Shinsky SA, Monteith KE, Viggiano S, Cosgrove MS. Biochemical reconstitution and phylogenetic comparison of human SET1 family core complexes involved in histone methylation. J Biol Chem. 2015;290:6361–75.
Wang S, Li W, Liu S, Xu J. RaptorX-Property: a web server for protein structure property prediction. Nucleic Acids Res. 2016;44:W430–5.
Meszaros B, Erdos G, Dosztanyi Z. IUPred2A: context-dependent prediction of protein disorder as a function of redox state and protein binding. Nucleic Acids Res. 2018;46:W329–37.
Lee JH, Skalnik DG. Rbm15-Mkl1 interacts with the Setd1b histone H3–Lys4 methyltransferase via a SPOC domain that is required for cytokine-independent proliferation. PLoS One. 2012;7:e42965.
Dharmarajan V, Lee JH, Patel A, Skalnik DG, Cosgrove MS. Structural basis for WDR5 interaction (Win) motif recognition in human SET1 family histone methyltransferases. J Biol Chem. 2012;287:27275–89.
Hyun K, Jeon J, Park K, Kim J. Writing, erasing and reading histone lysine methylations. Exp Mol Med. 2017;49:e324.
Krzyzewska IM, Maas SM, Henneman P, Lip K, Venema A, Baranano K, et al. A genome-wide DNA methylation signature for SETD1B-related syndrome. Clin Epigenetics. 2019;11:156.
Abay-Norgaard S, Attianese B, Boreggio L, Salcini AE. Regulators of H3K4 methylation mutated in neurodevelopmental disorders control axon guidance in Caenorhabditis elegans. Development. 2020;147:dev190637.
Hallson G, Hollebakken RE, Li T, Syrzycka M, Kim I, Cotsworth S, et al. dSet1 is the main H3K4 di- and tri-methyltransferase throughout Drosophila development. Genetics. 2012;190:91–100.
Karczewski KJ, Francioli LC, Tiao G, Cummings BB, Alföldi J, Wang Q, et al. The mutational constraint spectrum quantified from variation in 141,456 humans. Nature. 2020;581:434–43.
Hiraide T, Nakashima M, Yamoto K, Fukuda T, Kato M, Ikeda H, et al. De novo variants in SETD1B are associated with intellectual disability, epilepsy and autism. Hum Genet. 2018;137:95–104.
Hiraide T, Hattori A, Ieda D, Hori I, Saitoh S, Nakashima M, et al. De novo variants in SETD1B cause intellectual disability, autism spectrum disorder, and epilepsy with myoclonic absences. Epilepsia Open. 2019;4:476–81.
Den K, Kato M, Yamaguchi T, Miyatake S, Takata A, Mizuguchi T, et al. A novel de novo frameshift variant in SETD1B causes epilepsy. J Hum Genet. 2019;64:821–7.
Roston A, et al. SETD1B-associated neurodevelopmental disorder. J Med Genet. 2021;58:196–204.
Labonne JD, Lee KH, Iwase S, Kong IK, Diamond MP, Layman LC, et al. An atypical 12q24.31 microdeletion implicates six genes including a histone demethylase KDM2B and a histone methyltransferase SETD1B in syndromic intellectual disability. Hum Genet. 2016;135:757–71.
Qiao Y, Tyson C, Hrynchak M, Lopez-Rangel E, Hildebrand J, Martell S, et al. Clinical application of 2.7M Cytogenetics array for CNV detection in subjects with idiopathic autism and/or intellectual disability. Clin Genet. 2013;83:145–54.
Baple E, Palmer R, Hennekam RC. A microdeletion at 12q24.31 can mimic Beckwith-Wiedemann syndrome neonatally. Mol Syndromol. 2010;1:42–5.
Palumbo O, Palumbo P, Delvecchio M, Palladino T, Stallone R, Crisetti M, et al. Microdeletion of 12q24.31: report of a girl with intellectual disability, stereotypies, seizures and facial dysmorphisms. Am J Med Genet A. 2015;167A:438–44.
Sobreira N, Schiettecatte F, Valle D, Hamosh A. GeneMatcher: a matching tool for connecting investigators with an interest in the same gene. Hum Mutat. 2015;36:928–30.
Fokkema I, van der Velde KJ, Slofstra MK, Ruivenkamp C, Vogel MJ, Pfundt R, et al. Dutch genome diagnostic laboratories accelerated and improved variant interpretation and increased accuracy by sharing data. Hum Mutat. 2019;40:2230–3.
Richards S, Aziz N, Bale S, Bick D, Das S, Gastier-Foster J, et al. Standards and guidelines for the interpretation of sequence variants: a joint consensus recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Genet Med. 2015;17:405–24.
Waterhouse A, Bertoni M, Bienert S, Studer G, Tauriello G, Gumienny R, et al. SWISS-MODEL: homology modelling of protein structures and complexes. Nucleic Acids Res. 2018;46:W296–303.
Ashkenazy H, Abadi S, Martz E, Chay O, Mayrose I, Pupko T, et al. ConSurf 2016: an improved methodology to estimate and visualize evolutionary conservation in macromolecules. Nucleic Acids Res. 2016;44:W344–50.
Kumar M, Gouw M, Michael S, Sámano-Sánchez H, Pancsa R, Glavina J, et al. ELM—the eukaryotic linear motif resource in 2020. Nucleic Acids Res. 2020;48:D296–306.
Perenthaler E, Nikoncuk A, Yousefi S, Berdowski WM, Alsagob M, Capo I, et al. Loss of UGP2 in brain leads to a severe epileptic encephalopathy, emphasizing that biallelic isoform-specific start-loss mutations of essential genes can cause genetic diseases. Acta Neuropathol. 2020;139:415–42.
Sanderson LE, Lanko K, Alsagob M, Almass R, Al-Ahmadi N, Najafi M, et al. Biallelic variants in HOPS complex subunit VPS41 cause cerebellar ataxia and abnormal membrane trafficking. Brain. 2021;144:769–80.
Maris C, Dominguez C, Allain FH. The RNA recognition motif, a plastic RNA-binding platform to regulate post-transcriptional gene expression. FEBS J. 2005;272:2118–31.
Eichhorn CD, Yang Y, Repeta L, Feigon J. Structural basis for recognition of human 7SK long noncoding RNA by the La-related protein Larp7. Proc Natl Acad Sci U S A. 2018;115:E6457–66.
Sayou C, Millán-Zambrano G, Santos-Rosa H, Petfalski E, Robson S, Houseley J, et al. RNA binding by histone methyltransferases Set1 and Set2. Mol Cell Biol. 2017;37:e00165-17.
Zhang Y, Li N, Li C, Zhang Z, Teng H, Wang Y, et al. Genetic evidence of gender difference in autism spectrum disorder supports the female-protective effect. Transl Psychiatry. 2020;10:4.
Jacquemont S, Coe BP, Hersch M, Duyzend MH, Krumm N, Bergmann S, et al. A higher mutational burden in females supports a “female protective model” in neurodevelopmental disorders. Am J Hum Genet. 2014;94:415–25.
Bledau AS, Schmidt K, Neumann K, Hill U, Ciotta G, Gupta A, et al. The H3K4 methyltransferase Setd1a is first required at the epiblast stage, whereas Setd1b becomes essential after gastrulation. Development. 2014;141:1022–35.
Schmidt K, Zhang Q, Tasdogan A, Petzold A, Dahl A, Arneth BM, et al. The H3K4 methyltransferase Setd1b is essential for hematopoietic stem and progenitor cell homeostasis in mice. Elife. 2018;7:e27157.
Happ HC, Carvill GL. A 2020 view on the genetics of developmental and epileptic encephalopathies. Epilepsy Curr. 2020;20:90–96.
Kummeling J, Stremmelaar DE, Raun N, Reijnders MRF, Willemsen MH, Ruiterkamp-Versteeg M, et al. Characterization of SETD1A haploinsufficiency in humans and Drosophila defines a novel neurodevelopmental syndrome. Mol Psychiatry. 2020 Apr 28 [Epub ahead of print].
Nagahama K, Sakoori K, Watanabe T, Kishi Y, Kawaji K, Koebis M, et al. Setd1a insufficiency in mice attenuates excitatory synaptic function and recapitulates schizophrenia-related behavioral abnormalities. Cell Rep. 2020;32:108126.
Wang Q, Li M, Yang Z, Hu X, Wu HM, Ni P, et al. Increased co-expression of genes harboring the damaging de novo mutations in Chinese schizophrenic patients during prenatal development. Sci Rep. 2015;5:18209.
Yu X, Yang L, Li J, Li W, Li D, Wang R, et al. De novo and inherited SETD1A variants in early-onset epilepsy. Neurosci Bull. 2019;35:1045–57.
Hsu PL, Li H, Lau HT, Leonen C, Dhall A, Ong SE, et al. Crystal structure of the COMPASS H3K4 methyltransferase catalytic module. Cell. 2018;174:1106–16. e9
Acknowledgements
We thank all patients and families for participation in this study. Part of this research was made possible through access to the data and findings generated by the 100,000 Genomes Project. The 100,000 Genomes Project is managed by Genomics England Limited (a wholly owned company of the Department of Health and Social Care). The 100,000 Genomes Project is funded by the National Institute for Health Research and NHS England. The Wellcome Trust, Cancer Research UK, and the Medical Research Council have also funded research infrastructure. The 100,000 Genomes Project uses data provided by patients and collected by the National Health Service as part of their care and support. Family 2 was collected as part of the SYNaPS Study Group collaboration funded by The Wellcome Trust and strategic award (Synaptopathies) funding (WT093205 MA and WT104033aIA) and research was conducted as part of the Queen Square Genomics group at University College London, supported by the National Institute for Health Research University College London Hospitals Biomedical Research Centre. HH is funded by The MRC (MR/S01165X/1, MR/S005021/1, G0601943), The National Institute for Health Research University College London Hospitals Biomedical Research Centre, Rosetree Trust, Ataxia UK, MSA Trust, Brain Research UK, Sparks GOSH Charity, Muscular Dystrophy UK (MDUK), Muscular Dystrophy Association (MDA USA). G.M.M. was supported by Jordan’s Guardian Angels, the Brotman Baty Institute, and the Sunderland Foundation. J.R.L. acknowledges support by the Baylor Hopkins Center for Mendelian Genomics funded by the US National Human Genome Research Institute (UM1 HG006542). The DECODE-EE project (Health Research Call 2018, Tuscany Region) provided research funding to R.G. The Epilepsy Society supported this work, with funding to S.M.S. S.M.S. acknowledges that his work was partly carried out at NIHR University College London Hospitals Biomedical Research Centre, which receives a proportion of funding from the UK Department of Health’s NIHR Biomedical Research Centres funding scheme. A.J. is supported by Solve-RD. The Solve-RD project has received funding from the European Union’s Horizon 2020 research and innovation program under grant agreement number 779257. STA, R.R., K.J.C.L., K.A.P.G., and F.J.G.V. were supported by funding from King Abdullah University of Science and Technology (KAUST) through the baseline fund and award numbers FCC/1/1976-25 and REI/1/4446-01 from the Office of Sponsored Research (OSR). T.S.B.’s lab is supported by the Netherlands Organisation for Scientific Research (ZonMW Veni, grant 91617021), a NARSAD Young Investigator Grant from the Brain & Behavior Research Foundation, an Erasmus MC Fellowship 2017, and Erasmus MC Human Disease Model Award 2018.
Author information
Authors and Affiliations
Consortia
Contributions
Conceptualization: M.J.A.W., T.S.B.. Data curation: M.J.A.W., T.S.B.. Formal analysis: M.J.A.W., K.L., T.S.B. Funding acquisition: T.S.B.. Investigation: functional experiments: K.L.; methylation analysis: M.A., J.K., H.M.C., S.H., B.S.; computational modeling: F.J.G.V., R.R., K.J.C.L., K.A.P.G., S.T.A; patient recruitment, clinical, and diagnostic evaluations: A.J., Y.v.B., B.v.P., A.K., M.v.S., N.M.A., C.M.K., H.R., L.P., S.B., M.Z., S.E., H.H., I.J., L.L., T.M., I.S., S.M.L., C.A.L.R., D.Q.B.S., C.M.P.S., H.G., N.M., S.M.S., V.H., A.S., J.T., L.A.P., Y.L.T., I.K.C., J.R.L., A.G.F., J.M.G., L.Y., S.H., X.W., T.J., G.R., X.L., S.M., J.L.M., G.M.M., A.T., J.S., M.E., A.M.P., L.S., M.R.Z.R., A.V., M.M., R.G., D.C.K., T.M.M., M.T.P., K.L.M.B., J.P., Z.P., M.W., S.K., M.V., P.T., B.d.V., C.G., R.P., M.L., S.R.B., K.C.K., F.V., M.v.G., C.Q., M.A.D., J.A.H., A.M.P., R.B., L.W., S.A.W., J.B., R.E.P., K.G.M., A.C., J.K.R., A.R., H.Z.E., G.E.L., R.M., S.B. Methodology: M.J.A.W., K.L., T.S.B.. Writing—original draft: M.J.A.W., K.L., T.S.B.. Writing—review & editing: all authors.
Corresponding author
Ethics declarations
Competing interests
X.W. is employee of Cipher Gene, Ltd. R.E.P., K.G.M., A.C., J.K.R., A.R., and H.Z.E. are employees of GeneDx, Inc. The other authors declare no competing interests.
Ethics declaration
Individuals (and/or their legal guardians) recruited in a research setting gave informed consent for their research participation. Those individual research studies received approval from an institutional review board (IRB) (Supplementary Methods). Individuals (or their legal guardians) who were ascertained in diagnostic testing procedures gave informed consent for testing. Permission for inclusion of their anonymized medical data in this cohort, including photographs, was obtained using standard forms at each local site by the responsible referring physicians. For the Erasmus MC, use of genome-wide investigations in a diagnostic setting was IRB approved (METC-2012-387).
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Weerts, M.J.A., Lanko, K., Guzmán-Vega, F.J. et al. Delineating the molecular and phenotypic spectrum of the SETD1B-related syndrome. Genet Med 23, 2122–2137 (2021). https://doi.org/10.1038/s41436-021-01246-2
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41436-021-01246-2
This article is cited by
-
Epigenetic genes and epilepsy — emerging mechanisms and clinical applications
Nature Reviews Neurology (2022)