Abstract
p53 is a classic tumor suppressor that functions in maintaining genome stability by inducing either cell arrest for damage repair or cell apoptosis to eliminate damaged cells in response to different types of stress. Posttranslational modifications (PTMs) of p53 are thought to be the most effective way for modulating of p53 activation. Here, we show that SIRT5 interacts with p53 and suppresses its transcriptional activity. Using mass spectrometric analysis, we identify a previously unknown PTM of p53, namely, succinylation of p53 at Lysine 120 (K120). SIRT5 mediates desuccinylation of p53 at K120, resulting in the suppression of p53 activation. Moreover, using double knockout mice (p53−/−Sirt5−/−), we validate that the suppression of p53 target gene expression and cell apoptosis upon DNA damage is dependent on cellular p53. Our study identifies a novel PTM of p53 that regulates its activation as well as reveals a new target of SIRT5 acting as a desuccinylase.
Similar content being viewed by others
Introduction
The tumor suppressor p53 is situated at a central signaling node that mediates cellular responses to various stresses by inducing cell cycle arrest, senescence, and apoptosis [1]. As “the guardian of the genome,” p53 is mutated in over 50% of human cancers, and thus ranks first in all tumor suppressor genes [1,2,3,4]. Under normal conditions, p53 is tightly regulated, such that its protein product exists at low levels with a high turnover rate. However, under various types of stress, p53 is rapidly stabilized and its transcriptional activity in cells dramatically increases. Many mechanisms for the regulation of p53 function have been proposed; nonetheless, the precise mechanisms of p53 activation are still not fully understood [5,6,7,8,9,10]. Among these mechanisms, posttranslational modifications (PTMs) of p53 are generally thought to be the most effective, which include phosphorylation, ubiquitination, acetylation, methylation, SUMOylation, neddylation, O-GlcNAcylation, ADP-ribosylation, hydroxylation, and β-hydroxybutyrylation [11,12,13,14,15,16,17,18].
Succinylation is a relatively recently revealed novel PTM in which metabolically derived succinyl CoA modifies protein lysine groups, resulting in a protein flip from positive to negative and a relatively large increase in mass compared with other PTMs [19,20,21,22]. In the past few years, the proteins targeted by succinylation have been discovered [23,24,25,26,27,28]. Even though protein succinylation has garnered more attention, the enzyme responsible for succinylation within the mitochondria has yet to be identified [20, 21]. This has prompted scientists to hypothesize that the succinylation of proteins occurs non-enzymatically by direct reaction between succinyl-CoA and the modified protein, and the conditions of the mitochondria (abundance of succinyl CoA, PH) might be the key governing factors [20, 21]. However, SIRT5, a sirtuin family member that is predominantly located in the mitochondria and with barely detectable deacetylase activity, has been shown to contain potent desuccinylation activity on lysine residues [29,30,31,32].
SIRT5 has been shown to promote tumorigenesis through various mechanisms [33,34,35,36,37,38,39,40]. As a vital tumor suppressor, p53 loses its function in the majority of cancers [3, 41, 42]. This raises an intriguing question of whether SIRT5 also functions in tumorigenesis by modulating p53 activity. In addition, given that succinyl CoA is one of the metabolic intermediates, succinylation links metabolic intermediates to the modification of protein function [21]. However, it seems that putative succinyl transferases are not required for this reaction [20]. Interestingly, it is evident that SIRT5-mediated lysine desuccinylation influences various metabolic pathways [30, 31]. As a vital factor in modulating many physiological and pathological processes, p53 affects metabolism in several aspects [43,44,45,46,47,48]. Nevertheless, whether the metabolic intermediates such as succinyl CoA, or SIRT5-mediated lysine desuccinylation can modulate p53 function remains elusive. Further addressing these questions will not only give us a clear picture of the role of SIRT5 in tumorigenesis, but also provide us information about the relationship between the metabolites and p53 function.
In this study, to gain insights into the impact of SIRT5 on p53 function and the mechanisms by which SIRT5 affects p53 function, we examined the role of SIRT5 on p53 activity and checked whether the effects of SIRT5 on p53 function are mediated by its desuccinylase activity [29,30,31,32]. We found that SIRT5 inhibits p53 transcriptional activity. Further investigation shows that SIRT5 mediates desuccinylation of lysine 120 (K120) of p53, leading to impairment of p53 activation. These findings suggest a critical role of SIRT5 in suppressing p53 activity by desuccinylating p53 at K120.
Materials and methods
Reagents and antibodies
Anti-p53 (#sc-126), anti-TIGAR (#sc-166291), and anti-Myc (#sc-40) were purchased from Santa Cruz Biotechnology. Anti-p53 (#2524), anti-DYKDDDDK Tag (#14793), anti-SDHA (#11998), anti-Histone H3 (#4499), Acetyl-p53(Lys379) (#2570), Acetyl-p53(Lys382) (#2525), anti-HA tag (#3724), and goat anti-mouse IgG, light-chain specific antibody (HRP Conjugate) (#91196) were purchased from Cell Signaling Technology. Anti-SIRT5 (#HPA022002), ant-SIRT5 (HPA022992), anti-SIRT5 (HPA021798), and anti-Flag (#F1804) were purchased from Sigma. Anti-p21 (#A11877) and anti-ACTB (#AC026) were purchased from ABclonal. Anti-HA (#901515) was purchased from Covance. Anti-pan-succinyl-K (#PTM-401) was purchased from PTM Company. Alexa Fluor 488 goat anti-rabbit IgG (#A11008), Alexa Fluor 594 goat anti-mouse IgG (#A11005), anti-α-tubulin (#62204) were purchased from Thermo Fisher Scientific. Doxorubicin (hydrochloride) (#15007) and Actinomycin D (#11421) were purchased from Cayman chemical. 5-Fluorouracil (#F6627) and Etoposide (#E1383) were purchased from Sigma-Aldrich. Nutlin-3 (#S1061) was purchased from Selleckchem. SimpleChIP® Enzymatic Chromatin IP Kit (#9002) was purchased from Cell Signaling Technology. FITC Annexin V Apoptosis Detection Kit I (#556547) was purchased from BD Pharmingen. Dual-luciferase reporter assay system (#E194A) was purchased from Promega. Protein G Sepharose (#17-0618-01) was purchased from GE HealthCare Company.
Cell culture
HEK293T, H1299, and HCT116 cells originally obtained from American Type Culture Collection (ATCC) were cultured in Dulbeccos’ modified Eagle medium (DMEM) (HyClone) with 10% fetal bovine serum (FBS). Wild-type (Sirt5+/+p53+/+), Sirt5-null (Sirt5−/−p53+/+), p53-null (Sirt5+/+p53−/−), and Sirt5 & p53 double-null (Sirt5−/−p53−/−) mouse embryo fibroblast cells (MEFs) were established as described previously [49] and maintained in DMEM supplemented with sodium pyruvate (110 mg/L), 10% FBS, 1× nonessential amino acids (Sigma) and 1% penicillin–streptomycin. The cells were grown at 37 °C in a humidified incubator containing 5% CO2.
Mice
Sirt5 knockout (KO) mice (B6; 129 background) were purchased from the Jackson Laboratory (https://www.jax.org/strain/012757). p53-knockout (KO) mice (C57BL/6 background) (Strain name: C57BL/6-Trp53tm1/Bcgen) was originally obtained from Bcgen (Beijing Biocytogen Co., Ltd) (kindly provided by Dr. Zheng Ling at Wuhan University). The Sirt5-null mice were backcrossed seven generations onto a C57BL/6J background before conducting further study. Mice were housed (12-h light/dark cycle, 22 °C) and given unrestricted access to standard diet and tap water under specific pathogen-free conditions in Animal Research Center of Wuhan University. Sirt5-null mice were mated with p53-null mice to generate Sirt5+/−p53+/− mice, then Sirt5+/−p53+/− mice were self-crossed to obtain mice with different genetic genotypes, including Sirt5+/+p53+/+, Sirt5−/−p53+/+, Sirt5+/+p53−/−, and Sirt5−/−p53−/−. Mice were genotyped using the primer sets listed in Supplemental Table S1. Littermates of the same sex were randomly assigned to experimental groups. All the analyses were performed blindly. Male mice of 6 weeks of age were used for experiments unless noted otherwise.
Luciferase reporter assays
Cells were grown in 24-well plates and transfected with various amounts of plasmids by VigoFect (Vigorous Biotech, Beijing, China), as well as with pCMV-Renilla used as an internal control. After the cells were transfected for 18–24 h, the luciferase activity was determined by the dual-luciferase reporter assay system (Promega). Data were normalized to Renilla luciferase. Data are reported as mean ± SEM, which are representative of three independent experiments, each performed in triplicate.
Quantitative real-time PCR
Total RNAs were extracted using RNAiso Plus (TaKaRa Bio., Beijing, China) following the protocol provided by the manufacturer. cDNAs were synthesized using the Revert Aid First Strand cDNA Synthesis Kit (Thermo Scientific, Waltham, MA, USA). MonAmpTM SYBR® Green qPCR Mix (high Rox) (Monad Bio., Shanghai, China) was used for quantitative RT–PCR assays. The primers for quantitative RT- PCR assays are listed in Supplemental Table S1.
Immunoprecipitation and western blot
Co-immunoprecipitation and western blot analysis were performed as described previously [50]. Anti-Flag and anti-HA antibody-conjugated agarose beads were purchased from Sigma. Protein G-Sepharose beads were purchased from GE Company. The Fuji Film LAS4000 mini-luminescent image analyzer was used to photograph the blots. Image J software (National Institutes of Health) was used to quantify protein levels based on the band density obtained by western blot analysis.
Separation of cytosol, nuclei, and mitochondria
Preparation of cytoplasmic and nuclear extracts from mammalian cultured cells was conducted with the Nuclear and Cytoplasmic Extraction Kit (78833, Thermo Scientific). Preparation of mitochondrial extracts was conducted with Mitochondria Isolation Kit (89874, Thermo Scientific). The extracts were subjected to co-immunoprecipitation assay and analyzed by western blot analysis.
Generation of gene knockout cell line by CRISPR/Cas9
HCT116 SIRT5 knockout cell line (SIRT5−/−) was described as previously [49].
Lentivirus-mediated gene transfer
HEK293T cells were transfected with pHAGE-p53-K120R with the packaging vectors psPAX2 and pMD2.G. Eight hours later, the medium was changed with fresh medium containing 10% FBS, 1% streptomycin–penicillin, and 10 μM β-mercaptoethanol. Forty hours later, supernatants were harvested to infect SIRT5-intact (SIRT5+/+) and SIRT5-null (SIRT5−/−) HCT116 cells.
Immunofluorescence confocal microscopy
Cells seeded on glass coverslips were fixed in 4% paraformaldehyde in PBS for 30 min at 25 °C. After washing for three times by PBS, the slides were blocked in the blocking buffer (5% goat serum, 2 mg/mL BSA, 0.1%Triton X-100 in PBS) for 1 h followed by incubation with primary antibodies overnight at 4 °C. Then, the slides were incubated with Alexa Fluor 488 goat anti-rabbit IgG or Alexa Fluor 594 goat anti-mouse IgG for 1 h at 25 °C. Subsequently, the slides were mounted with VECTASHIELD® mounting medium containing DAPI, and imaged under Leica SP8 laser scanning confocal fluorescence microscope.
In vitro succinylation assay
HA-tagged p53 was purified from HEK293T cells and subjected to reactions contain succinylation buffer [20 mmol/L pH 8.0 HEPES, 1 mmol/L dithiothreitol, 1 mmol/L phenylmethyl sulfonyl fluoride, and 0.1 mg/mL BSA], and different concentrations of succinyl-CoA (S1129, Sigma-Aldrich). Reaction mixture was incubated at 30 °C for 15 min. The reaction was stopped by adding loading buffer and subjected to SDS-PAGE. Proteins were analyzed by Western blot analysis.
In vitro desuccinylation assay
HA-tagged p53 was purified from HEK293T cells and subjected to in vitro succinylation assay, and then washed with lysis buffer and eluted with HA peptide to obtain succinylated p53 proteins (Su-p53). Su-p53 were incubated with recombinant SIRT5 (ab101134, Abcam) in the desuccinylation buffer [1 mmol/L NAD+, 50 mmol/L Tris-HCl (pH 8.0), 20% glycerol, 8 mmol/L MgCl2,100 mmol/L NaCl, 1 mmol/L phenylmethyl sulfonyl fluoride, 1 mmol/L dithiothreitol, 0.1 mg/mL BSA]. Reaction mixture was incubated at 30 °C for 1 h. The reaction was stopped by adding loading buffer and subjected to SDS-PAGE. Proteins were analyzed by western blot analysis.
Identification of p53 succinylation site (s) by mass spectrometry
HEK293T cells were transfected with HA-p53 plasmid. Cell lysate was immunoprecipitated with anti-HA Ab–conjugated agarose beads overnight. Immunoprecipitated p53 proteins were subjected to 8% SDS-PAGE gel, and p53 bands were excised from the gel and analyzed by mass spectrometry (MS) in Protein Gene Biotech, Wuhan, Hubei, China.
Generation of anti-Su-p53-K120 antibody and anti-Ac-p53-K120 antibody
p53-K120 site-specific succinylation antibody (anti Su-p53-K120) was generated by using a human p53 succinylated peptide (SGTA(succ-K)SVTC) as an antigen (ABclonal). After purifying the antibodies with excess unmodified peptide (SGTAKSVTC), the specificity of anti-Su-p53-K120 antibody was verified by dot blot analysis. p53 K120 site-specific acetylation antibody (anti Ac-p53-K120) was generated by using a human p53 acetylated peptide (SGTA(ac-K)SVTC) as an antigen (ABclonal). After purifying the antibodies with excess unmodified peptide (SGTAKSVTC), the specificity of anti-Ac-p53-K120 antibody was verified by dot blot analysis.
Flow cytometry assay
HCT116 and MEF cells were treated with DNA damage regents as indicated. Subsequently, the cells were harvested and washed twice with PBS, then resuspended in 1 ×Binding Buffer at a concentration of 1 × 106 cells/ml. In all, 100 µl of the solution (1 × 105 cells) were transferred to a 5 ml culture tube, 5 µl of FITC Annexin V, and 5 µl PI were added. The cells were gently vortexed and incubated for 15 min at 25 °C in the dark. In total, 400 µl of 1X Binding Buffer were added to each tube and then analyzed using Beckman CytoFLEX S within 1 h. The data were analyzed with CytExpert software.
Chromatin Immunoprecipitation assay
Chromatin Immunoprecipitation (ChIP) assay was performed according to the protocol with some modification [51]. HCT116 cells were incubated in culture media containing 1% formaldehyde with gentle shaking for 10 min at room temperature, and crosslinking was stopped by addition of 2.5 M glycine to a final concentration of 0.125 M. Then the procedure was performed according to the protocol of the SimpleChIP® Enzymatic Chromatin IP Kit. The purified DNA was analysed by quantitative real-time PCR (qPCR) and the primers are listed in Supplemental Table S1.
Tissue TUNEL staining and Immunofluorescent staining
Six-week-old male mice were treated with 5Gy IR using an RS 2000 Pro x-ray irradiator (Rad Source Technologies, USA). Untreated and treated animals were killed at 24 h following IR treatment by cervical dislocation. Target organs (testis, spleen, intestine, and liver) were immediately harvested. In Situ Cell Death Detection Kit (Roche, 11684817910) or anti-Su-p53-K120 antibody was used for histological analysis of indicated tissues. Images were captured by fluorescence microscopy as described above. The apoptotic cell number was scored with Image J software.
Quantification and statistical analysis
GraphPad Prism software (7.0) was used for all statistical analysis. Results with error bars express mean ± SEM. Statistical analysis was performed by using Student’s two-tailed t-test. A P value <0.05 was considered significant. Statistical significance is represented as follows: *p < 0.05, **p < 0.01, ***p < 0.001, ****p < 0.0001.
Results
SIRT5 suppresses p53 transcriptional activity
To determine whether SIRT5 has an impact on p53, we used promoter assays to evaluate whether SIRT5 could influence p53 transcriptional activity. Overexpression of SIRT5 dramatically suppressed the activity of the p21-promoter luciferase reporter, BAX-promoter luciferase reporter, and MDM2-promoter luciferase reporter that were induced by ectopic expression of p53 in p53-deficient H1299 cells (Fig. 1A–C). Consistently, the expression of p53 target genes, including p21, PUMA, TIGAR, MDM2, PIG3, and SFN, induced by ectopic expression of p53 was inhibited by overexpression of SIRT5 in H1299 cells (Fig. 1D–I).
A p21 promoter activity in co-transfection of HA-p53 together with or without Flag-SIRT5 in H1299 cells for 24 h, Flag empty vector was used as a control (−). Data show mean ± SEM; Student’s two-tailed t-test; Data from three independent experiments. B BAX promoter activity in co-transfection of HA-p53 together with or without Flag-SIRT5 in H1299 cells for 24 h, Flag empty vector was used as a control (−). Data show mean ± SEM; Student’s two-tailed t-test; Data from three independent experiments. C MDM2 promoter activity in co-transfection of HA-p53 together with or without Flag-SIRT5 in H1299 cells for 24 h, Flag empty vector was used as a control (−). Data show mean ± SEM; Student’s two-tailed t-test; Data from three independent experiments. D–I Quantitative real-time PCR (qPCR) analysis of p21 (D), PUMA (E), TIGAR (F), MDM2 (G), PIG3 (H), and SFN (I) mRNA in co-transfection of HA-p53 together with or without Flag-SIRT5 in H1299 cells for 24 h, Flag empty vector was used as a control (−). Data show mean ± SEM; Student’s two-tailed t-test; Data from three independent experiments.
These findings suggest that SIRT5 could suppress p53 transcriptional activity.
SIRT5 suppresses p53 target gene expression in response to DNA damage
As the guardian of the genome, p53 is stabilized and activated in response to DNA damage to maintain genome stability [52]. We investigated whether SIRT5 regulates p53 stability and activation in response to DNA damage. In HCT116 cells harboring a wild-type (WT) TP53 gene (p53+/+), treatment with the chemotherapeutic agent doxorubicin (Dox) [53] dramatically increased the mRNA levels of p21 and MDM2 compared with the control treatment (DMSO) (Fig. 2A, B). However, overexpression of SIRT5 significantly suppressed the mRNA levels of these genes (Fig. 2A, B). Concordant to this notion, by western blot analysis, p53 was stabilized, p21 and TIGAR were induced after treatment with Dox, but p21 and TIGAR were downregulated when SIRT5 was overexpressed (Fig. 2C).
A, B Overexpression of SIRT5 in HCT116 cells caused a reduction of p21 (A) and MDM2 (B) mRNA upon Dox (Doxorubicin) treatment (1 μM, 24 h), as revealed by qPCR analysis. DMSO was used as control. Data show mean ± SEM; Student’s two-tailed t-test; Data from three independent experiments. C Overexpression of SIRT5 in HCT116 cells caused a reduction of the protein levels of p21 and TIGAR upon Dox treatment (1 μM, 24 h), as revealed by Western blot analysis. DMSO was used as control. D–G qPCR analysis of p21 (D), PUMA (E), TIGAR (F), and MDM2 (G) mRNA in SIRT5-deficient or SIRT5-intact HCT116 cells (SIRT5−/− or SIRT5+/+) treated with or without Dox (1 μM, 24 h). DMSO was used as control. Data show mean ± SEM; Student’s two-tailed t-test; Data from three independent experiments. H Western blot analysis of p21 and TIGAR protein level in SIRT5-deficient or SIRT5-intact HCT116 cells (SIRT5−/− or SIRT5+/+) treated with or without Dox (1 μM, 24 h). DMSO was used as control. I, J qPCR analysis of p21 (I), and MDM2 (J) mRNA in SIRT5-deficient or SIRT5-intact HCT116 cells (SIRT5−/− or SIRT5+/+) treated with or without Nutlin-3 (5 μM, 24 h). DMSO was used as control. Data show mean ± SEM; Student’s two-tailed t-test; Data from three independent experiments. K Western blot analysis of p21 and TIGAR protein level in SIRT5-deficient or SIRT5-intact HCT116 cells (SIRT5−/− or SIRT5+/+) treated with or without Nutlin-3 (5 μM, 24 h). DMSO was used as control. L, M qPCR analysis of p21 (L), and MDM2 (M) mRNA in SIRT5-deficient or SIRT5-intact HCT116 cells (SIRT5−/− or SIRT5+/+) treated with or without 5-Fu (5-fluorouracil) (5 μg/ml, 24 h). DMSO was used as control. Data show mean ± SEM; Student’s two-tailed t-test; Data from three independent experiments. N Western blot analysis of p21 and TIGAR protein level in SIRT5-deficient or SIRT5-intact HCT116 cells (SIRT5−/− or SIRT5+/+) treated with or without 5-Fu (5 μg/ml, 24 h). DMSO was used as control.
In contrast, disruption of SIRT5 in HCT116 cells (SIRT5−/−) significantly enhanced mRNA levels of p21, PUMA, TIGAR, and MDM2 and protein levels of p21 and TIGAR after DNA damage (Fig. 2D–H).
Subsequently, we tested the effect of Nutlin-3, a small-molecule antagonist of MDM2 that induces p53 stabilization and activation [53,54,55]. As expected, treatment with Nutlin-3 significantly increased the mRNA levels of p21 and MDM2 compared with the control treatment (DMSO) (Fig. 2I, J). Moreover, compared with those in SIRT5-intact HCT116 cells (SIRT5+/+), the mRNA levels of p21 and MDM2 and protein levels of p21 and TIGAR were higher in SIRT5-null HCT116 cells (SIRT5−/−) (Fig. 2I–K).
Furthermore, we examined the effect of another genotoxic agent, 5-fluorouracil (5-FU) [53]. Consistently, treatment with 5-FU significantly increased p21 and MDM2 mRNA levels compared with the control treatment (DMSO) (Fig. 2L, M). However, compared with those in SIRT5-intact HCT116 cells (SIRT5+/+), the mRNA levels of p21 and MDM2 and protein levels of p21 and TIGAR were higher in SIRT5-null HCT116 cells (SIRT5−/−) (Fig. 2L–N).
In addition, U2OS and MEF cells harboring a WT TP53 gene (p53+/+) were used to validate the effects of SIRT5 on p53. In U2OS cells, treatment with Dox dramatically increased the mRNA levels of p21, TIGAR, and PIG3 compared with the control treatment (DMSO), while disruption of SIRT5 in U2OS cells (SIRT5−/−) significantly enhanced mRNA levels of p21, TIGAR, and PIG3 after DNA damage (Fig. S1A–C). In MEF cells, treatment with Dox (Fig. S1D–F) or 5-Fu (Fig. S1G–I) dramatically increased the mRNA levels of p21, Puma and Mdm2 compared with the control treatment (DMSO), while disruption of Sirt5 in MEF cells (Sirt5−/−) significantly enhanced mRNA levels of p21, Puma and Mdm2 after DNA damage (Fig. S1D–I).
Together, these findings suggest that SIRT5 suppresses p53 target gene expression in response to DNA damage.
p53 is succinylated at lysine 120 (K120) and desuccinylated by SIRT5
The observations that SIRT5 suppresses p53 transcriptional activity and p53 target gene expression prompted us to determine whether SIRT5 influences p53 function through protein–protein interaction. First, we examined their co-localization by immunofluorescence staining using anti-p53 and anti-SIRT5 antibodies. Fluorescence confocal microscopy showed that SIRT5 could co-localize with p53 in the nucleus in both HCT116 cells and U2OS cells in response to DNA damage (Figs. 3A and S2–S4). Without staining with anti-p53 and anti-SIRT5 antibodies, no signal was detected in cells after Dox treatment (Fig. S5), ruling out non-specific red signals caused by Dox absorbance. Co-immunoprecipitation assays showed that ectopically expressed SIRT5 pulled down ectopically expressed p53 in HEK293T cells and vice versa (Fig. 3B, C). In HCT116 cells, endogenous p53 was also co-immunoprecipitated with endogenous SIRT5 after DNA damage (Fig. 3D). Furthermore, we separated cell components into mitochondria, cytosol, and nuclei, and performed co-immunoprecipitation assays respectively. Endogenous SIRT5 interacted with endogenous p53 only in nuclei but not in mitochondria and cytosol after DNA damage (Fig. 3E). However, SIRT5 did not influence protein stability of p53 revealed by either overexpression of SIRT5 in H1299 cells, knockout of SIRT5 in HCT116 cells and U2OS cells, or reconstitution SIRT5 in SIRT5-deficient HCT116 cells (SIRT5−/−) (Fig. 3F–I).
A Confocal microscopy image of endogenous SIRT5 co-localized with endogenous p53 in HCT116 cells. The cells were treated with or without Dox (1 μM, 24 h) and detected by immunofluorescence staining using anti-SIRT5 and anti-p53 antibodies. Mito, Mitotracker; Scale bar in blue = 25 µm; Scale bar in green = 5 µm. DMSO was used as control. B, C Co-immunoprecipitation of Flag-SIRT5 with HA-p53 and vice versa. HEK293T cells were co-transfected with indicated plasmids for 24 h. Anti-Flag (B) or anti-HA (C) antibody-conjugated agarose beads were used for immunoprecipitation and the interaction was detected by immunoblotting with the indicated antibodies. TCL, total cell lysate; IP, immunoblotting.  , antibody light chain. D Endogenous interaction between p53 and SIRT5 in HCT116 cells with Dox (1 μM, 24 h) treatment. Anti-p53 antibody was used for immunoprecipitation, and normal mouse IgG was used as a control. TCL, total cell lysate; IP: immunoblotting. E Co-immunoprecipitation of SIRT5 and p53 in mitochondria (Mito), cytosol (Cyto), and nuclear fractions (Nuc) before and after Dox (Doxorubicin) treatment (1 μM, 24 h). SE shorter exposure, LE longer exposure. F Immunoblotting of exogenous HA-p53 expression in H1299 cells transfected with an increasing amount of Flag-SIRT5 expression plasmid. G Immunoblotting of endogenous p53 expression in SIRT5-deficient or SIRT5-intact HCT116 cells (SIRT5−/− or SIRT5+/+). H Immunoblotting of endogenous p53 expression in SIRT5-deficient or SIRT5-intact U2OS cells (SIRT5−/− or SIRT5+/+). I Immunoblotting of endogenous p53 expression in SIRT5-deficient HCT116 cells transfected with an increasing amount of Flag-SIRT5 expression plasmid.
, antibody light chain. D Endogenous interaction between p53 and SIRT5 in HCT116 cells with Dox (1 μM, 24 h) treatment. Anti-p53 antibody was used for immunoprecipitation, and normal mouse IgG was used as a control. TCL, total cell lysate; IP: immunoblotting. E Co-immunoprecipitation of SIRT5 and p53 in mitochondria (Mito), cytosol (Cyto), and nuclear fractions (Nuc) before and after Dox (Doxorubicin) treatment (1 μM, 24 h). SE shorter exposure, LE longer exposure. F Immunoblotting of exogenous HA-p53 expression in H1299 cells transfected with an increasing amount of Flag-SIRT5 expression plasmid. G Immunoblotting of endogenous p53 expression in SIRT5-deficient or SIRT5-intact HCT116 cells (SIRT5−/− or SIRT5+/+). H Immunoblotting of endogenous p53 expression in SIRT5-deficient or SIRT5-intact U2OS cells (SIRT5−/− or SIRT5+/+). I Immunoblotting of endogenous p53 expression in SIRT5-deficient HCT116 cells transfected with an increasing amount of Flag-SIRT5 expression plasmid.
Moreover, overexpression of SIRT5 suppressed Mdm2 and PIG3 expression induced by Dox treatment in SIRT5-deficient HCT116 cells (SIRT5−/−) (Fig. 4A, B; column 5 versus column 4). However, overexpression of SIRT5-H158Y, an enzyme-deficient mutant of SIRT5, showed no obvious effect on suppression of Mdm2 and PIG3 expression induced by Dox treatment in SIRT5-deficient HCT116 cells (SIRT5−/−) (Fig. 4A, B; column 6 versus column 4), indicating that the enzymatic activity of SIRT5 was required for the suppressive function of SIRT5 on p53.
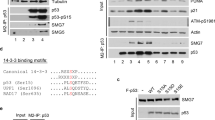
A qPCR analysis of MDM2 mRNA in SIRT5-deficient HCT116 cells (SIRT5−/−) transfected with the control plasmid (Flag empty) or the plasmid expressing Flag–SIRT5 (Flag-SIRT5) or its enzyme-deficient mutant H158Y (Flag-SIRT5-H158Y) for 24 h, followed by treatment with or without Dox (1 μM, 24 h). DMSO was used as control. Data show mean ± SEM; Student’s two-tailed t-test; Data from three independent experiments. B qPCR analysis of PIG3 mRNA in SIRT5-deficient HCT116 cells (SIRT5−/−) transfected with the control plasmid (Flag empty) or the plasmid expressing Flag–SIRT5 (Flag-SIRT5) or its enzyme-deficient mutant H158Y (Flag-SIRT5-H158Y) for 24 h, followed by treatment with or without Dox (1 μM, 24 h). DMSO was used as control. Data show mean ± SEM; Student’s two-tailed t-test; Data from three independent experiments. C p53 was succinylated in vitro. HA-tagged p53 protein purified from HEK293T cells was incubated with the indicated concentrations of succinyl-CoA. Protein succinylation was detected with anti-pan-succinyl-lysine antibody; HA-p53 was detected with anti-HA antibody. D p53 is desuccinylated by SIRT5 in vitro. Succinylated p53 was incubated with purified SIRT5. Protein succinylation was detected with anti-pan-succinyl-lysine antibody. E Disruption of SIRT5 in HCT116 cells enhanced succinylation of p53. The cell lysates from SIRT5-deficient (SIRT5−/−) or SIRT5-intact (SIRT+/+) HCT116 cells treated with Dox (1 μM, 24 h) were immunoprecipitated with anti-p53 antibody, followed by immunoblotting with anti-pan-succinyl-K antibody. TCL total cell lysate, IP immunoprecipitating. F Reconstitution of wild-type SIRT5 (Myc-SIRT5) in SIRT5-deficient HCT116 cells (SIRT5−/−) caused a reduction of p53 succinylation; but overexpression of the enzymatic-deficient SIRT5 (Myc-dSIRT5-H158Y) in SIRT5−/− HCT116 cells did not cause a reduction of p53 succinylation. The SIRT5−/− HCT116 cells were transfected with Myc-SIRT5 or Myc-SIRT5-H158Y and treated with Dox (1 μM, 24 h), followed by immunoprecipitating with anti-p53 antibody, and immunoblotting with anti-pan-succinyl-K antibody. TCL total cell lysate, IP immunoprecipitating. G The succinylated residue in p53 identified by mass spectrometry analysis. HEK293T cells were transfected with HA-p53 plasmid. Cell lysate was immunoprecipitated with anti-HA Ab–conjugated agarose beads overnight. Immunoprecipitated p53 proteins were subjected to 8% SDS-PAGE gel, and p53 bands were excised from the gel and analyzed by mass spectrometry. H Sequence alignment of partial p53 proteins (116–132 amino acids) from human, macaque, cow, pig, dog, mouse, and zebrafish. The red box indicates a conserved lysine (K120). I Dot-blot assay for validating the specificity of anti-Su-p53-K120 antibody. Equal amounts of succinyl-peptides or the control peptides were immunoblotted with the indicated dilutions of anti-Su-p53-K120 antibody. J Disruption of SIRT5 in HCT116 cells caused an enhancement of p53 succinylation at Lys 120 compared with that in the SIRT5-intact HCT116 cells (SIRT5+/+) treated with or without Dox. The cell lysates from SIRT5-deficient (SIRT5−/−) or SIRT5-intact (SIRT5+/+) HCT116 cells treated with or without Dox (1 μM, 24 h) were immunoprecipitated with anti-p53 antibody, followed by immunoblotting with anti-Su-p53-K120 antibody. DMSO was used as a control. TCL total cell lysate, IP immunoprecipitating. K Disruption of SIRT5 in HCT116 cells caused an enhancement of p53 succinylation at K120 compared with that in the SIRT5-intact HCT116 cells (SIRT5+/+) treated with or without 5-Fu. The cell lysates from SIRT5-deficient (SIRT5−/−) or SIRT5-intact (SIRT5+/+) HCT116 cells treated with or without 5-Fu (5 μg/ml, 24 h) were immunoprecipitated with anti-p53 antibody, followed by immunoblotting with anti-Su-p53-K120 antibody. DMSO was used as a control. TCL total cell lysate, IP immunoprecipitating.
Given a well-defined function of SIRT5 in desuccinylation, we sought to determine whether SIRT5 suppresses p53 transcriptional activity through its desuccinylase activity. We initially examined whether p53 was succinylated, by using a pan-succinyl-K antibody (PTM Biolab Co. Ltd. #PTM401) [30, 49]. Using in vitro succinylation assays, we found that p53 expressed in HEK293T cells could be succinylated by adding succinyl-CoA in a dose-dependent manner (Fig. 4C), suggesting that p53 could be succinylated. Subsequently, we examined whether succinylated p53 could be desuccinylated by SIRT5 in vitro. In the presence of NAD+, purified SIRT5 (ab101134, Abcam) induced desuccinylation of succinylated p53 obviously (Fig. 4D).
Compared with the SIRT5-intact HCT116 cells (SIRT5+/+), the SIRT5-null HCT116 cells (SIRT5−/−) showed greater p53 succinylation (Fig. 4E). Reconstitution of SIRT5 in SIRT5-deficient HCT116 cells (SIRT5−/−) caused a significant reduction in succinylation of p53 (Fig. 4F; lane 2 versus lane 1). However, overexpression of SIRT5-H158Y (an enzyme-deficient mutant of SIRT5) [49] had no effect on the reduction of p53 succinylation (Fig. 4F; lane 3 versus lane 1).
Subsequently, we determined the succinylation site(s) on p53 through MS analysis. One succinylation site (lysine 120) in p53 was identified (Fig. 4G). Lysine 120 (K120) of p53 is evolutionary conserved across species (Fig. 4H). To further confirm this succinylated site in p53, we developed a specific antibody against succinylated K120 of p53 (anti-Su-p53-K120 antibody). The specificity of this antibody was validated by dot blot assay (Fig. 4I). Subsequently, we confirmed that p53 could be succinylated at K120 in vitro (Fig. S6A). In addition, in the presence of NAD+, purified SIRT5 (ab101134, Abcam) could indeed induce desuccinylation of succinylated p53 at K120 (Fig. S6B).
In SIRT5-deficient HCT116 cells (SIRT5−/−), succinylation of p53 at K120 was higher than in the SIRT5-intact HCT116 cells (SIRT5+/+) treated with (Fig. 4J; line 4 versus line 3) or without Dox treatment (Fig. 4J; lane 2 versus lane 1).
Similar to treatment with Dox, as expected, succinylation of p53 at K120 was higher in SIRT5-deficient HCT116 cells (SIRT5−/−) than in the SIRT5-intact HCT116 cells (SIRT5+/+) with (Fig. 4K; line 4 versus line 3) or without 5-FU treatment (Fig. 4K; line 2 versus line 1).
Taken together, these findings suggest that SIRT5 might catalyze the desuccinylation of p53 at lysine 120.
SIRT5 has no obvious impact on p53 deacetylation at lysine 120
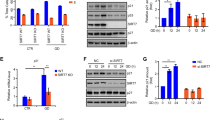
It is reported that p53 acetylation at lysine 120 (K120) within the DNA-binding domain is required for p53-mediated apoptosis and K120 is acetylated by acetyltransferases including Tip60, hMOF, MOZ, and NAT10 [56,57,58,59]. Given that SIRT5 belongs to the sirtuin family of NAD+-dependent deacetylases [60], SIRT5 might also have deacetylase activity even though it is barely detected [61]. To determine whether SIRT5 suppresses p53 transcriptional activity by catalyzing p53 deacetylation at K120, we generated a specific antibody against acetylated K120 of p53 (anti-Ac-p53-K120 antibody). The specificity of this antibody was validated by dot blot assay (Fig. 5A). As shown in Fig. 5B, p53 acetylation at K120 was lower in SIRT5-deficient HCT116 cells (SIRT5−/−) than in the SIRT5-intact HCT116 cells (SIRT5+/+) with (line 4 versus line 3) or without 5-FU treatment (line 2 versus line 1). These results suggest that disruption of SIRT5 does not enhance p53 acetylation at K120, thereby indicating that the deacetylase activity of SIRT5 has no impact on p53 function. Moreover, disruption of SIRT5 also had on effect on p53 acetylation at K379 and K382 (Figure S6C).
A Dot-blot assay for validating the specificity of anti-Ac-p53-K120 antibody. Equal amounts of acetyl-peptides or the control peptides were immunoblotted with the indicated dilutions of anti-Ac-p53-K120 antibody. B Disruption of SIRT5 in HCT116 cells caused a reduction of acetylation of p53 at K120 compared with that in the SIRT5-intact HCT116 cells (SIRT5+/+) treated with or without 5-Fu. The cell lysates from SIRT5-deficient (SIRT5−/−) or SIRT5-intact (SIRT5+/+) HCT116 cells treated with or without 5-Fu (5 μg/ml, 24 h) were immunoprecipitated with anti-p53 antibody, followed by immunoblotting with anti-Ac-p53-K120 antibody. The ratios of Ac-p53/p53 are indicated in blue fonts (DMSO control) or red fonts (5-Fu treatment) respectively. DMSO was used as control. TCL total cell lysate, IP immunoprecipitating. C, D p21 promoter activity (C), and BAX promoter activity (D) in H1299 cells co-transfected Myc-SIRT5 together with HA-p53 or HA-p53-K120R. Data show mean ± SEM; Student’s two-tailed t-test; Data from three independent experiments. E, F Disruption of SIRT5 in HCT116 cells caused more apoptotic cells compared with that in the SIRT5-intact HCT116 cells (SIRT5+/+) treated with 5-Fu (5 μg/ml, 24 h). Representative flow cytometry histograms of apoptotic cells (E) and quantitative analysis (F). SIRT5-deficient (SIRT5−/−) or SIRT5-intact (SIRT5+/+) HCT116 cells treated with or without 5-Fu (5 μg/ml, 24 h) and subsequently stained by annexin V-PI. Percentages in the LR and UR quadrants represent the early and late apoptotic populations, respectively. Data show mean ± SEM; Student’s two-tailed t-test; Data from three independent experiments. G, H Reconstitution of p53-K120R in SIRT5-intact HCT116 cells (SIRT5+/+) and SIRT5-deficient HCT116 cells (SIRT5−/−) counteracted the effect of SIRT5 disruption on apoptotic induction treated with 5-Fu (5 μg/ml, 24 h). Representative flow cytometry histograms of apoptotic cells (G) and quantitative analysis (H). Data show mean ± SEM; Student’s two-tailed t-test; Data from three independent experiments.
Subsequently, we examined the effect of SIRT5 on the transcriptional activity of p53-K120R mutant using promoter assays. The ability of p53-K120R to activate both p21-promoter reporter and BAX-promoter reporter was dramatically reduced compared with that of wild-type (WT) p53 (Fig. 5C, D; column 5 versus column 3), indicating the importance of K120 modification in affecting p53 transcriptional activity [55,56,57,58,59, 62]. Overexpression of SIRT5 caused a significant reduction in WT p53 activity (Fig. 5C, D; column 4 versus column 3), but had no obvious effect on the activity of the p53-K120R mutant (Fig. 5C, D; column 6 versus. column 5). These results suggest that SIRT5 suppresses p53 transcriptional activity by catalyzing p53 desuccinylation at K120.
p53 is a key mediator of apoptosis in DNA damage response [4, 63, 64], and thus, to further determine the function of SIRT5 as mediated by p53, we compared the ratio of apoptotic cells between SIRT5-intact HCT116 cells (SIRT5+/+) and SIRT5-null HCT116 (SIRT5−/−) cells by flow cytometry assays after treatment with 5-FU. More apoptotic cells were detected in SIRT5−/− HCT116 cells (Fig. 5E, F), suggesting that SIRT5 impairs apoptosis in response to DNA damage. However, reconstitution of p53-K120R in SIRT5-intact HCT116 cells (SIRT5+/+) and SIRT5-null HCT116 (SIRT5−/−) cells counteracted the effect of SIRT5 impairment on apoptosis in response to DNA damage (Fig. 5G, H), further suggesting that suppression of p53 by SIRT5 is K120 modification-dependent in cells.
Disruption of SIRT5 promotes p53 enrichment at its target gene promoters in response to DNA damage
Given that lysine 120 (K120) is located within the DNA-binding domain of p53, and SIRT5 does not affect p53-K120R activity, we sought to determine whether SIRT5 has an impact on p53 enrichment in the promoters of p53 target genes in response to DNA damage. As shown in Fig. 6A–D, after cells were treated with Dox, p53 was significantly enriched in the promoters of p21, PUMA, BAX, and MDM2 in SIRT5-null HCT116 cells (SIRT5−/−) compared with SIRT5-intact HCT116 cells (SIRT5+/+) as revealed by qRT-PCR followed by ChIP assays. Similarly, in response to treatment with 5-FU or actinomycin D (ActD), another DNA damaging agent, p53 was significantly enriched in the promoters of p21, PUMA, BAX, and MDM2 in SIRT5-null HCT116 cells (SIRT5−/−) compared with SIRT5-intact HCT116 cells (SIRT5+/+) (Fig. 6E–L).
A–D Disruption of SIRT5 in HCT116 cells (SIRT5−/−) promoted p53 accumulation at the region encompassing p53 consensus binding sites of p21 promoter (A), PUMA promoter (B), BAX promoter (C) and MDM2 promoter (D) in response to Dox treatment. SIRT5-deficient (SIRT5−/−) or SIRT5-intact (SIRT5+/+) HCT116 cells treated with Dox (1 μM, 12 h) or DMSO (control) were crosslinked and harvested for chromatin immunoprecipitation assays (ChIP). Lysates from each condition were divided into equal aliquots and incubated with either p53 antibody or mouse IgG. Precipitated DNA was recovered and analyzed by qRT-PCR with primers that amplify the region encompassing the p53-binding sites within the promoter indicated. Data show mean ± SEM; *p < 0.05, **p < 0.01, ****p < 0.0001, Student’s two-tailed t-test; Data from three independent experiments. E–H Disruption of SIRT5 in HCT116 cells (SIRT5−/−) promoted p53 accumulation at the region encompassing p53 consensus binding sites of p21 promoter (E), PUMA promoter (F), BAX promoter (G), and MDM2 promoter (H) in response to 5-Fu treatment. SIRT5-deficient (SIRT5−/−) or SIRT5-intact (SIRT5+/+) HCT116 cells treated with 5-Fu (5 μg/ml, 12 h) or DMSO (control) were crosslinked and harvested for ChIP. Lysates from each condition were divided into equal aliquots and incubated with either p53 antibody or mouse IgG. Precipitated DNA was recovered and analyzed by qRT-PCR with primers that amplify the region encompassing the p53-binding sites within the promoter indicated. Data show mean ± SEM; *p < 0.05, ****p < 0.0001, Student’s two-tailed t-test; Data from three independent experiments. I–L Disruption of SIRT5 in HCT116 cells (SIRT5−/−) promoted p53 accumulation at the region encompassing p53 consensus binding sites of p21 promoter (I), PUMA promoter (J), BAX promoter (K), and MDM2 promoter (L) in response to ActD (Actinomycin D) treatment. SIRT5-deficient (SIRT5−/−) or SIRT5-intact (SIRT5+/+) HCT116 cells treated with ActD (10 nM, 12 h) or DMSO (control) were crosslinked and harvested for ChIP. Lysates from each condition were divided into equal aliquots and incubated with either p53 antibody or mouse IgG. Precipitated DNA was recovered and analyzed by qRT-PCR with primers that amplify the region encompassing the p53-binding sites within the promoter indicated. Data show mean ± SEM; **p < 0.01, ***p < 0.001, ****p < 0.0001, Student’s two-tailed t-test; Data from three independent experiments.
These results suggest that p53 succinylation facilitates the binding of p53 to the promoters of p53 target genes, and desuccinylation of p53 at K120 that is mediated by SIRT5 impairs p53 enrichment in its target gene promoters, resulting in the suppression of p53 transcriptional activity.
Suppression of p53 function by SIRT5 is dependent on p53 status
To determine whether suppression of p53 function by SIRT5 in response to DNA damage is indeed mediated by p53, we sought to get genetic evidences by taking advantage of p53-knockout mice and Sirt5-knockout mice. Through crossbreeding, we obtained mice as well as mouse embryonic fibroblast (MEF) cells with different genotypes, including Sirt5+/+p53+/+, Sirt5−/−p53+/+, Sirt5+/+p53−/−, and Sirt5−/−p53−/−. Initially, we used MEF cells to examine the effect of Sirt5 on p53 function. When p53 was intact, disruption of Sirt5 caused a significant increase in the expression of p53 target genes, including Puma, Mdm2, and Apaf1 after treatment with Dox or Eto (Etoposide) (Fig. 7A–F). However, in the absence of p53, disruption of Sirt5 had no obvious effect on the expression of p53 target genes (Fig. 7A–F).
A–C qPCR analysis of Puma (A), Mdm2 (B), and Apaf1 (C) mRNA in Sirt5-intact & p53-intact (Sirt5+/+p53+/+), Sirt5-deficient & p53-intact (Sirt5−/−p53+/+), Sirt5-intact and p53-deficient (Sirt5+/+p53−/−), or Sirt5-deficient and p53-deficient (Sirt5−/−p53−/−), treated with or without Dox (1 μM, 12 h). DMSO were used as control. Data show mean ± SEM; Student’s two-tailed t-test; Data from three independent experiments. D–F qPCR analysis of Puma (D), Mdm2 (E), and Apaf1 (F) mRNA in Sirt5-intact & p53-intact (Sirt5+/+p53+/+), Sirt5-deficient & p53-intact (Sirt5−/−p53+/+), Sirt5-intact & p53-deficient (Sirt5+/+p53−/−), or Sirt5-deficient and p53-deficient (Sirt5−/−p53−/−), treated with or without Eto (Etoposide) (5 μM, 12 h). DMSO were used as control. Data show mean ± SEM; Student’s two-tailed t-test; Data from three independent experiments. G, H Flow cytometry analysis of apoptotic cells in Sirt5-intact & p53-intact (Sirt5+/+p53+/+), Sirt5-deficient & p53-intact (Sirt5−/−p53+/+), Sirt5-intact & p53-deficient (Sirt5+/+p53−/−), or Sirt5-deficient and p53-deficient (Sirt5−/−p53−/−) MEF cells treated with or without Dox (1 μM, 12 h). DMSO was used as control. Quantitation is shown H. Data show mean ± SEM; Student’s two-tailed t-test; Data from three independent experiments. I p53 succinylation in Sirt5-intact & p53-intact (Sirt5+/+p53+/+), Sirt5-deficient & p53-intact (Sirt5−/−p53+/+), Sirt5-intact & p53-deficient (Sirt5+/+p53−/−), or Sirt5-deficient and p53-deficient (Sirt5−/−p53−/−) MEF cells were validated by Western blot analysis.
Given that disruption of SIRT5 promotes cell apoptosis (Fig. 5E), and p53 is a key mediator of apoptosis in DNA damage response, we examined whether the enhancement of apoptosis by Sirt5 is mediated by p53. In the presence of p53, more apoptotic cells were detected in Sirt5-deficient MEF cells compared with Sirt5-intact MEF cells after Dox treatment (Fig. 7G, H). However, in the absence of p53, no significant difference in apoptosis was observed between Sirt5-intact and Sirt5-deficient MEF cells (Fig. 7G, H). The disruption of Sirt5 and p53 and the effect of Sirt5 on p53 desuccinylation in the context of double knockout MEF cells were confirmed by western blot analysis (Fig. 7I).
To further determine whether suppression of p53 function by Sirt5 in response to DNA damage is mediated by p53 in vivo, we examined apoptosis in mice after challenging with ionizing radiation (IR). Sirt5+/+p53+/+, Sirt5−/−p53+/+, Sirt5+/+p53−/−, and Sirt5−/−p53−/− littermates were treated with IR (5 Gray), then their testis, spleen and intestine were analyzed by Terminal deoxynucleotidyl transferase dUTP Nick End Labeling (TUNEL) staining, using liver as negative control [65, 66]. We observed a remarkable increase in apoptosis levels in tissues from Sirt5−/−p53+/+ mice relative to the Sirt5+/+p53+/+ mice. However, no significant difference in apoptosis levels in tissues was detected between Sirt5+/+p53−/− mice and Sirt5−/−p53−/− mice (Fig. 8A–F). In these conditions, no obvious apoptotic effects were observed in the livers from mice with four different genotypes (Fig. 8G). Moreover, succinylation of p53 at K120 in mice after challenging with IR was also determined by anti-Su-p53-K120 antibody (Fig. S8). In consistent with apoptosis levels, a remarkable higher succinylated p53 at K120 was seen in intestine and testis but not in liver of Sirt5−/−p53+/+ mice compared with those of Sirt5+/+p53+/+ mice (Fig. S8).
A, B Testicular tissue from Sirt5+/+p53+/+, Sirt5−/−p53+/+, Sirt5+/+p53−/−, and Sirt5−/−p53−/− mice untreated or after 5 Gy (Gray) IR (Ionizing radiation) treatment. TUNEL staining was used for detecting apoptotic cells. Quantitation is shown B. Data show mean ± SEM; Student’s two-tailed t-test. Scale bar = 50 µm. C, D Spleen tissue from Sirt5+/+p53+/+, Sirt5−/−p53+/+, Sirt5+/+p53−/−, and Sirt5−/−p53−/− mice untreated or after 5 Gy IR treatment. TUNEL staining was used for detecting apoptotic cells. Quantitation is shown (D). Data show mean ± SEM; Student’s two-tailed t-test. Scale bar = 150 µm. E, F Intestine tissue from Sirt5+/+p53+/+, Sirt5−/−p53+/+, Sirt5+/+p53−/−, and Sirt5−/−p53−/− mice untreated or after 5Gy IR treatment. TUNEL staining was used for detecting apoptotic cells. Quantitation is shown (F). Data show mean ± SEM; Student’s two-tailed t-test. Scale bar = 200 µm. G Liver tissue from Sirt5+/+p53+/+, Sirt5−/−p53+/+, Sirt5+/+p53−/−, and Sirt5−/−p53−/− mice untreated or after 5 Gy IR treatment. TUNEL staining was used for detecting apoptotic cells. Scale bar = 150 µm.
Collectively, these data suggest that the suppressive role of SIRT5 on p53 function is mediated by p53, further validating the impact of SIRT5 on p53 function.
Discussion
In the current study, we found that p53 is succinylated at K120, and SIRT5 mediates its desuccinylation. Furthermore, we demonstrate that SIRT5 suppresses p53 function, including suppression of p53 target gene expression and of p53-mediated apoptosis. These findings not only reveal a novel PTM of p53, but also identify a new target of SIRT5, that is, acting as a desuccinylase.
It is noteworthy that SIRT5 localizes mostly or exclusively to the mitochondrial matrix [61]. In line with this notion, SIRT5-catalyzed desuccinylation of mitochondrial proteins has been uncovered [31, 49]. p53 is a classic transcriptional factor that mainly transactivates gene expression in nucleus even though its other roles outside the nucleus have also been demonstrated [67,68,69,70,71]. Here, we observed that SIRT5 co-localizes with p53 in the nucleus and suppresses p53 transcriptional activity, implicating that SIRT5 could also act its function extra-mitochondrially. Virtually, SIRT5 has been detected in the nuclear fraction, in which it catalyses desuccinylation of nuclear proteins [30, 72].
K120 within the DNA-binding domain of p53 is evolutionary conserved in all of the species known to encode p53 and frequently mutated in human tumors. K120 appears to be one of the key p53 residues that modulates its essential functions [55,56,57,58,59, 62]. K120 of p53 can be acetylated by Tip60/hMOF/NAT10 [56, 57, 59, 73]. It has been proposed that K120 acetylation is likely to be crucial for p53-mediated apoptosis, but has no obvious effect on cell cycle arrest, thus defining a unique function of K120 acetylation [56, 57, 62, 74]. Here, we determined that p53 is succinylated at K120, and SIRT5 catalyses its desuccinylation. Our findings uncover another modification of K120 and reveal its functional importance. Interestingly, based on proteomic analysis of protein modification, lysine succinylation has been found to extensively overlap with acetylation in prokaryotes and eukaryotes [25]. Our findings provide a real example of functional overlap between acetylation and succinylation. It appears that K120 acetylation and succinylation simultaneously enhance p53 transcriptional activity. Nevertheless, differences are observed in the acetylation and succinylation of p53 K120. With the exception of apoptosis-related genes targeted by p53, such as PUMA, BAX, and PIG3 [57], other p53 target genes, including p21, MDM2, TIGAR, and SFN, are also suppressed by SIRT5-mediated desuccinylation of p53. SIRT5-mediated desuccinylation is likely to affect the expression of the majority of p53 target genes. Notably, the activation of the p53-K120R mutant on p21-promoter and BAX-promoter is significantly reduced compared with that of WT p53, but SIRT5 has no obvious effect on the activation of p53-K120R mutant, suggesting that K120 modification can influence the expression of genes related to apoptosis and cell cycle arrest, and SIRT5 suppresses p53 target gene expression by catalyzing desuccinylation of p53 K120.
p53-K120R mutant still showed a slightly reduced ability to activate p21 expression although this was not as significant as that on activating PUMA expression [56, 62]. In addition, a recent study on NAT10-mediating p53 acetylation at K120 also indicated that acetylation of 53 at K120 can affect the expression of p21, PUMA, and MDM2 [59]. Therefore, K120 modification not only affects p53-mediated apoptosis but also influences other functions of p53. Of note, p53-K120R mutant could specifically activate NFAT signaling [55]. Taken together, our findings further highlight the importance of K120 of p53 in modulating p53 function.
In this study, we only identify that p53 is succinylated at K120, and SIRT5 catalyses its desuccinylated, resulting in the modulation of p53 target gene expression. As MS analysis has limited accuracy, we cannot rule out other lysine residues in p53 can also be succinylated, and SIRT5 mediates desuccinylation of these sites. Nonetheless, our findings strongly suggest that K120 of p53 is the key site for succinylation, and is desuccinylated by SIRT5.
The crosstalk between different modifications, which synergistically or antagonistically affects target function, has been extensively investigated [12, 14]. K120 of p53 is not only modified by acetylation but also by succinylation, which seems to simultaneously enhance p53 transcriptional activity, but as one falls, another rises. It will be very interesting to further address how the acetylases/deacetylases and succinylases/desuccinylases cooperate to modulate p53 function by modifying K120. Based on the previous studies [56, 57, 59], it is evident that acetylation on p53 K120 promotes apoptosis. Here, we showed that disruption of SIRT5 induced apoptosis, but it actually caused the enhancement of succinylation and the reduction of acetylation on p53 at K120. It appears that succinylation resulted from SIRT5-disruption competes with acetylation on p53 at K120 to reduce acetylation of p53 K120, resulted in apoptosis. Perhaps, the enhancement of succinylation rather than the reduction of acetylation on p53 at K120 by SIRT5 disruption accounts for the main cause of apoptosis induction. However, it still cannot rule out the possibility that acetylation on p53 at K120 catalyzed by other factors can also induce apoptosis. This inconsistency might be resulted from different levels of modification existed between succinylation and acetylation or different systems employed by these studies. Further investigations will get insights into the underlying mechanisms.
Increasing evidence indicates that SIRT5 plays an oncogenic role [33, 34, 36, 37, 39]. Various mechanisms have been proposed to describe its role in promoting tumorigenesis. As a well-defined tumor suppressor, p53 inhibits tumor initiation and progression through multiple ways even though K120R mutation does not seem to contribute to early-onset tumor formation [8, 74]. Here, we provide evidence showing that suppression of p53 target gene expression and of p53-induced apoptosis by SIRT5 is actually dependent on p53 status. Therefore, SIRT5 possibly promotes tumorigenesis by suppressing p53 function. These findings reveal a probably unique mechanism by which SIRT5 exhibits its oncogenic function.
Data availability
Further information and requests for resources and reagents should be directed to and will be fulfilled by W.X.
References
Vousden KH, Lu X. Live or let die: the cell’s response to p53. Nat Rev Cancer. 2002;2:594–604.
Vogelstein B, Lane D, Levine AJ. Surfing the p53 network. Nature. 2000;408:307–10.
Bykov VJN, Eriksson SE, Bianchi J, Wiman KG. Targeting mutant p53 for efficient cancer therapy. Nat Rev Cancer. 2018;18:89–102.
Lane DP. Cancer. p53, guardian of the genome. Nature. 1992;358:15–6.
Muller PAJ, Vousden KH. Mutant p53 in Cancer: New Functions and Therapeutic Opportunities. Cancer Cell. 2014;25:304–17.
Grochola LF, Zeron-Medina J, Meriaux S, Bond GL. Single-nucleotide polymorphisms in the p53 signaling pathway. Cold Spring Harb Perspect Biol. 2010;2:a001032.
Saldana-Meyer R, Recillas-Targa F. Transcriptional and epigenetic regulation of the p53 tumor suppressor gene. Epigenetics. 2011;6:1068–77.
Vieler M, Sanyal S. p53 isoforms and their implications in cancer. Cancers. 2018;10:288.
Walerych D, Kudla G, Gutkowska M, Wawrzynow B, Muller L, King FW, et al. Hsp90 chaperones wild-type p53 tumor suppressor protein. J Biol Chem. 2004;279:48836–45.
Liang SH, Clarke MF. Regulation of p53 localization. Eur J Biochem. 2001;268:2779–83.
Brooks CL, Gu W. Ubiquitination, phosphorylation and acetylation: the molecular basis for p53 regulation. Curr Opin Cell Biol. 2003;15:164–71.
Dai C, Gu W. p53 post-translational modification: deregulated in tumorigenesis. Trends Mol Med. 2010;16:528–36.
Gu B, Zhu WG. Surf the Post-translational Modification Network of p53 Regulation. Int J Biol Sci. 2012;8:672–84.
Liu YQ, Tavana O, Gu W. p53 modifications: exquisite decorations of the powerful guardian. J Mol Cell Biol. 2019;11:564–77.
Hafner A, Bulyk ML, Jambhekar A, Lahav G. The multiple mechanisms that regulate p53 activity and cell fate. Nat Rev Mol Cell Biol. 2019;20:199–210.
Kastenhuber ER, Lowe SW. Putting p53 in context. Cell. 2017;170:1062–78.
Joerger AC, Fersht AR. The p53 pathway: origins, inactivation in cancer, and emerging therapeutic approaches. Annu Rev Biochem. 2016;85:375–404.
Kruiswijk F, Labuschagne CF, Vousden KH. p53 in survival, death and metabolic health: a lifeguard with a licence to kill. Nat Rev Mol Cell Biol. 2015;16:393–405.
Zhang ZH, Tan MJ, Xie ZY, Dai LZ, Chen Y, Zhao YM. Identification of lysine succinylation as a new post-translational modification. Nat Chem Biol. 2011;7:58–63.
Alleyn M, Breitzig M, Lockey R, Kolliputi N. The dawn of succinylation: a posttranslational modification. Am J Physiol Cell Physiol. 2018;314:C228–C32.
Yang Y, Gibson GE. Succinylation links metabolism to protein functions. Neurochem Res. 2019;44:2346–59.
Weinert BT, Iesmantavicius V, Wagner SA, Scholz C, Gummesson B, Beli P, et al. Acetyl-phosphate is a critical determinant of lysine acetylation in E. coli. Mol Cell. 2013;51:265–72.
Colak G, Xie ZY, Zhu AY, Dai LZ, Lu ZK, Zhang Y, et al. Identification of lysine succinylation substrates and the succinylation regulatory enzyme CobB in Escherichia coli. Mol Cell Proteomics. 2013;12:3509–20.
Hirschey MD, Zhao YM. Metabolic regulation by lysine malonylation, succinylation, and glutarylation. Mol Cell Proteomics. 2015;14:2308–15.
Weinert BT, Scholz C, Wagner SA, Iesmantavicius V, Su D, Daniel JA, et al. Lysine succinylation is a frequently occurring modification in prokaryotes and eukaryotes and extensively overlaps with acetylation. Cell Rep. 2013;4:842–51.
Xie ZY, Dai JBA, Dai LZ, Tan MJ, Cheng ZY, Wu YM, et al. Lysine succinylation and lysine malonylation in histones. Mol Cell Proteomics. 2012;11:100–7.
Xu H, Chen XY, Xu XL, Shi RY, Suo SS, Cheng KY, et al. Lysine acetylation and succinylation in hela cells and their essential roles in response to UV-induced stress. Sci Rep. 2016;6:30212.
Gibson GE, Xu H, Chen HL, Chen W, Denton TT, Zhang S. Alpha-ketoglutarate dehydrogenase complex-dependent succinylation of proteins in neurons and neuronal cell lines. J Neurochem. 2015;134:86–96.
Du JT, Zhou YY, Su XY, Yu JJ, Khan S, Jiang H, et al. Sirt5 is a NAD-dependent protein lysine demalonylase and desuccinylase. Science. 2011;334:806–9.
Park J, Chen Y, Tishkoff DX, Peng C, Tan MJ, Dai LZ, et al. SIRT5-mediated lysine desuccinylation impacts diverse metabolic pathways. Mol Cell. 2013;50:919–30.
Rardin MJ, He WJ, Nishida Y, Newman JC, Carrico C, Danielson SR, et al. SIRT5 regulates the mitochondrial lysine succinylome and metabolic networks. Cell Metab. 2013;18:920–33.
Wang F, Wang K, Xu W, Zhao SM, Ye D, Wang Y, et al. SIRT5 desuccinylates and activates pyruvate kinase M2 to block macrophage IL-1 beta production and to prevent DSS-induced colitis in mice. Cell Rep. 2017;19:2331–44.
Shi L, Yan H, An S, Shen M, Jia W, Zhang R, et al. SIRT5-mediated deacetylation of LDHB promotes autophagy and tumorigenesis in colorectal cancer. Mol Oncol. 2019;13:358–75.
Greene KS, Lukey MJ, Wang XY, Blank B, Druso JE, Lin MCJ, et al. SIRT5 stabilizes mitochondrial glutaminase and supports breast cancer tumorigenesis. Proc Natl Acad Sci USA. 2019;116:26625–32.
Zhang R, Wang C, Tian Y, Yao Y, Mao J, Wang H, et al. SIRT5 promotes hepatocellular carcinoma progression by regulating mitochondrial apoptosis. J Cancer. 2019;10:3871–82.
Ma Y, Qi Y, Wang L, Zheng Z, Zhang Y, Zheng J. SIRT5-mediated SDHA desuccinylation promotes clear cell renal cell carcinoma tumorigenesis. Free Radic Biol Med. 2019;134:458–67.
Yang X, Wang Z, Li X, Liu B, Liu M, Liu L, et al. SHMT2 desuccinylation by SIRT5 drives cancer cell proliferation. Cancer Res. 2018;78:372–86.
Chen XF, Tian MX, Sun RQ, Zhang ML, Zhou LS, Jin L, et al. SIRT5 inhibits peroxisomal ACOX1 to prevent oxidative damage and is downregulated in liver cancer. EMBO Rep. 2018;19:e45124.
Wang YQ, Wang HL, Xu J, Tan J, Fu LN, Wang JL, et al. Sirtuin5 contributes to colorectal carcinogenesis by enhancing glutaminolysis in a deglutarylation-dependent manner. Nat Commun. 2018;9:545.
Greene KS, Lukey MJ, Wang X, Blank B, Druso JE, Lin M-CJ, et al. SIRT5 stabilizes mitochondrial glutaminase and supports breast cancer tumorigenesis. Proceedings of the National Academy of Sciences 2019;11626625–32. https://doi.org/10.1073/pnas.1911954116.
Feng M, Pan y, Kong R, Shu S. Therapy of Primary Liver Cancer. Innovation. 2020;1100032. https://doi.org/10.1016/j.xinn.2020.100032.
Jiao D, Yang S. Overcoming Resistance to Drugs Targeting KRAS Mutation. The Innovation 2020;1100035. https://doi.org/10.1016/j.xinn.2020.100035.
Liu J, Zhang C, Hu WW, Feng ZH. Tumor suppressor p53 and metabolism. J Mol Cell Biol. 2019;11:284–92.
Li L, Mao YX, Zhao LN, Li LJ, Wu JJ, Zhao MJ, et al. p53 regulation of ammonia metabolism through urea cycle controls polyamine biosynthesis. Nature. 2019;567:253.
Basu S, Gnanapradeepan K, Barnoud T, Kung CP, Tavecchio M, Scott J, et al. Mutant p53 controls tumor metabolism and metastasis by regulating PGC-1 alpha. Gene Dev. 2018;32:230–43.
Mai WX, Gosa L, Daniels VW, Ta L, Tsang JE, Higgins B, et al. Cytoplasmic p53 couples oncogene-driven glucose metabolism to apoptosis and is a therapeutic target in glioblastoma. Nat Med. 2017;23:1342.
Humpton TJ, Vousden KH. Regulation of cellular metabolism and hypoxia by p53. Col Spring Harb Perspect Med. 2016;6:a026146.
Bensaad K, Tsuruta A, Selak MA, Vidal MN, Nakano K, Bartrons R, et al. TIGAR, a p53-inducible regulator of glycolysis and apoptosis. Cell. 2006;126:107–20.
Liu X, Zhu C, Zha H, Tang J, Rong F, Chen X, et al. SIRT5 impairs aggregation and activation of the signaling adaptor MAVS through catalyzing lysine desuccinylation. EMBO J. 2020;39:e103285.
Wang J, Zhang D, Du J, Zhou C, Li Z, Liu X, et al. Tet1 facilitates hypoxia tolerance by stabilizing the HIF-alpha proteins independent of its methylcytosine dioxygenase activity. Nucleic Acids Res. 2017;45:12700–14.
Liu X, Chen Z, Xu C, Leng X, Cao H, Ouyang G, et al. Repression of hypoxia-inducible factor α signaling by Set7-mediated methylation. Nucleic Acids Res. 2015;43:5081–98.
Vousden KH, Prives C. Blinded by the light: the growing complexity of p53. Cell. 2009;137:413–31.
Castellini L, Moon EJ, Razorenova OV, Krieg AJ, von Eyben R, Giaccia AJ. KDM4B/JMJD2B is a p53 target gene that modulates the amplitude of p53 response after DNA damage. Nucleic Acids Res. 2017;45:3674–92.
Vassilev LT, Vu BT, Graves B, Carvajal D, Podlaski F, Filipovic Z, et al. In vivo activation of the p53 pathway by small-molecule antagonists of MDM2. Science. 2004;303:844–8.
Shinmen N, Koshida T, Kumazawa T, Sato K, Shimada H, Matsutani T, et al. Activation of NFAT signal by p53-K120R mutant. FEBS Lett. 2009;583:1916–22.
Tang Y, Luo J, Zhang W, Gu W. Tip60-dependent acetylation of p53 modulates the decision between cell-cycle arrest and apoptosis. Mol Cell. 2006;24:827–39.
Sykes SM, Mellert HS, Holbert MA, Li K, Marmorstein R, Lane WS, et al. Acetylation of the p53 DNA-binding domain regulates apoptosis induction. Mol Cell. 2006;24:841–51.
Rokudai S, Laptenko O, Arnal SM, Taya Y, Kitabayashi I, Prives C. MOZ increases p53 acetylation and premature senescence through its complex formation with PML. Proc Natl Acad Sci USA. 2013;110:3895–900.
Liu X, Tan Y, Zhang C, Zhang Y, Zhang L, Ren P, et al. NAT10 regulates p53 activation through acetylating p53 at K120 and ubiquitinating Mdm2. EMBO Rep. 2016;17:349–66.
Finkel T, Deng CX, Mostoslavsky R. Recent progress in the biology and physiology of sirtuins. Nature. 2009;460:587–91.
Anderson KA, Green MF, Huynh FK, Wagner GR, Hirschey MD. SnapShot: mammalian sirtuins. Cell. 2014;159:956–e1.
Li T, Kon N, Jiang L, Tan M, Ludwig T, Zhao Y, et al. Tumor suppression in the absence of p53-mediated cell-cycle arrest, apoptosis, and senescence. Cell. 2012;149:1269–83.
Reinhardt HC, Schumacher B. The p53 network: cellular and systemic DNA damage responses in aging and cancer. Trends Genet. 2012;28:128–36.
Lakin ND, Jackson SP. Regulation of p53 in response to DNA damage. Oncogene. 1999;18:7644–55.
Erster S, Mihara M, Kim RH, Petrenko O, Moll UM. In vivo mitochondrial p53 transloclation triggers a rapid first wave of cell death in response to DNA damage that can precede p53 target gene activation. Mol Cell Biol. 2004;24:6728–41.
Castelli M, Piobbico D, Chiacchiaretta M, Brunacci C, Pieroni S, Bartoli D, et al. HOPS/TMUB1 retains p53 in the cytoplasm and sustains p53-dependent mitochondrial apoptosis. EMBO Rep. 2020;21:e48073.
Comel A, Sorrentino G, Capaci V, Sal GD. The cytoplasmic side of p53’s oncosuppressive activities. FEBS Lett. 2014;588:2600–9.
Marchenko ND, Moll UM. Mitochondrial death functions of p53. Mol Cell Oncol. 2014;1:e955995.
Mihara M, Erster S, Zaika A, Petrenko O, Chittenden T, Pancoska P, et al. p53 has a direct apoptogenic role at the mitochondria. Mol Cell. 2003;11:577–90.
Chipuk JE, Kuwana T, Bouchier-Hayes L, Droin NM, Newmeyer DD, Schuler M, et al. Direct activation of Bax by p53 mediates mitochondrial membrane permeabilization and apoptosis. Science. 2004;303:1010–4.
Castelli M, Piobbico D, Chiacchiaretta M, Brunacci C, Pieroni S, Bartoli D, et al. HOPS/TMUB1 retains p53 in the cytoplasm and sustains p53-dependent mitochondrial apoptosis. EMBO Rep. 2020;21:e48073.
Shahidian LZ, Haas M, Gras SL, Nitsch S, Mourão A, Geerlof A, et al. Succinylation of H3K122 destabilizes nucleosomes and enhances transcription. EMBO reports 2021;22:e51009. https://doi.org/10.15252/embr.202051009.
Li X, Wu L, Corsa CA, Kunkel S, Dou Y. Two mammalian MOF complexes regulate transcription activation by distinct mechanisms. Mol Cell. 2009;36:290–301.
Tang Y, Zhao WH, Chen Y, Zhao YM, Gu W. Acetylation is indispensable for p53 activation (vol 133, pg 612, 2008). Cell. 2008;133:1290.
Acknowledgements
We are grateful to Drs. Bert Vogelstein, Moshe Oren, Scott Lowe, and John Reed for reagents and also grateful to Yan Wang for helping in flow cytometry assay and Fang Zhou for helping in fluorescent microscope. This work was supported by National Natural Science Foundation of China Grant 31830101, 31721005; the Strategic Priority Research Program of the Chinese Academy of Sciences Grant XDA24010308, and National Key R & D Program of China 2018YFD0900602, and Youth Innovation Promotion Association (CAS).
Author information
Authors and Affiliations
Contributions
X.L., F.R., and W.X. designed the projects and analysed the data. X.L. and F.R. performed the experiments. J.T., C.Z., X.C., S.J., Z.W., X.S., H.D., H.Z., and G.O. helped with the experiments or provided reagents. X.L., F.R., and W.X. prepared the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Ethical approval
All animal protocols were approved by the Institutional Animal Care and Use Committee (IACUC) at Institute of Hydrobiology, Chinese Academy of Science.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Edited by M Oren
Rights and permissions
About this article
Cite this article
Liu, X., Rong, F., Tang, J. et al. Repression of p53 function by SIRT5-mediated desuccinylation at Lysine 120 in response to DNA damage. Cell Death Differ 29, 722–736 (2022). https://doi.org/10.1038/s41418-021-00886-w
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41418-021-00886-w
This article is cited by
-
Sirtuins in kidney diseases: potential mechanism and therapeutic targets
Cell Communication and Signaling (2024)
-
SIRT5-mediated ME2 desuccinylation promotes cancer growth by enhancing mitochondrial respiration
Cell Death & Differentiation (2024)
-
Histidine re-sensitizes pediatric acute lymphoblastic leukemia to 6-mercaptopurine through tetrahydrofolate consumption and SIRT5-mediated desuccinylation
Cell Death & Disease (2024)
-
DNA Damage-driven Inflammatory Cytokines: Reprogramming of Tumor Immune Microenvironment and Application of Oncotherapy
Current Medical Science (2024)
-
SIRT5-related desuccinylation modification of AIFM1 protects against compression-induced intervertebral disc degeneration by regulating mitochondrial homeostasis
Experimental & Molecular Medicine (2023)