Abstract
Similar to DNA epigenetic modifications, multiple reversible chemical modifications on RNAs have been uncovered in a new layer of epigenetic modification. N6-methyladenosine (m6A), a modification that occurs in ~30% transcripts, is dynamically regulated by writer complex (methylase) and eraser (RNA demethylase) proteins, and is recognized by reader (m6A-binding) proteins. The effects of m6A modification are reflected in the functional modulation of mRNA splicing, export, localization, translation, and stability by regulating RNA structure and interactions between RNA and RNA-binding proteins. This modulation is involved in a variety of physiological behaviors, including neurodevelopment, immunoregulation, and cellular differentiation. The disruption of m6A modulations impairs gene expression and cellular function and ultimately leads to diseases such as cancer, psychiatric disorders, and metabolic disease. This review focuses on the mechanisms and functions of m6A modification in a variety of physiological behaviors and diseases.
Similar content being viewed by others
Facts
-
A new research field: m6A has been known since the 1970s and serves as a new layer of epigenetic modification with the discovery of demethylases in recent years. m6A plays a broad and crucial role in almost all aspects of RNA metabolism.
-
m6A exerts important roles in physiological regulation and its disruption may impair gene expression and cellular function and is involved in many diseases such as cancer, psychiatric disorders, and metabolic disease.
-
Many enzymes concerning m6A may have not yet been identified. It is unclear how m6A changes the secondary structure of RNA and promotes the binding of RNA to proteins. Even the functions of the known m6A-related enzymes are not known.
Open questions
-
Are other enzymes related to m6A still to be discovered?
-
How can m6A change the secondary structure of RNA and promote the binding of RNA to proteins?
-
Can m6A be used as a biomarker for the early screening, diagnosis and treatment of cancer?
-
Is m6A involved in a variety of pathways, and how can drugs that target specific m6A sites be developed to reduce unwanted side effects?
Introduction
Epigenetics is one of the most intensely studied research fields, encompassing modifications that include DNA methylation, histone, and chromatin modifications. It is well known that in the central dogma of molecular biology, genetic information is transferred from DNA to RNA and then to proteins. Since there are reversible chemical modifications on DNA that can control the expression of genes, researchers suspect that similar modifications to RNA could be functional mediators of gene expression. In fact, multiple chemical modifications, such as m6A, 5-methylcytosine, and pseudouridine have been detected in a large subset of eukaryotic mRNAs. m6A is one of the most deeply researched modifications and plays a broad and crucial role in almost all aspects of physiological behavior. In the 1970s, RNA was found to possess complex base-methyl nucleoside patterns, and the distribution of these patterns in mRNA consists predominantly of m6A1. By treating HeLa cells with radioactive [methyl-3H] methionine, researchers found about one-third of the radioactivity was present in m6A in HeLa cell mRNA2. After the same treatment, approximately three-quarters of [3H] methyl label was in m6A in cytoplasmic simian-virus-40-specific RNA3. Most m6A residues are enriched at specific transcript landmarks, especially at the start of the last exon, in the 3′ UTRs and near stop codons4,5.
In the past few decades, the biological significance of m6A remained elusive. Only in recent years have several groundbreaking studies suggested that m6A plays a vital role in various aspects of RNA metabolism, including splicing, export, localization, translation, and stability6,7,8,9,10.These results promoted research into the biological significance of m6A. Lineage restriction of the yeast Saccharomyces cerevisiae during nutrient limitation depends on the mRNA methyltransferase activity of Ime4, suggesting a broad role of m6A in cell fate11. Spenito, a novel bona fide subunit of the methyltransferase complex, modulates neuronal functions and sex determination in Drosophila, suggesting a crucial role for m6A during the development of complex organisms12.
The methods used to detect m6A sites have been classified according to different purposes. LC-MS/MS is bases on liquid mass spectrometry with tandem mass spectrometry and is used to detect the overall m6A level on mRNA, revealed as molecular ion peaks and fragment ion peaks, and is a tool for performing qualitative and quantitative analysis of bases simultaneously13. The colorimetric method is similar to that of LC-MS/MS, but its procedure is simpler. Researchers can extract total RNA or enrich mRNAs with oligodT magnetic beads. m6A is then detected using specific capture and detection antibodies14. MiCLIP-seq and MeRIP-seq are high-throughput sequencing methods, but the former maps m6A sites by using anti-m6A antibodies and UV cross-linking techniques15, and it can be used to identify m6A sites at single-base resolution, in contrast to the latter. For MeRIP-seq, anti-m6A antibodies are incubated with RNA fragments to precipitate them for sequenceing16. m6A-IP-qPCR is used to quantify enriched RNA directly17, while dot blotting is used to detect the overall m6A methylation level in a more rapid and inexpensive way18. Additionally, there are some drawbacks to these different methods. For example, some MeRIP-seq data do not conform with validated data19.
m6A controls cell fate transition in mammalian embryonic stem cells (ESCs), and its disruption impairs ESC exit from self-renewal and ESC differentiation into several lineages20. These findings revealed that m6A is involved in a variety of physiological behaviors, and its dysfunction may be involved in the mechanisms associated with various diseases. In this review, we discuss the functions and biological consequences of m6A methylation, including physiology and disease, and the prospects for using m6A methylation as a new diagnostic biomarker and potential therapeutic target in disease.
m6A writers, erasers, and readers
Methyltransferases/writers
Researchers first identified a nucleic acid methyltransferase complex that comprises three components and is separable under nondenaturing conditions. The complex recognizes a highly conserved consensus site and specifically methylates only the N6 amino group of adenosine21. The stable heterodimeric complex of METTL3-METTL14 is the core of the methyltransferase complex and functions in cellular m6A deposition on mammalian nuclear RNAs22. Both the METTL3 and METTL14 proteins contain methyltransferase domains, but METTL3 is the catalytically active subunit that transfers a methyl group to RNA, and METTL14 plays a structural and noncatalytic role in substrate recognition, maintaining complex integrity and substrate RNA binding. METTL3 associates with chromatin and localizes to the transcriptional start sites (TSSs) of active genes, suggesting an independent role in transcription23. METTL3 alters the expression of splicing regulators, leading to unexpected splicing events, including insufficient DNA repair24. In addition, METTL3 promotes translation, and its depletion reduces m6A levels on the mRNA of the histone methyltransferase Ezh2, downregulating its expression at the translational level25. However, researchers have recently found that METTL3 can directly enhance the translation of certain mRNAs by recruiting eIF3 to the translation initiation complex independently of its methyltransferase activity and of downstream m6A reader proteins26.
The other subunit of the methyltransferase is Wilms’ tumor 1-associating protein (WTAP), a mammalian splicing factor that can interact with the METTL3-METTL14 complex and is essential for its localization to nuclear speckles and for its catalytic activity. The loss of WTAP reduces the RNA-binding capability of METTL3, suggesting that WTAP may promote the recruitment of the m6A methyltransferase complex to mRNA targets27,28. Further studies have demonstrated that KIAA1429 and RBM15 are also required for methylation29,30. The latter has been suggested to function in m6A modification in the long noncoding RNA X-inactive specific transcript (XIST) and in cellular mRNAs by binding and recruiting the m6A-methylation complex to specific sites in RNA.
Recently, researchers have found that METTL16, an active m6A methyltransferase, binds to the U6 snRNA and other ncRNAs, as well as numerous lncRNAs and pre-mRNAs, and is responsible for the m6A modification of A43 of the U6 snRNA, which base pairs with 50 pre-mRNA splice sites during splicing, expanding the mechanisms by which m6A is deposited on RNAs31,32. METTL16 was presumed to be a rRNA methyltransferase, as the ybiN gene in E.coli encodes the N6 position of A1618 in 23S rRNA specifically33. Interestingly, METTL16 also interacts with MALAT1 (metastasis-associated lung adenocarcinoma transcript 1) ENE+A (element for nuclear expression with a downstream A-rich tract)34. METTL16 regulates the expression of S-adenosylmethionine (SAM) synthetase MAT2A transcripts to modulate SAM homeostasis, which facilitates mouse embryonic development35,36. METTL5 was defined as the methyltransferase of 18S rRNA, ZCCHC4 was defined as the methyltransferase of 28S rRNA, and TRMT112 acts as a methyltransferase activator to stabilize METTL5 in cells37. METTL5 was also reported to be involved in the pluripotency and differentiation potential of mouse embryonic stem cells and the fly behavior of Drosophila38,39.
Demethylases/erasers
The discovery of demethylases suggests that m6A modification is dynamic and reversible. Fat mass and obesity-associated protein (FTO), which was first found to be linked to obesity in population studies, partially colocalizes with nuclear speckles and exhibits efficient oxidative demethylation of abundant m6A in RNA40,41. Further studies have demonstrated that FTO is involved in the formation of two additional modifications derived from the prevalent m6A in mRNA, N6-hydroxymethyladenosine (hm6A), and N6-formyladenosine (f6A), which may regulate gene expression by affecting RNA-protein interactions7. Additionally, FTO regulates poly(A) sites and 3′ UTR length, and knocking it out results in substantial changes in pre-mRNA splicing, with a prevalence of exon skipping events42. In addition, the debate over m6Am continues. According to one report, m6A but not m6Am plays an important role in leukemia43, and the function of FTO is related to the location of m6A. In the nucleus, FTO is critical for m6A and m6Am in snRNAs, while in the cytoplasm, FTO is critical for cap m6Am in poly-A RNA44. Recently, FTO was shown to demethylate m6Am during snRNA biogenesis, which provides new insight into the classification of snRNA45. It is difficult to say whether FTO plays an oncogenic role in leukemia or is related to snRNA. ALKB homolog 5 (ALKBH5), another demethylase, oxidatively reverses m6A to affect mRNA export, metabolism, and the assembly of mRNA processing factors in nuclear speckles46. Unlike the mechanism by which methylases recognize their target transcripts via conserved consensus recognition sites, m6A, as a conformational marker, induces different conformational outcomes in RNAs depending on the sequence context, which promotes substrate recognition by the demethylases FTO and ALKBH547.
Readers
With methyltransferase and demethylases identified as the writers and erasers of m6A on mRNA, researchers focused on the readers of the m6A modification. The YT521-B homology (YTH) domain family has five members: YTHDF1, YTHDF2, YTHDF3, YTHDC1, and YTHDC2. All of these members can bind to m6A-modified RNA at the RRm6ACH consensus sequence through a conserved m6A-binding domain. The human YTH domain family 2 (YTHDF2) protein can selectively recognize and bind m6A-containing mRNA through a conserved core motif, G(m6A)C, to promote mRNA degradation. Its C-terminal domain is responsible for binding to m6A-modified mRNA, and its N-terminal domain promotes the localization of the YTHDF2-mRNA complex to cellular RNA decay sites48. Another m6A reader protein YTHDF1 actively interacts with translation machinery, increases translation efficiency and ultimately promotes protein synthesis, which enables fast changes in gene expression and controllable protein production9. Several studies show that YTHDF3 promotes protein synthesis in synergy with YTHDF1 and regulates YTHDF2-mediated methylated mRNA decay10. These three YTHDF proteins suggest a dynamic and multidimensional mechanism by which m6A regulates mRNA degradation and translation.
The nuclear m6A reader YTHDC1 can recruit and promote the interaction of pre-mRNA splicing factors with target mRNAs. For example, YTHDC1 recruits the pre-mRNA splicing factor SRSF3 (SRp20) and blocks SRSF10 (SRp38) mRNA binding, promoting exon inclusion in target mRNAs49. In addition, YTHDC1 interacts with SRSF3 and facilitates target RNA binding to both SRSF3 and NXF1, which promotes the export of methylated mRNA from the nucleus to the cytoplasm50.
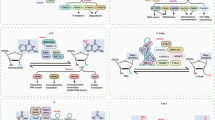
The heterogeneous nuclear ribonucleoprotein (HNRNP) family and insulin-like growth factor 2 mRNA-binding proteins (IGF2BPs, including IGF2BP1/2/3) also bind m6A-bearing RNAs and serve as m6A readers. HNRNPA2B1 directly binds a set of nuclear m6A-methylated transcripts and regulates their alternative splicing in a similar manner as METTL351. m6A can alter the local structure of mRNAs to promote the binding of transcripts to HNRNPG and HNRNPC, which affects the abundance and alternative splicing of target mRNAs52,53. In contrast to the mRNA-decay-promoting function of YTHDF2, IGF2BPs, a distinct family of m6A readers, bind mRNA transcripts through the consensus sequence GG(m6A)C and promote the stability and storage of their target mRNA54. In addition, eukaryotic initiation factor 3 (eIF3) directly binds 5′ UTR m6A to initiate translation without the cap-binding factor eIF4E, which suggests eIF3 is an m6A reader55. Recent studies have found that FMR1 and LRPPRC can also read m6A modifications, serving as m6A readers56. The mechanism is included in Fig. 1.
M6A is added, removed and recognized by its writers, erasers and readers. METTL3-METTL14 is the core of the methyltransferase and functions in cellular m6A deposition on nuclear RNAs. WTAP is a subunit of the methyltransferase, which promotes the recruitment of the m6A methyltransferase complex to mRNA targets. KIAA1429 and RBM15 (RNA binding motif protein 15) are also required for the above process. FTO and ALKBH5 are two enzymes capable of removing m6A, exhibiting efficient oxidative demethylation activity of abundant m6A in RNA. YTH domain family, HNRNP protein family, IGF2BPs and eIF3 bind to m6A-modified RNA through conserved m6A-binding domains and play different roles in RNA metabolism.
m6A and normal physiological behaviors
m6A and neurophysiology
Neurodevelopment
All known m6A enzymes and readers have been found in major brain cell types including neurons and neuroglia and their subtypes. m6A is extensively involved in neurodevelopment regulation which involves a complex system with multiple mechanisms. Studies have shown that m6A is important in the temporal control of mammalian cortical neurogenesis, promoting neurogenesis and neuronal development. Mettl14 knockout in embryonic mouse brains results in a prolonged radial glial cell (RGC) cycle, extending cortical neurogenesis into postnatal stages. METTL3 depletion in adult neural stem cells (aNSCs) not only inhibits the proliferation of aNSCs but also inhibits neuronal development and the morphological maturation of newborn neurons. Mechanistically, m6A depletion affects the decay of a set of transcripts related to transcription factors, neurogenesis, the cell cycle, and neuronal differentiation57. Meanwhile, m6A depletion on the mRNA of the histone methyltransferase Ezh2 downregulates its protein expression and consequent H3K27me3 levels, ultimately causes defects of neuronal development25. This modification of the Btg2 transcription factor also increases the efficiency of induced neuronal cell generation58.
m6A also regulates axon guidance via the translational control of the axon guidance receptor Robo3.1, which plays a key role in the midline crossing of spinal commissural axons. Mechanistically, YTHDF1 binds to endogenous m6A-modified Robo3.1 mRNA, upregulates its translation without affecting mRNA levels, and ultimately controls the guidance of pre-crossing commissural axons in the embryonic spinal cord59. A new m6A reader, proline rich coiled-coil 2A (Prrc2a), plays a crucial role in oligodendrocyte progenitor cell (OPC) proliferation and oligodendrocyte fate determination. Prrc2a can bind Olig2, a critical gene in oligodendrocyte development, and stabilize its mRNA. These results reveal Prrc2a as a novel m6A reader and provide a new pathway for therapies for hypomyelination-related neurological diseases60. Additionally, researchers found a new m6A reader fragile X mental retardation protein (FMRP), which functions during neural progenitor differentiation. FMRP preferentially binds m6A-modified mRNAs related to the regulation of neural differentiation to facilitate their nuclear export by the nuclear export protein CRM1, and its loss results in delayed neural progenitor cell cycle progression61.
The type and amount of m6A enzymes and binding proteins may exhibit distinct regional and subcellular distribution patterns in specific cell types. The complex neurodevelopment regulation system is likely to be based on this distribution, because of the long-distance distribution of mRNAs and proteins across axons and dendrites and the high cellular compartmentalization of the neuron (For a summary see Table 1).
Learning and memory
Studies have demonstrated that m6A is dynamically upregulated in the mouse medial prefrontal cortex (mPFC) in response to behavioral training, suggesting a close association with behavioral adaptation. The mechanism likely involves m6A increasing the targeting of plasticity-related genes and promoting their efficient translation and rapid degradation. Therefore, FTO knockdown in the mPFC promotes the consolidation of cued fear memory62,63. m6A also plays a significant role in reward learning by regulating related protein expression in the DA signaling pathway. FTO depletion impairs D2/3R (dopamine receptor type 2 and type 3) signaling in the midbrain of mice through the disruption of demethylation on specific mRNAs related to DA transmission. and related neuronal activity and behavioral responses64. In addition, FTO variants modulate the connectivity in a basic reward circuit of the neostriata-prefrontal regions, showing that genetic predisposition can also affect other disorders with altered D2R-dependent impulse control, such as addiction65. FTO is strongly associated with learning, memory, and behavioral training. However, the multiple underlying neurobiological mechanisms by which FTO influences the brain and behavior through m6A remains unknown,
Recent studies also show that YTHDF1 promotes learning and memory in response to neuronal stimuli by facilitating the translation of targeted transcripts in the adult mouse hippocampus. Its deletion impairs hippocampal synaptic transmission and long-term potentiation, causing learning and memory defects66.
m6A and immunoregulation
m6A and T-cell homeostasis
Recent studies have demonstrated that m6A plays a crucial role in controlling T-cell homeostasis. m6A modification in IL-7-induced naïve T cells promotes the degradation of the suppressor of cytokine signaling (SOCS) genes, which are involved in inhibiting JAK-STAT signaling. IL-7-STAT5 signaling is activated and initiates naïve T cell reprogramming for proliferation and differentiation67.
Inflammatory response
Dental pulp inflammation is a typical inflammatory disease that is characterized by the partial accumulation of inflammatory mediators. Researchers identified lnc-Dpf3, which suppresses CCR7-mediated DC migration. Mechanistically, CCR7 stimulation decreases the m6A levels of lnc-Dpf3, relieves m6A-dependent degradation, and ultimately upregulates its expression. lnc-Dpf3 feedback suppresses the HIF-1a-dependent transcription of the glycolytic gene Ldha by directly binding to HIF-1a, subsequently inhibiting DC glycolytic metabolism and migratory capacity68.
m6A and stem cell fate
ESCs are cells isolated from the early embryo or original gonad that have two main features: self-renewal and multidirectional differentiation potential. Recent studies have found m6A promotes the resolution of murine naïve pluripotency toward differentiation by reducing the stability of key naïve pluripotency-promoting transcripts, including the core pluripotency regulator Nanog20,69,70. The results reflected the effect of m6A on the differentiation of ESCs, however, many studies show conflicting results. Zc3h13-WTAP-Virilizer-Hakai is an evolutionarily conserved complex that contributes to the regulation of RNA m6A methylation. ZC3H13, a zinc-finger protein, maintains mouse embryonic stem cell (mESC) self-renewal by anchoring the above complex in the nucleus and and facilitating m6A methylation. Zc3h13 depletion impairs self-renewal and triggers mESC differentiation71. The vast majority of large intergenic noncoding RNAs (lincRNAs) are necessary for the maintenance of ESC pluripotency72. The role of m6A in stem cell fate determination has also been confirmed in hematopoietic stem cells (HSCs).
The interaction of m6A with stem cell fate has been found in NSCs, glioblastoma (GBM), and leukemia, and the relevant mechanisms will be described in the following sections.
m6A plays conflicting and dual roles in stem cell fate, especially ESCs. Many studies have found that m6A reduces pluripotency and promotes differentiation, whereas others have shown the opposite. This dual role is likely due to the widespread presence of m6A in pluripotency regulators and developmental regulators. Meanwhile, abundant RNA-binding proteins including m6A reader proteins and non-reader proteins directly or indirectly interacts with m6A-modified RNA, forming a complex network that regulates stem cell fate. This network allows stem cells to choose to self-renew or differentiate at the right time, which would make m6A play a different or even opposite role. The existence of above readers such as eIF3, LRPPRC, FMR1, and Prrc2a are also likely involved in regulating stem cell fate.
m6A and gametogenesis
By analyzing the m6A mRNA methylomes of mouse spermatogenic cells at five different developmental stages, researchers found dynamic changes in m6A abundance during different developmental stages of spermatogenesis, suggesting crucial role of m6A in spermatogenesis.
Recent studies indicate the enrichment of m6A RNA modification in most key regulators of spermatogonial stem cell progenitor cells, including Plzf, Id4, Dnmt3b, and Sohlh2. m6A could provide a marker to these transcripts to modulate their coordinated translation (For a summary see Table 1).
m6A and disease
m6A and cancer
m6A and cancer stem cell pluripotency
Cancer stem cells (CSCs) constitute a rare subclass of neoplastic cells within tumors that have a stem cell-like capacity to self-renew and undergo multidirectional differentiation. We have described the role of m6A in determining the fate of stem cells above, and discuss the role of m6A in cancer stem cells as a new research direction here.
The abnormal or blocked differentiation of HSCs is an important feature of myeloid hematological malignancies. Because HSCs have well-defined cell differentiation trajectories and serve as ideal model systems, we chose to focus on HSCs to discuss the relationship between m6A and cancer stem cell pluripotency. Acute myeloid leukemia (AML) is a cancer of haematopoietic progenitor cells characterized by the proliferation of blast cells and loss of normal haematopoiesis73. Many studies have shown that METTL3 is highly expressed and that the m6A levels of transcripts are increased, including myelocytomatosis (MYC), B cell lymphoma 2 (BCL2), and phosphatase and tensin homolog (PTEN) transcripts, in human AML. Upregulated m6A levels promote the translation of these mRNAs and thereby retain pluripotency properties and inhibit cell differentiation23,74. Similar to METTL3, METTL14 targets MYB and MYC mRNAs and increases their expression, suggesting crucial roles in myelopoiesis and leukemogenesis75. Interestingly, the FTO-mediated decrease in m6A may play oncogenic roles in certain types of AML. A suppressor of cytokine signaling box-2 (ASB2) and retinoic acid receptor alpha (RARA), which promote normal hematopoiesis and the ATRA-induced APL differentiation of leukemia cells, are downregulated by FTO in an m6A-dependent manner76. Related to the role of FTO, R-2-hydroxyglutarate (R-2HG) increases the m6A levels of MYC/CEBPA mRNAs through inhibition of FTO, reduces their stability and downregulates their expression, exhibiting anti-proliferation effects in leukemia43. Through the analysis of these results, we found that m6A plays a dual function in AML cell pluripotency by regulating the expression of key genes. This regulatory network is complex. Widespread m6A does drive cells in a specific direction, it is a part of a comprehensive regulation system, and the final result is determined by various factors.
Intratumoral hypoxia is a critical factor that drives breast cancer progression. ZNF217, an m6A methyltransferase inhibitor targeting METTL3, is upregulated in hypoxia-induced breast cancer cells. ZNF217 increases pluripotency factor KLF4 and NANOG expression in an m6A-dependent manner, promoting pluripotency factor expression and breast cancer stem cell (BCSC) specification77. Glioblastoma stem-like cells (GSCs) are a class of self-renewing cells related to GBM origin, growth, invasion, and recurrence. Studies found that METTL3 overexpression suppresses GSC proliferation and self-renewal by increasing m6A levels and decreasing the expression of ADAM19, which has critical biological functions in GSCs78. Other studies demonstrated that ALKBH5 is highly expressed in GBM and promotes GSC tumorigenicity. ALKBH5 decreases m6A on FOXM1 nascent transcript and increases the expression of FOXM1, which plays a pivotal role in GSC proliferation and self-renewal, thereby increasing its expression79. CSCs represent a reservoir of self-sustaining cells that cause many types of cancers. The role of m6A in CSCs is complex and interesting, and relevant studies are not described here. We believe that the role of m6A in CSC is a critical cancer research direction.
m6A is involved in cancer cell migration and tumor metastasis
The studies that show m6A regulating cancer cell migration and metastasis have been an important and full-fledged aspects of cancer research. Here, we discuss many studies demonstrating the possibility of inhibiting tumor migration through the regulation of m6A.
Hepatocellular carcinoma (HCC) is the major type of primary hepatic carcinoma. METTL3 is prominently upregulated and promotes the migration of HCC cells by targeting suppressor of cytokine signaling 2 (SOCS2) causing its degradation. Downregulated SOCS2 cannot effectively serve as a tumor suppressor in HCC80. However, as another m6A writer, METTL14 shows decreased expression in tissues and advanced metastasis capability in HCC. METTL14 increases the m6A abundance of pri-miR126 and then promotes its interaction with DGCR8, positively modulating the pri-miR-126 process which has been found to suppress metastasis76.
In pancreatic cancer, ALKBH5 is weakly expressed and inhibits cell migration and invasion by demethylating the lncRNA KCNK15-AS1 and regulating KCNK15-AS1-mediated cell motility81. Additionally, YTHDF2 has the dual effect of promoting the proliferation and inhibiting the migration and invasion of pancreatic cancer cells; this effect is called the “migration proliferation dichotomy”. YTHDF2 also regulates the epithelial–mesenchymal transition (EMT) via the downregulation of total yes-associated protein (YAP) mRNA which has been reported to be closely related to the EMT in pancreatic cancer cells. Previous studies found that YAP has two m6A sites in the CDS and exon region. It is reasonable to think that YTHDF2 might directly bind to YAP mRNA to decrease its stability82.
Many studies have been carried out on the function of m6A in cancer cell migration and tumor metastasis, but many precise mechanisms such as the dynamic regulation of the co-transcriptional installation of m6A RNA methylation remain unknown. Meanwhile, due to different target mRNA and reader, m6A act antipodal roles in HCC, suggesting a complex regulator control system that needs further research.
m6A regulates cancer cell proliferation
In addition to abnormal cell migration, uncontrolled proliferation is a main characteristic of tumor growth. m6A has been shown to regulate cancer cell proliferation in many kinds of cancers.
In breast cancer, METTL3 participates in a positive feedback loop comprising HBXIP/let-7g/METTL3/HBXIP that promotes proliferation. HBXIP, a type of oncoprotein associated with the aggressiveness of breast cancer, upregulates METTL3 by suppressing miRNA let-7g, and then METTL3 promotes the expression of HBXIP in an m6A-dependent manner83.
Researchers found that METTL14 mutations or METTL3 downregulation in endometrial cancer increases cell proliferation and tumorigenicity. Mechanistically, a low abundance of m6A downregulates the negative AKT regulator PHLPP2 and upregulates the positive AKT regulator mTORC2. The AKT signaling pathway is thus significantly activated and promotes the growth of endometrial cancer17.
In prostate cancer (PCa), YTHDF2 is frequently upregulated and promotes the proliferation of cancer cells. miR-493-3p is negatively correlated with YTHDF2 and suppresses tumor cell proliferation. YTHDF2 decreases m6A levels, and miR-493-3p increases m6A levels in PCa. These two crucial m6A regulators are involved in the progression of PCa by indirectly modulating m6A levels. In addition, miR-493-3p directly targets the 3′-UTR of YTHDF2 and reduces its expression84. These results preliminarily indicate a role for m6A in regulating PCa, but the specific mechanism remains to be studied.
m6A and chemoradiotherapy resisitance
FTO enhances the chemoradiotherapy resistance of cervical squamous cell carcinoma (CSCC) by positively regulating β-catenin expression via mRNA demethylation and in turn increasing excision repair cross-complementation group 1 (ERCC1) activity85. In GSCs, METTL3 is upregulated and confers radio resistance to GSCs. Mechanistically, METTL3 directly targets the 3′UTR of the SOX2 transcript, enhancing its stability. SOX2 has been shown to be associated with radiation resistance in various cancers. METTL3 causes radio resistance of GSCs through SOX2-dependent enhanced DNA repair86. There are few studies on m6A and chemoradiotherapy resistance, but research results may provide potential molecular targets for cancer therapy.
m6A and antitumor immunity
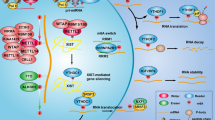
Recently, researchers demonstrated that m6A controls antitumor immunity via YTHDF1 in dendritic cells (DCs). The depletion of YTHDF1 in classical DCs can enhance the cross-presentation of tumor antigens and the cross-priming of CD8+T cells. Mechanistically, m6A-modified transcripts encoding lysosomal proteases are recognized and bound by YTHDF1, which promotes their translation in DCs and the subsequent suppression of the cross-presentation of DCs. These results reveal a previously unrecognized mechanism of immune evasion and show that YTHDF1 may be a potential therapeutic target for immunotherapy87. FTO also increases resistance to anti-PD-1 blockade immunotherapy by decreasing the number of m6A-modified mRNA transcripts, thus reducing their decay in melanoma88 (For a summary see Fig. 2).
M6A plays diverse roles in different cancer, and even plays the opposite roles in a type of cancer. On the on hand, m6A promotes tumor progression by increasing oncogene expression and decreasing tumor suppressor gene expression. On the other hand, m6A suppresses tumor progression in opposite ways. Specific functions of m6A in the main text.
m6A and neuronal disorders
Nerve injury and malformation
Peripheral axon injury mostly increases m6A levels in the adult mouse dorsal root ganglion (DRG), with an enrichment of mRNA related to encoding regeneration-association genes (RAGs) and translational machinery, suggesting that m6A plays an essential role in the axonal regeneration of adult DRG neurons. Mettl14 and YTHDF1 are required for SNL-induced global protein synthesis and the robust axonal regeneration of DRG neurons; the depletion of these proteins reduces injury-induced protein translation in adult DRGs and negatively impacts functional axon regeneration in the peripheral nervous system in vivo. Mettl14 is also required in the central nervous system for the PTEN-deletion-induced robust axonal regeneration of adult retinal ganglion neurons. These results reveal that m6A methylation serves a crucial role in normal physiology and in responses to pathological stimuli in the adult mammalian nervous system89. METTL5 is abundant in the nucleus and synapses of hippocampal neurons, and its deletion leads to microcephaly in zebrafish90.
Psychiatric disorders
The FTO rs9939609 A variant may be connected with a lower risk of depression independently of its effect on BMI, and its effect on major depression (MDD) differ across MDD subtypes91,92. The ALKBH5 rs12936694 variant also showed an allelic association and a genotypic association with MDD93. Further studies have shown that the FTO SNP rs8050136 is involved in modulating the risk for attention-deficit/hyperactivity disorder (ADHD), especially in children who are not exposed to maternal smoking during pregnancy (MSDP). These results may provide a possible link between the physiopathology of ADHD and obesity94. Many transcripts modified by m6A are related to mental disorders such as autism and schizophrenia95. Elevated METTL3 and decreased FTO expression were associated with synaptic and neuron development in Alzheimer’s disease (AD)96. Arsenite induced elevated m6A modifications with deficiency of dopaminergic neurotransmission, and FTO participated in the process97. In Parkinson’s disease (PD), decreased m6A modification mediated by FTO overexpression led to N-methyl-d-aspartate (NMDA) receptor 1 expression, which promoted oxidative stress and induced dopaminergic neuron apoptosis98.
m6A and osteoporosis
Bone-marrow-derived mesenchymal stem cells (BMSCs) have been demonstrated to differentiate into different cell lineages99. An imbalance in the differentiation of adipocytes and osteoblasts from BMSCs is an important factor leading to osteoporosis100. Recently, studies have revealed the potential involvement of m6A in bone homeostasis and osteoporosis. Growth differentiation factor 11 (GDF11) is a key factor in the development of osteoporosis101. Researchers have found that GDF11 controls the shift in osteoporotic MSC fate to adipocytes and inhibits bone formation during osteoporosis in an m6A-dependent manner. GDF11 upregulates FTO in a C/EBPα-dependent manner in osteoporotic BMSCs. FTO can reduce m6A levels in the mRNA of peroxisome proliferator-activated receptor gamma (Pparg) and subsequently promote its expression, which has been demonstrated to promote adipocyte differentiation from BMSCs102. These findings identify a novel axis for adipocyte and osteoblast differentiation, as well as osteoporosis. Unlike the role of GDF11, miR-149-3p has been suggested to inhibit the adipogenic differentiation of BMSCs and promote osteogenic differentiation and osteoblast extracellular matrix maturation and mineralization by targeting FTO103. METTL3 has also been found to regulate the fate of BMSCs and osteoporosis. Its overexpression prevents mice from developing estrogen-deficiency-induced osteoporosis, and its loss induces the pathological features of osteoporosis in mice. Mechanistically, METTL3 depletion reduces the translation efficiency of Pth1r (parathyroid hormone receptor-1), which regulates osteogenic and adipogenic responses in vivo and ultimately results in bone impairment and marrow fat accumulation104,105. These findings may provide many novel strategies for the treatment of osteoporosis. The role of m6A in osteoporosis is mainly reflected in the regulation of BMSCs, suggesting the importance of the complex and fine-tuned regulation of m6A on stem cell fate. These results also highlight the far-reaching implications of m6A in disease treatment.
m6A and metabolic disease
Obesity and lipid metabolism
FTO was first found to be linked to obesity in multiple human populations and ethnic groups in population studies41,106,107. There is a common variant, rs9939609, in the first intron of the FTO gene that is associated with elevated body mass index (BMI) and leads to childhood and adult obesity108. Obesity is one of the main risk factors for the development of cardiovascular disease (CVD) and hypertension. A meta-analysis revealed that this FTO variant is significantly associated with the risk of CVD and hypertension109,110. Many studies have revealed that the polymorphisms in the first intron of FTO control the expression of RPGRIP1 similar to RPGRIP1L and iroquois-related homeobox 3 (IRX3). RPGRIP1L, a ciliary gene near the FTO locus, is related to diminished AcIII-positive cilia and the impaired assembly of the leptin receptor and is probably responsible for the obesity susceptibility signal at the FTO locus111. IRX3, whose expression is associated with obesity-associated SNPs, directly regulates body mass and composition with browning of white adipose tissue112. However, most effects caused by FTO are hard to distinguish from the function of IRX3. The status of FTO in obesity alone does not reveal an effect, especially when the level of IRX3 is changed through FTO knockdown or overexpression.
Results from loss-of-function studies and overexpression studies in mice have revealed that FTO plays an important role in controlling body weight and fat mass and functionally regulates energy homeostasis by controlling energy expenditure. The inactivation of the FTO gene results in postnatal growth retardation and reduced adipose tissue and lean body mass in mice113. A missense mutation (I367F) within the C-terminal domain of FTO leads to a reduction in fat mass and an increase in energy expenditure with unchanged physical activity114. By contrast, FTO overexpression increases food intake and results in a dose-dependent increase in the body and the fat mass of mice independently of their receiving a standard or a high-fat diet115.
Further studies have found that FTO inhibits the adipogenesis of pre-adipocytes by controlling cell cycle progression at the early stage of adipogenesis. Mechanistically, FTO depletion significantly upregulates the m6A levels of CCNA2 and CDK2 mRNA, causes them to be recognized and degraded by YTHDF2 and ultimately prolongs cell cycle progression to suppress adipogenesis116.
m6A mRNA methylation is involved in hepatic lipid metabolism. BMAL1 is a key component of the mammalian clock gene regulatory network associated with regulating metabolism117. FTO promotes the mitochondrial recruitment of STAT3 at the expense of its nuclear localization, affecting oxidative metabolism and the expression of leptin-targeted genes118. Further studies have found that FTO mRNA and protein levels were significantly increased in nonalcoholic fatty liver disease (NAFLD), which enhances lipogenesis and oxidative stress119.
Glucose metabolism
The association of genetic variations in FTO with the risk of type 2 diabetes in multiple human populations and ethnic groups reveals that FTO plays an important role in glucose metabolism120,121,122. Reported findings support the hypothesis that hepatic FTO is involved in the regulation of glucose homeostasis by inhibiting gluconeogenic gene expression in the liver under the of effects of glucose and insulin123 (For a summary see Table 2).
m6A and viral infection
The cytosolic RIG-I-like receptors (RLRs) play a crucial role in activating innate immune signaling by recognizing and binding to Invading pathogen nucleic acids. Recent studies have found that viral transcripts modified by m6A poorly bind to RIG-I and cannot effectively stimulate RIG-I-mediated antiviral signaling124. Interestingly, studies have revealed an opposite role for m6A in antiviral immunity in which m6A is required for the propagation of human cytomegalovirus (HCMV). METTL3/METTL14 and YTHDF2/YTHDC1 are upregulated in primary human foreskin fibroblasts infected by HCMV. METTL3 depletion decreases the m6A levels of IFNB mRNA, enhances its stability and sustains IFN-β production125. Further studies have also demonstrated that m6A-modifying enzymes regulate responses to nonmicrobial dsDNA in uninfected cells, which could shape host immunity and lead to autoimmune disease126.
Recently, researchers identified multiple m6A sites on prototypic polyomavirus simian virus 40 (SV40) mRNAs, which play a positive role in the regulation of SV40 gene expression. The inactivation of these m6A sites or endogenous YTHDF2/METTL3 inhibits SV40 replication in BSC40 cells. m6A sites present in the VP1 open reading frame (ORF) in the SV40 late region promote VP1 mRNA translation. The drug 3-deazaadenosine (DAA), a global inhibitor of methylation, can inhibit viral replication via the depletion of m6A127 (Summarized in Table 2).
Concluding remarks and future perspectives
As increasing evidence suggests that m6A plays a crucial role in cancer, using m6A as a target for the early screening, diagnosis, and treatment of cancer seems to be feasible. For instance, R-2HG exhibits antitumor activity in AML and glioma by inhibiting FTO, but its roles in cancer are complex. For certain genes, m6A may promote the development of cancer, but for other genes, modification may serve as a suppressor of cancer. The modification occurs not only on eukaryotic mRNAs but also on noncoding RNAs. It is difficult to define a uniquely promoting or suppressing role of m6A in the development of human diseases. Among the enzymes participating in m6A modification, METTL3 and FTO seem to play more important roles in the progression of different diseases, and their function may serve as blueprints for translational research and therapeutics. Ubiquitination and SUMOylation have been reported to affect their demethylase or methyltransferase activity. Further studies are needed before m6A can be used in clinical therapies.
m6A is also involved in a variety of physiological behaviors such as neurodevelopment, T cell homeostasis, glucolipid metabolism and gametogenesis, and its disruption leads to various diseases, including addiction, autoimmune disease, metabolic disease, and infertility. For example, FTO is essential for neurodevelopment, as its depletion leads to the reduced proliferation and neuronal differentiation of NSCs, which ultimately reduces the number of NSCs in both the SGZ and SVZ regions. Mechanistically, the loss of FTO alters the m6A modification of key mRNAs and regulates their expression, especially affecting genes involved in the BDNF pathway128. In the last few years, there have been many breakthroughs in m6A, which has become an attractive target for therapy. For example, the drug 3-deazaadenosine (DAA) can inhibit viral replication via the depletion of m6A, but many queries of the mechanism remain. DAA acts as an inhibitor of S-adenosylhomocysteine hydrolase and it has anti-HIV activity. The reason of the inhibited level of m6A modification remains unclear. The expression of methyltransferases may be suppressed and that of demethylases may be promoted. However, the mechanism explaining theimbalance of m6A is also unclear. There may be many enzymes functionally related to m6A that have not yet been identified. It is unclear how m6A changes the secondary structure of RNA and promotes the binding of RNA to proteins. Even the functions of the known m6A-related enzymes are not known. Moreover, m6A is involved in a variety of pathways, and m6A-related drugs could likely cause unwanted side effects. Relevant studies on m6A are still focused on mechanisms and functions, and there is still a long way to go before clinical applications and drug development get processed.
Whole-transcriptome m6A sequencing of major human fetal tissues has been conducted, revealing a positive correlation between m6A level and gene expression homeostasis. The findings show the enrichment of m6A for genes with CpG-rich promoters, and these promoters and gene variations regulate m6A modification to drive human development and disease129. For co-transcription regulation, m6A mediation of YTHDC1 recruits KDM3B to m6A-associated chromatin regions, leading to H3K9me2 demethylation and enhanced gene expression130. Future research will focus on the genetic regulation of m6A and its role and mechanism in human health and diseases.
References
Desrosiers, R., Friderici, K. & Rottman, F. Identification of methylated nucleosides in messenger RNA from Novikoff hepatoma cells. Proc. Natl Acad. Sci. USA 71, 3971–3975 (1974).
Furuichi, Y. et al. Methylated, blocked 5 termini in HeLa cell mRNA. Proc. Natl Acad. Sci. USA 72, 1904–1908 (1975).
Lavi, S. & Shatkin, A. J. Methylated simian virus 40-specific RNA from nuclei and cytoplasm of infected BSC-1 cells. Proc. Natl Acad. Sci. USA 72, 2012–2016 (1975).
Meyer, K. D. et al. Comprehensive analysis of mRNA methylation reveals enrichment in 3′ UTRs and near stop codons. Cell 149, 1635–1646 (2012).
Ke, S. et al. A majority of m6A residues are in the last exons, allowing the potential for 3′ UTR regulation. Genes Dev. 29, 2037–2053 (2015).
Wang, P., Doxtader, K. A. & Nam, Y. Structural basis for cooperative function of Mettl3 and Mettl14 methyltransferases. Mol. Cell 63, 306–317 (2016).
Fu, Y. et al. FTO-mediated formation of N6-hydroxymethyladenosine and N6-formyladenosine in mammalian RNA. Nature Communications 4, 1798 (2013).
Sledz, P. & Jinek, M. Structural insights into the molecular mechanism of the m(6)A writer complex. Elife 5, e18434 (2016).
Wang, X. et al. N(6)-methyladenosine modulates messenger RNA translation efficiency. Cell 161, 1388–1399 (2015).
Vu, L. P. et al. The N6-methyladenosine (m6A)-forming enzyme METTL3 controls myeloid differentiation of normal hematopoietic and leukemia cells. Nat. Med. 23, 1369–1376 (2017).
Agarwala, S. D., Blitzblau, H. G., Hochwagen, A. & Fink, G. R. RNA methylation by the MIS complex regulates a cell fate decision in yeast. PLoS Genet. 8, e1002732 (2012).
Lence, T. et al. m6A modulates neuronal functions and sex determination in Drosophila. Nature 540, 242–247 (2016).
Hsu, P. J. et al. The RNA-binding protein FMRP facilitates the nuclear export of N (6)-methyladenosine-containing mRNAs. J. Biol. Chem. 294, 19889–19895 (2019).
Zhang, C. et al. Hypoxia induces the breast cancer stem cell phenotype by HIF-dependent and ALKBH5-mediated m(6)A-demethylation of NANOG mRNA. Proc. Natl Acad. Sci. USA 113, E2047–E2056 (2016).
Linder, B. et al. Single-nucleotide-resolution mapping of m6A and m6Am throughout the transcriptome. Nat. Methods 12, 767–772 (2015).
Wang, X. et al. N6-methyladenosine-dependent regulation of messenger RNA stability. Nature 505, 117–120 (2014).
Liu, J. et al. m(6)A mRNA methylation regulates AKT activity to promote the proliferation and tumorigenicity of endometrial cancer. Nat. Cell Biol. 20, 1074–1083 (2018).
Nagarajan, A., Janostiak, R. & Wajapeyee, N. Dot blot analysis for measuring global N(6)-methyladenosine modification of RNA. Methods Mol. Biol. 1870, 263–271 (2019).
McIntyre, A. B. R. et al. Limits in the detection of m(6)A changes using MeRIP/m(6)A-seq. Sci. Rep. 10, 6590 (2020).
Batista, P. J. et al. m(6)A RNA modification controls cell fate transition in mammalian embryonic stem cells. Cell Stem Cell 15, 707–719 (2014).
Bokar, J. A., Rath-Shambaugh, M. E., Ludwiczak, R., Narayan, P. & Rottman, F. Characterization and partial purification of mRNA N6-adenosine methyltransferase from HeLa cell nuclei. Internal mRNA methylation requires a multisubunit complex. J. Biol. Chem. 269, 17697–17704 (1994).
Liu, J. et al. A METTL3-METTL14 complex mediates mammalian nuclear RNA N6-adenosine methylation. Nat. Chem. Biol. 10, 93–95 (2014).
Barbieri, I. et al. Promoter-bound METTL3 maintains myeloid leukaemia by m(6)A-dependent translation control. Nature 552, 126–131 (2017).
Taketo, K. et al. The epitranscriptome m6A writer METTL3 promotes chemo- and radioresistance in pancreatic cancer cells. Int. J. Oncol. 52, 621–629 (2018).
Chen, J. et al. m(6)A regulates neurogenesis and neuronal development by modulating histone methyltransferase Ezh2. Genom. Proteom. Bioinform. 17, 154–168 (2019).
Lin, S., Choe, J., Du, P., Triboulet, R. & Gregory, R. I. The m(6)A methyltransferase METTL3 promotes translation in human cancer cells. Mol. Cell 62, 335–345 (2016).
Ping, X. L. et al. Mammalian WTAP is a regulatory subunit of the RNA N6-methyladenosine methyltransferase. Cell Res. 24, 177–189 (2014).
Horiuchi, K. et al. Identification of Wilms’ tumor 1-associating protein complex and its role in alternative splicing and the cell cycle. J. Biol. Chem. 288, 33292–33302 (2013).
Schwartz, S. et al. Perturbation of m6A writers reveals two distinct classes of mRNA methylation at internal and 5′ sites. Cell Rep. 8, 284–296 (2014).
Patil, D. P. et al. m(6)A RNA methylation promotes XIST-mediated transcriptional repression. Nature 537, 369–373 (2016).
Warda, A. S. et al. Human METTL16 is a N(6)-methyladenosine (m(6)A) methyltransferase that targets pre-mRNAs and various non-coding RNAs. EMBO Rep. 18, 2004–2014 (2017).
Pendleton, K. E. et al. The U6 snRNA m(6)A methyltransferase METTL16 regulates SAM synthetase intron retention. Cell 169, 824–835 (2017).
Sergiev, P. V., Serebryakova, M. V., Bogdanov, A. A. & Dontsova, O. A. The ybiN gene of Escherichia coli encodes adenine-N6 methyltransferase specific for modification of A1618 of 23 S ribosomal RNA, a methylated residue located close to the ribosomal exit tunnel. J. Mol. Biol. 375, 291–300 (2008).
Brown, J. A., Kinzig, C. G., DeGregorio, S. J. & Steitz, J. A. Methyltransferase-like protein 16 binds the 3′-terminal triple helix of MALAT1 long noncoding RNA. Proc. Natl Acad. Sci. USA 113, 14013–14018 (2016).
Doxtader, K. A. et al. Structural basis for regulation of METTL16, an S-Adenosylmethionine homeostasis factor. Mol. Cell 71, 1001–1011 (2018).
Mendel, M. et al. Methylation of structured RNA by the m(6)A writer METTL16 is essential for mouse embryonic development. Mol. Cell 71, 986–1000 (2018). e1011.
van Tran, N. et al. The human 18S rRNA m6A methyltransferase METTL5 is stabilized by TRMT112. Nucleic Acids Res. 47, 7719–7733 (2019).
Ignatova, V. V. et al. The rRNA m(6)A methyltransferase METTL5 is involved in pluripotency and developmental programs. Genes Dev. 34, 715–729 (2020).
Leismann, J. et al. The 18S ribosomal RNA m(6) A methyltransferase Mettl5 is required for normal walking behavior in Drosophila. EMBO Rep. 21, e49443 (2020).
Jia, G. et al. N6-methyladenosine in nuclear RNA is a major substrate of the obesity-associated FTO. Nat. Chem. Biol. 7, 885–887 (2011).
Fox, C. S. et al. Genome-wide association for abdominal subcutaneous and visceral adipose reveals a novel locus for visceral fat in women. PLoS Genet. 8, e1002695 (2012).
Bartosovic, M. et al. N6-methyladenosine demethylase FTO targets pre-mRNAs and regulates alternative splicing and 3′-end processing. Nucleic Acids Res. 45, 11356–11370 (2017).
Su, R. et al. R-2HG exhibits anti-tumor activity by targeting FTO/m(6)A/MYC/CEBPA signaling. Cell 172, 90–105 (2018).
Wei, J. et al. Differential m(6)A, m(6)Am, and m(1)A demethylation mediated by FTO in the cell nucleus and cytoplasm. Mol. Cell 71, 973–985 e975 (2018).
Mauer, J. et al. FTO controls reversible m(6)Am RNA methylation during snRNA biogenesis. Nat. Chem. Biol. 15, 340–347 (2019).
Zheng, G. et al. ALKBH5 is a mammalian RNA demethylase that impacts RNA metabolism and mouse fertility. Mol. Cell 49, 18–29 (2013).
Zou, S. et al. N(6)-Methyladenosine: a conformational marker that regulates the substrate specificity of human demethylases FTO and ALKBH5. Sci. Rep. 6, 25677 (2016).
Zhao, X. et al. FTO-dependent demethylation of N6-methyladenosine regulates mRNA splicing and is required for adipogenesis. Cell Res. 24, 1403–1419 (2014).
Xiao, W. et al. Nuclear m(6)A reader YTHDC1 regulates mRNA splicing. Mol. Cell 61, 507–519 (2016).
Roundtree, I. A. et al. YTHDC1 mediates nuclear export of N(6)-methyladenosine methylated mRNAs. Elife 6, 31311 (2017).
Alarcon, C. R. et al. HNRNPA2B1 is a mediator of m(6)A-dependent nuclear RNA processing events. Cell 162, 1299–1308 (2015).
Liu, N. et al. N(6)-methyladenosine-dependent RNA structural switches regulate RNA-protein interactions. Nature 518, 560–564 (2015).
Liu, N. et al. N6-methyladenosine alters RNA structure to regulate binding of a low-complexity protein. Nucleic Acids Res. 45, 6051–6063 (2017).
Huang, H. et al. Recognition of RNA N6-methyladenosine by IGF2BP proteins enhances mRNA stability and translation. Nat. Cell Biol. 20, 285–295 (2018).
Meyer, K. D. et al. 5′ UTR m(6)A promotes cap-independent translation. Cell 163, 999–1010 (2015).
Arguello, A. E., DeLiberto, A. N. & Kleiner, R. E. RNA chemical proteomics reveals the N(6)-methyladenosine (m(6)A)-regulated protein-RNA interactome. J. Am. Chem. Soc. 139, 17249–17252 (2017).
Yoon, K. J. et al. Temporal control of mammalian cortical neurogenesis by m(6)A methylation. Cell 171, 877–889 (2017).
Choi, H. et al. Epitranscriptomic N(6)-methyladenosine modification is required for direct lineage reprogramming into neurons. ACS Chem. Biol. 15, 2087–2097 (2020).
Zhuang, M. et al. The m6A reader YTHDF1 regulates axon guidance through translational control of Robo3.1 expression. Nucleic Acids Res. 47, 4765–4777 (2019).
Wu, R. et al. A novel m(6)A reader Prrc2a controls oligodendroglial specification and myelination. Cell Res. 29, 23–41 (2019).
Edens, B. M. et al. FMRP modulates neural differentiation through m(6)A-dependent mRNA nuclear export. Cell Rep. 28, 845–854 (2019).
Walters, B. J. et al. The role of the RNA demethylase FTO (Fat Mass and Obesity-Associated) and mRNA methylation in hippocampal memory formation. Neuropsychopharmacology 42, 1502–1510 (2017).
Widagdo, J. et al. Experience-dependent accumulation of N6-methyladenosine in the prefrontal cortex is associated with memory processes in mice. J. Neurosci. 36, 6771–6777 (2016).
Hess, M. E. et al. The fat mass and obesity associated gene (Fto) regulates activity of the dopaminergic midbrain circuitry. Nat. Neurosci. 16, 1042–1048 (2013).
Sevgi, M. et al. An obesity-predisposing variant of the FTO gene regulates D2R-dependent reward learning. J. Neurosci. 35, 12584–12592 (2015).
Shi, H. et al. m(6)A facilitates hippocampus-dependent learning and memory through YTHDF1. Nature 563, 249–253 (2018).
Li, H. B. et al. m(6)A mRNA methylation controls T cell homeostasis by targeting the IL-7/STAT5/SOCS pathways. Nature 548, 338–342 (2017).
Liu, J. et al. CCR7 chemokine receptor-inducible lnc-Dpf3 restrains dendritic cell migration by inhibiting HIF-1α-mediated glycolysis. Immunity 50, 600–615 (2019).
Geula, S. et al. Stem cells. m6A mRNA methylation facilitates resolution of naive pluripotency toward differentiation. Science 347, 1002–1006 (2015).
Bertero, A. et al. The SMAD2/3 interactome reveals that TGFbeta controls m(6)A mRNA methylation in pluripotency. Nature 555, 256–259 (2018).
Wen, J. et al. Zc3h13 regulates nuclear RNA m(6)A methylation and mouse embryonic stem cell self-renewal. Mol. Cell 69, 1028–1038 (2018).
Guttman, M. et al. lincRNAs act in the circuitry controlling pluripotency and differentiation. Nature 477, 295–300 (2011).
Chen, E. C., Fathi, A. T. & Brunner, A. M. Reformulating acute myeloid leukemia: liposomal cytarabine and daunorubicin (CPX-351) as an emerging therapy for secondary AML. Onco Targets Ther. 11, 3425–3434 (2018).
Vu, L. P. et al. The N(6)-methyladenosine (m(6)A)-forming enzyme METTL3 controls myeloid differentiation of normal hematopoietic and leukemia cells. Nat. Med. 23, 1369–1376 (2017).
Weng, H. et al. METTL14 inhibits hematopoietic stem/progenitor differentiation and promotes leukemogenesis via mRNA m(6)A modification. Cell Stem Cell 22, 191–205 (2018).
Ma, J. Z. et al. METTL14 suppresses the metastatic potential of hepatocellular carcinoma by modulating N(6)-methyladenosine-dependent primary MicroRNA processing. Hepatology 65, 529–543 (2017).
Zhang, C. et al. Hypoxia-inducible factors regulate pluripotency factor expression by ZNF217- and ALKBH5-mediated modulation of RNA methylation in breast cancer cells. Oncotarget 7, 64527–64542 (2016).
Cui, Q. et al. m(6)A RNA methylation regulates the self-renewal and tumorigenesis of glioblastoma stem cells. Cell Rep. 18, 2622–2634 (2017).
Zhang, S. et al. m(6)A demethylase ALKBH5 maintains tumorigenicity of glioblastoma stem-like cells by sustaining FOXM1 expression and cell proliferation program. Cancer Cell 31, 591–606 (2017).
Chen, M. et al. RNA N6-methyladenosine methyltransferase-like 3 promotes liver cancer progression through YTHDF2-dependent posttranscriptional silencing of SOCS2. Hepatology 67, 2254–2270 (2018).
He, Y. et al. ALKBH5 inhibits pancreatic cancer motility by decreasing long non-coding RNA KCNK15-AS1 methylation. Cell Physiol. Biochem. 48, 838–846. (2018).
Chen, J. et al. YTH domain family 2 orchestrates epithelial-mesenchymal transition/proliferation dichotomy in pancreatic cancer cells. Cell Cycle 16, 2259–2271 (2017).
Cai, X. et al. HBXIP-elevated methyltransferase METTL3 promotes the progression of breast cancer via inhibiting tumor suppressor let-7g. Cancer Lett. 415, 11–19 (2018).
Li, J. et al. Downregulation of N(6)-methyladenosine binding YTHDF2 protein mediated by miR-493-3p suppresses prostate cancer by elevating N(6)-methyladenosine levels. Oncotarget 9, 3752–3764 (2018).
Zhou, S. et al. FTO regulates the chemo-radiotherapy resistance of cervical squamous cell carcinoma (CSCC) by targeting beta-catenin through mRNA demethylation. Mol. Carcinog. 57, 590–597 (2018).
Visvanathan, A. et al. Essential role of METTL3-mediated m(6)A modification in glioma stem-like cells maintenance and radioresistance. Oncogene 37, 522–533 (2018).
Han, D. et al. Anti-tumour immunity controlled through mRNA m(6)A methylation and YTHDF1 in dendritic cells. Nature 566, 270–274 (2019).
Yang, S. et al. m(6)A mRNA demethylase FTO regulates melanoma tumorigenicity and response to anti-PD-1 blockade. Nat. Commun. 10, 2782 (2019).
Weng, Y. L. et al. Epitranscriptomic m(6)A regulation of axon regeneration in the adult mammalian nervous system. Neuron 97, 313–325 (2018).
Richard, E. M. et al. Bi-allelic variants in METTL5 cause autosomal-recessive intellectual disability and microcephaly. Am. J. Hum. Genet. 105, 869–878 (2019).
Samaan, Z. et al. The protective effect of the obesity-associated rs9939609 A variant in fat mass- and obesity-associated gene on depression. Mol. Psychiatry 18, 1281–1286 (2013).
Milaneschi, Y. et al. The effect of FTO rs9939609 on major depression differs across MDD subtypes. Mol. Psychiatry 19, 960–962 (2014).
Du, T. et al. An association study of the m6A genes with major depressive disorder in Chinese Han population. J. Affect Disord. 183, 279–286 (2015).
Choudhry, Z. et al. Association between obesity-related gene FTO and ADHD. Obesity 21, E738–E744 (2013).
Angelova, M. T. et al. The emerging field of epitranscriptomics in neurodevelopmental and neuronal disorders. Front. Bioeng. Biotechnol. 6, 46 (2018).
Han, M. et al. Abnormality of m6A mRNA methylation is involved in Alzheimer’s disease. Front. Neurosci. 14, 98 (2020).
Bai, L. et al. m6A demethylase FTO regulates dopaminergic neurotransmission deficits caused by arsenite. Toxicol. Sci. 165, 431–446 (2018).
Chen, X. et al. Down-regulation of m6A mRNA methylation is involved in dopaminergic neuronal death. ACS Chem. Neurosci. 10, 2355–2363 (2019).
Chen, X., Zhi, X., Wang, J. & Su, J. RANKL signaling in bone marrow mesenchymal stem cells negatively regulates osteoblastic bone formation. Bone Res. 6, 34 (2018).
Shen, G. et al. GOLM1 stimulation of glutamine metabolism promotes osteoporosis via inhibiting osteogenic differentiation of BMSCs. Cell Physiol. Biochem. 50, 1916–1928 (2018).
Liu, W. et al. GDF11 decreases bone mass by stimulating osteoclastogenesis and inhibiting osteoblast differentiation. Nat. Commun. 7, 12794 (2016).
Shen, G. S. et al. The GDF11-FTO-PPARgamma axis controls the shift of osteoporotic MSC fate to adipocyte and inhibits bone formation during osteoporosis. Biochim. Biophys. Acta Mol. Basis Dis. 1864, 3644–3654 (2018).
Li, Y. et al. miR-149-3p regulates the switch between adipogenic and osteogenic differentiation of BMSCs by targeting FTO. Mol. Ther. Nucleic Acids 17, 590–600 (2019).
Wu, Y. et al. Mettl3-mediated m(6)A RNA methylation regulates the fate of bone marrow mesenchymal stem cells and osteoporosis. Nat. Commun. 9, 4772 (2018).
Fan, Y. et al. Parathyroid hormone directs bone marrow mesenchymal cell fate. Cell Metab. 25, 661–672 (2017).
Renstrom, F. et al. Replication and extension of genome-wide association study results for obesity in 4923 adults from northern Sweden. Hum. Mol. Genet. 18, 1489–1496 (2009).
Qureshi, S. A., Mumtaz, A., Shahid, S. U. & Shabana, N. A. rs3751812, a common variant in fat mass and obesity-associated (FTO) gene, is associated with serum high- and low-density lipoprotein cholesterol in Pakistani individuals. Nutrition 39–40, 92–95 (2017).
Frayling, T. M. et al. A common variant in the FTO gene is associated with body mass index and predisposes to childhood and adult obesity. Science 316, 889–894 (2007).
Liu, C., Mou, S. & Pan, C. The FTO gene rs9939609 polymorphism predicts risk of cardiovascular disease: a systematic review and meta-analysis. PLoS ONE 8, e71901 (2013).
Meng, J. et al. A protocol for RNA methylation differential analysis with MeRIP-Seq data and exomePeak R/Bioconductor package. Methods 69, 274–281 (2014).
Stratigopoulos, G. et al. Hypomorphism for RPGRIP1L, a ciliary gene vicinal to the FTO locus, causes increased adiposity in mice. Cell Metab. 19, 767–779 (2014).
Smemo, S. et al. Obesity-associated variants within FTO form long-range functional connections with IRX3. Nature 507, 371–375 (2014).
Fischer, J. et al. Inactivation of the Fto gene protects from obesity. Nature 458, 894–898 (2009).
Church, C. et al. A mouse model for the metabolic effects of the human fat mass and obesity associated FTO gene. PLoS Genet. 5, e1000599 (2009).
Church, C. et al. Overexpression of Fto leads to increased food intake and results in obesity. Nat. Genet. 42, 1086–1092 (2010).
Wu, R. et al. FTO regulates adipogenesis by controlling cell cycle progression via m(6)A-YTHDF2 dependent mechanism. Biochim. Biophys. Acta Mol. Cell Biol. Lipids 1863, 1323–1330 (2018).
Jia, X., Nie, Q., Lamont, S. J. & Zhang, X. Variation in sequence and expression of the avian FTO, and association with glucose metabolism, body weight, fatness and body composition in chickens. Int. J. Obes. 36, 1054–1061 (2012).
Bravard, A. et al. FTO contributes to hepatic metabolism regulation through regulation of leptin action and STAT3 signalling in liver. Cell Commun. Signal. 12, 4 (2014).
Guo, J. et al. Fat mass and obesity-associated gene enhances oxidative stress and lipogenesis in nonalcoholic fatty liver disease. Dig. Dis. Sci. 58, 1004–1009 (2013).
Li, H. et al. Association of genetic variation in FTO with risk of obesity and type 2 diabetes with data from 96,551 East and South Asians. Diabetologia 55, 981–995 (2012).
Hertel, J. K. et al. FTO, type 2 diabetes, and weight gain throughout adult life: a meta-analysis of 41,504 subjects from the Scandinavian HUNT, MDC, and MPP studies. Diabetes 60, 1637–1644 (2011).
Yajnik, C. S. et al. FTO gene variants are strongly associated with type 2 diabetes in South Asian Indians. Diabetologia 52, 247–252 (2009).
Mizuno, T. M., Lew, P. S., Luo, Y. & Leckstrom, A. Negative regulation of hepatic fat mass and obesity associated (Fto) gene expression by insulin. Life Sci. 170, 50–55 (2017).
Durbin, A. F., Wang, C., Marcotrigiano, J. & Gehrke, L. RNAs containing modified nucleotides fail to trigger RIG-I conformational changes for innate immune signaling. mBio 7 (2016).
Winkler, R. et al. m(6)A modification controls the innate immune response to infection by targeting type I interferons. Nat. Immunol. 20, 173–182 (2019).
Rubio, R. M., Depledge, D. P., Bianco, C., Thompson, L. & Mohr, I. RNA m(6) A modification enzymes shape innate responses to DNA by regulating interferon beta. Genes Dev. 32, 1472–1484 (2018).
Flemington, E. K., Tsai, K., Courtney, D. G. & Cullen, B. R. Addition of m6A to SV40 late mRNAs enhances viral structural gene expression and replication. PLOS Pathogens 14 (2018).
Li, L. et al. Fat mass and obesity-associated (FTO) protein regulates adult neurogenesis. Hum. Mol. Genet 26, 2398–2411 (2017).
Xiao, S. et al. The RNA N(6)-methyladenosine modification landscape of human fetal tissues. Nat. Cell Biol. 21, 651–661 (2019).
Li, Y. et al. N(6)-Methyladenosine co-transcriptionally directs the demethylation of histone H3K9me2. Nat. Genet. 52, 870–877 (2020).
Wang, C. X. et al. METTL3-mediated m6A modification is required for cerebellar development. PLoS Biol. 16, e2004880 (2018).
Tong, J. et al. m(6)A mRNA methylation sustains Treg suppressive functions. Cell Res 28, 253–256 (2018).
Wang, H. et al. Mettl3-mediated mRNA m(6)A methylation promotes dendritic cell activation. Nat. Commun. 10, 1898 (2019).
Feng, Z., Li, Q., Meng, R., Yi, B. & Xu, Q. METTL3 regulates alternative splicing of MyD88 upon the lipopolysaccharide-induced inflammatory response in human dental pulp cells. J. Cell. Mol. Med. 22, 2558–2568 (2018).
Yang, D. et al. N6-Methyladenosine modification of lincRNA 1281 is critically required for mESC differentiation potential. Nucleic Acids Res. 46, 3906–3920 (2018).
Chen, T. et al. m(6)A RNA methylation is regulated by microRNAs and promotes reprogramming to pluripotency. Cell Stem Cell 16, 289–301 (2015).
Li, Z. et al. Suppression of m(6)A reader Ythdf2 promotes hematopoietic stem cell expansion. Cell Res. 28, 904–917 (2018).
Wang, H. et al. Loss of YTHDF2-mediated m(6)A-dependent mRNA clearance facilitates hematopoietic stem cell regeneration. Cell Res. 28, 1035–1038 (2018).
Zhang, C. et al. m(6)A modulates haematopoietic stem and progenitor cell specification. Nature 549, 273–276 (2017).
Lin, Z. et al. Mettl3-/Mettl14-mediated mRNA N(6)-methyladenosine modulates murine spermatogenesis. Cell Res. 27, 1216–1230 (2017).
Xu, K. et al. Mettl3-mediated m(6)A regulates spermatogonial differentiation and meiosis initiation. Cell Res. 27, 1100–1114 (2017).
Tang, C. et al. ALKBH5-dependent m6A demethylation controls splicing and stability of long 3′-UTR mRNAs in male germ cells. Proc. Natl Acad. Sci. USA 115, E325–E333 (2018).
Huang, T. et al. Meclofenamic acid represses spermatogonial proliferation through modulating m(6)A RNA modification. J. Anim. Sci. Biotechnol. 10, 63 (2019).
Hsu, P. J. et al. Ythdc2 is an N(6)-methyladenosine binding protein that regulates mammalian spermatogenesis. Cell Res. 27, 1115–1127 (2017).
Bailey, A. S. et al. The conserved RNA helicase YTHDC2 regulates the transition from proliferation to differentiation in the germline. Elife 6, 26116 (2017).
Jain, D. et al. Ketu mutant mice uncover an essential meiotic function for the ancient RNA helicase YTHDC2. Elife 7, 30919 (2018).
Soh, Y. Q. S. et al. Meioc maintains an extended meiotic prophase I in mice. PLoS Genet 13, e1006704 (2017).
Kasowitz, S. D. et al. Nuclear m6A reader YTHDC1 regulates alternative polyadenylation and splicing during mouse oocyte development. PLoS Genet 14, e1007412 (2018).
Ivanova, I. et al. The RNA m(6)A reader YTHDF2 is essential for the post-transcriptional regulation of the maternal transcriptome and oocyte competence. Mol. Cell 67, 1059–1067 (2017).
Zhong, X. et al. Circadian Clock Regulation of Hepatic Lipid Metabolism by Modulation of m(6)A mRNA Methylation. Cell Rep. 25, 1816–1828 (2018).
Guo, F. et al. Fatmass and obesity associated (FTO) gene regulates gluconeogenesis in chicken embryo fibroblast cells. Comp. Biochem Physiol. A Mol. Integr. Physiol. 179, 149–156 (2015).
Zhou, J. et al. N6-Methyladenosine Guides mRNA Alternative Translation during Integrated Stress Response. Mol. Cell 69, 636–647. e637 (2018).
Seo, J. et al. Atf4 regulates obesity, glucose homeostasis, and energy expenditure. Diabetes 58, 2565–2573 (2009).
Xiao, G. et al. ATF4 protein deficiency protects against high fructose-induced hypertriglyceridemia in mice. J. Biol. Chem. 288, 25350–25361 (2013).
Zheng, Q., Hou, J., Zhou, Y., Li, Z. & Cao, X. The RNA helicase DDX46 inhibits innate immunity by entrapping m(6)A-demethylated antiviral transcripts in the nucleus. Nat. Immunol. 18, 1094–1103 (2017).
Lichinchi, G. et al. Dynamics of the human and viral m(6)A RNA methylomes during HIV-1 infection of T cells. Nat. Microbiol. 1, 16011 (2016).
Hesser, C. R., Karijolich, J., Dominissini, D., He, C. & Glaunsinger, B. A. N6-methyladenosine modification and the YTHDF2 reader protein play cell type specific roles in lytic viral gene expression during Kaposi’s sarcoma-associated herpesvirus infection. PLoS Pathog. 14, e1006995 (2018).
Acknowledgements
This work was supported by the National Natural Science Foundation of China (81773037), Science and Technology Foundation of Chongqing (cstc2015jcyjBX0021 and cstc2017jcyjAX0421).
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Edited by F. Pentimalli
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Yang, C., Hu, Y., Zhou, B. et al. The role of m6A modification in physiology and disease. Cell Death Dis 11, 960 (2020). https://doi.org/10.1038/s41419-020-03143-z
Received:
Revised:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41419-020-03143-z
This article is cited by
-
xWAS analysis in neuropsychiatric disorders by integrating multi-molecular phenotype quantitative trait loci and GWAS summary data
Journal of Translational Medicine (2024)
-
An association study of m6A methylation with major depressive disorder
BMC Psychiatry (2024)
-
Intrafamily heterooligomerization as an emerging mechanism of methyltransferase regulation
Epigenetics & Chromatin (2024)
-
The epigenetic downregulation of LncGHRLOS mediated by RNA m6A methylase ZCCHC4 promotes colorectal cancer tumorigenesis
Journal of Experimental & Clinical Cancer Research (2024)
-
Homozygous EPRS1 missense variant causing hypomyelinating leukodystrophy-15 alters variant-distal mRNA m6A site accessibility
Nature Communications (2024)