Abstract
Background
Traditional histopathology performed by pathologists through naked eyes is insufficient for accurate survival prediction of clear cell renal cell carcinoma (ccRCC).
Methods
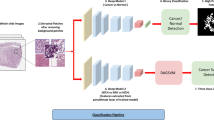
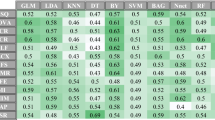
A total of 483 whole slide images (WSIs) data from three patient cohorts were retrospectively analyzed. We performed machine learning algorithm to identify optimal digital pathological features and constructed machine learning-based pathomics signature (MLPS) for ccRCC patients. Prognostic performance of the prognostic model was also verified in two independent validation cohorts.
Results
MLPS could significantly distinguish ccRCC patients with high survival risk, with hazard ratio of 15.05, 4.49 and 1.65 in three independent cohorts, respectively. Cox regression analysis revealed that the MLPS could act as an independent prognostic factor for ccRCC patients. Integration nomogram based on MLPS, tumour stage system and tumour grade system improved the current survival prediction accuracy for ccRCC patients, with area under curve value of 89.5%, 90.0%, 88.5% and 85.9% for 1-, 3-, 5- and 10-year disease-free survival prediction.
Discussion
The machine learning-based pathomics signature could act as a novel prognostic marker for patients with ccRCC. Nevertheless, prospective studies with multicentric patient cohorts are still needed for further verifications.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 24 print issues and online access
$259.00 per year
only $10.79 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout



Similar content being viewed by others
Data availability
Data supporting the findings of this study are available within the supplementary information and are also available from the authors upon reasonable request.
Code availability
The code used for slide image process and feature extraction had been made publicly available at https://qupath.github.io/ [17].
References
Siegel RL, Miller KD, Fuchs HE, Jemal A. Cancer statistics, 2021. CA Cancer J Clin. 2021;71:7–33.
Humphrey PA, Moch H, Cubilla AL, Ulbright TM, Reuter VE. The 2016 WHO classification of tumours of the urinary system and male genital organs-Part B: prostate and bladder tumours. Eur Urol. 2016;70:106–19.
Liu L, Qi L, Li Y, Chen M, Li C, Cheng X, et al. Retroperitoneoscopic partial nephrectomy for moderately complex ventral hilar tumors: surgical technique and trifecta outcomes from a single institution in China. J Laparoendosc Adv Surg Tech Part A. 2017;27:812–7.
Yi X, Xiao Q, Zeng F, Yin H, Li Z, Qian C, et al. Computed tomography radiomics for predicting pathological grade of renal cell carcinoma. Front Oncol. 2020;10:570396.
Inamura K, Fujiwara T, Hoshida Y, Isagawa T, Jones MH, Virtanen C, et al. Two subclasses of lung squamous cell carcinoma with different gene expression profiles and prognosis identified by hierarchical clustering and non-negative matrix factorization. Oncogene. 2005;24:7105–13.
Chen ST, Zhang N, Jiang LR, Gao F, Shao JL, Wang T, et al. Clinical use of a machine learning histopathological image signature in diagnosis and survival prediction of clear cell renal cell carcinoma. Int J Cancer. 2021;148:780–90.
Delahunt B, Cheville JC, Martignoni G, Humphrey PA, Magi-Galluzzi C, McKenney J, et al. The International Society of Urological Pathology (ISUP) grading system for renal cell carcinoma and other prognostic parameters. Am J Surg Pathol. 2013;37:1490–504.
Klimov S, Miligy IM, Gertych A, Jiang Y, Toss MS, Rida P, et al. A whole slide image-based machine learning approach to predict ductal carcinoma in situ (DCIS) recurrence risk. Breast Cancer Res. 2019;21:83.
Louis DN, Gerber GK, Baron JM, Bry L, Dighe AS, Getz G, et al. Computational pathology: an emerging definition. Arch Pathol Lab Med. 2014;138:1133–8.
Gertych A, Ing N, Ma Z, Fuchs TJ, Salman S, Mohanty S, et al. Machine learning approaches to analyze histological images of tissues from radical prostatectomies. Comput Med Imaging Graph. 2015;46:197–208.
Ehteshami Bejnordi B, Veta M, Johannes van Diest P, van Ginneken B, Karssemeijer N, Litjens G, et al. Diagnostic assessment of deep learning algorithms for detection of lymph node metastases in women with breast cancer. JAMA. 2017;318:2199–210.
Yu KH, Zhang C, Berry GJ, Altman RB, Re C, Rubin DL, et al. Predicting non-small cell lung cancer prognosis by fully automated microscopic pathology image features. Nat Commun. 2016;7:12474.
Kickingereder P, Isensee F, Tursunova I, Petersen J, Neuberger U, Bonekamp D, et al. Automated quantitative tumour response assessment of MRI in neuro-oncology with artificial neural networks: a multicentre, retrospective study. Lancet Oncol. 2019;20:728–40.
Brinker TJ, Hekler A, Enk AH, Berking C, Haferkamp S, Hauschild A, et al. Deep neural networks are superior to dermatologists in melanoma image classification. Eur J Cancer. 2019;119:11–7.
Cao R, Yang F, Ma SC, Liu L, Zhao Y, Li Y, et al. Development and interpretation of a pathomics-based model for the prediction of microsatellite instability in Colorectal Cancer. Theranostics. 2020;10:11080–91.
Clark K, Vendt B, Smith K, Freymann J, Kirby J, Koppel P, et al. The Cancer Imaging Archive (TCIA): maintaining and operating a public information repository. J Digit Imaging. 2013;26:1045–57.
Bankhead P, Loughrey MB, Fernández JA, Dombrowski Y, McArt DG, Dunne PD, et al. QuPath: Open source software for digital pathology image analysis. Sci Rep. 2017;7:16878.
Kulkarni PM, Robinson EJ, Sarin Pradhan J, Gartrell-Corrado RD, Rohr BR, Trager MH, et al. Deep learning based on standard H&E images of primary melanoma tumors identifies patients at risk for visceral recurrence and death. Clin Cancer Res. 2020;26:1126–34.
Friedman J, Hastie T, Tibshirani R. Regularization paths for generalized linear models via coordinate descent. J Stat Softw. 2010;33:1–22.
Schuettfort VM, Pradere B, Rink M, Comperat E, Shariat SF. Pathomics in urology. Curr Opin Urol. 2020;30:823–31.
Jiang Y, Jin C, Yu H, Wu J, Chen C, Yuan Q, et al. Development and validation of a deep learning CT signature to predict survival and chemotherapy benefit in gastric cancer: a multicenter, retrospective study. Ann Surg. 2021;274:e1153–61.
Esteva A, Kuprel B, Novoa RA, Ko J, Swetter SM, Blau HM, et al. Dermatologist-level classification of skin cancer with deep neural networks. Nature. 2017;542:115–8.
Zhao K, Li Z, Yao S, Wang Y, Wu X, Xu Z, et al. Artificial intelligence quantified tumour-stroma ratio is an independent predictor for overall survival in resectable colorectal cancer. EBioMedicine. 2020;61:103054.
Beer DG, Kardia SL, Huang CC, Giordano TJ, Levin AM, Misek DE, et al. Gene-expression profiles predict survival of patients with lung adenocarcinoma. Nat Med. 2002;8:816–24.
Acknowledgements
We appreciate the QuPath digital pathology software for its contribution. Open data used in this publication were generated by the National Cancer Institute Clinical Proteomic Tumor Analysis Consortium (CPTAC) and TCGA database.
Funding
This work was supported by the National Natural Science Foundation of China (81972393 and 82002665). The funding sources had no role in the design of the study; collection, analysis or interpretation of the data; writing of the report; the decision to submit for publication.
Author information
Authors and Affiliations
Contributions
NZ, XW, JZ and SC participated in the study conception and design, data collection, data analysis, reviewed the paper and approved the final draft for submission. LJ, FG, EZ and TW participated in data collection, reviewed the paper and approved the final draft for submission.
Corresponding authors
Ethics declarations
Ethics approval and consent to participate
The ethical approval of this study has been approved from the Research Ethics Committee of Shanghai General Hospital (approval number: 2021SQ121). Not further ethical approval was required since all the slice images from the CPTAC cohort and the TCGA cohort were publicly available for research purposes.
Consent to publish
Not applicable.
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
About this article
Cite this article
Chen, S., Jiang, L., Gao, F. et al. Machine learning-based pathomics signature could act as a novel prognostic marker for patients with clear cell renal cell carcinoma. Br J Cancer 126, 771–777 (2022). https://doi.org/10.1038/s41416-021-01640-2
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41416-021-01640-2
This article is cited by
-
Association of the pathomics-collagen signature with lymph node metastasis in colorectal cancer: a retrospective multicenter study
Journal of Translational Medicine (2024)
-
Demographic bias in misdiagnosis by computational pathology models
Nature Medicine (2024)
-
The Molecular Twin artificial-intelligence platform integrates multi-omic data to predict outcomes for pancreatic adenocarcinoma patients
Nature Cancer (2024)
-
Application of radiomics-based multiomics combinations in the tumor microenvironment and cancer prognosis
Journal of Translational Medicine (2023)
-
Development and validation of a radiopathomic model for predicting pathologic complete response to neoadjuvant chemotherapy in breast cancer patients
BMC Cancer (2023)