Abstract
Celastrol is a triterpene derived from the traditional Chinese medicine Tripterygium wilfordii Hook f, which displays potential anticancer activity. In the present study, we investigated the anticancer effects of celastrol against clear cell renal cell carcinoma (ccRCC) and the underlying mechanisms. Using Cancer Genome Atlas (TCGA) database and genotype-tissue expression (GTEx) database we conducted a bioinformatics analysis, which showed that the mRNA levels of liver-X receptors α (LXRα) and ATP-binding cassette transporter A1 (ABCA1) in ccRCC tissues were significantly lower than those in adjacent normal tissues. This result was confirmed by immunoblotting analysis of 4 ccRCC clinical specimens, which showed that the protein expression of LXRα and ABCA1 was downregulated. Similar results were obtained in a panel of ccRCC cell lines (786-O, A498, SN12C, and OS-RC-2). In 786-O and SN12C cells, treatment with celastrol (0.25–2.0 μM) concentration-dependently inhibited the cell proliferation, migration, and invasion as well as the epithelial-mesenchymal transition (EMT) process. Furthermore, we demonstrated that celastrol inhibited the invasion of 786-O cells through reducing lipid accumulation; celastrol concentration-dependently promoted autophagy to reduce lipid storage. Moreover, we revealed that celastrol dramatically activated LXRα signaling, and degraded lipid droplets by inducing lipophagy in 786-O cells. Finally, celastrol promoted cholesterol efflux from 786-O cells via ABCA1. In high-fat diet-promoted ccRCC cell line 786-O xenograft model, administration of celastrol (0.25, 0.5, 1.0 mg·kg−1·d−1, for 4 weeks, i.p.) dose-dependently inhibited the tumor growth with upregulated LXRα and ABCA1 protein in tumor tissue. In conclusion, this study reveals that celastrol triggers lipophagy in ccRCC by activating LXRα, promotes ABCA1-mediated cholesterol efflux, suppresses EMT progress, and ultimately inhibits cell proliferation, migration, and invasion as well as tumor growth. Thus, our study provides evidence that celastrol can be used as a lipid metabolism-based anticancer therapeutic approach.
Similar content being viewed by others
Introduction
As the most commonly occurring form of primary renal tumor, renal cell carcinoma (RCC) is a malignancy with a high mortality rate [1]. Clear cell renal cell carcinoma (ccRCC) is the most prevalent histological subtype of RCC, accounting for ~75% of RCCs [2]. Nearly 35% of ccRCC patients who have local invasion or distant metastasis after nephrectomy need further chemotherapy or radiotherapy. Unfortunately, ccRCC is not sensitive to current chemotherapy and radiotherapy modalities [3]. Therefore, it is critical to recognize the underlying pathogenesis of ccRCC, as this understanding can facilitate the development of reliable biomarkers and more effective therapeutic strategies.
Our previous study revealed that abnormal lipid metabolism promoted carcinogenesis, invasion, and metastasis [4]. A defining morphological hallmark of ccRCC is abnormal cytoplasmic accumulation of lipids, including cholesterol, cholesterol esters, and neutral lipids, a characteristic leading to the name “clear cell” [5]. Excess lipids in cancer cells are stored in lipid droplets (LDs), and high levels of LDs are considered a hallmark of cancer aggressiveness [6, 7]. Thus, targeting lipid metabolism may play a significant role in ccRCC. Accumulating evidence indicates that lipid metabolism is also an important regulator of epithelial-mesenchymal transition (EMT) [8]. Moreover, inhibition of autophagy promotes EMT and alters the metabolic phenotype of gastric cancer cells [9]. “Lipophagy” is a type of autophagy in which LDs are selectively delivered for lysosomal degradation [10]. The regulatory and functional similarities between autophagy and lipolysis, along with the capability of lysosomes to degrade lipids, indicate that autophagy may contribute to LD degradation.
Liver-X receptor α (LXRα, also called NR1H3) has been shown to be an important regulator in several cancers, including breast cancer and prostate cancer, through inhibition of tumorigenic phenotypes such as proliferation and survival [11,12,13]. LXRα is a member of the nuclear hormone receptor family of transcription factors. LXRα plays a critical role in maintaining lipid homeostasis through regulation of the expression of various target genes involved in lipid uptake, storage, catabolism, and transport [14]. In particular, LXRα activates the expression of ATP-binding cassette transporter A1 (ABCA1), which accelerates lipid efflux [13, 15, 16]. Ouimet et al. clarified that autophagy-mediated lipid efflux is closely linked to ABCA1, and ABCA1 is linked to endosomal/lysosomal cholesterol pools [17].
In recent years, the triterpene celastrol, an active ingredient initially isolated from the roots of the traditional Chinese herb Tripterygium wilfordii Hook f., has attracted widespread attention, especially for its potential antitumor properties in various cancers, including gastric carcinoma, nasopharyngeal carcinoma, and hepatocellular carcinoma [18,19,20]. Celastrol has been identified as a leptin sensitizer and potential novel treatment for obesity [21]. Celastrol-treated mice exhibit a decrease in liver weight due to reduced triglyceride (TG) accumulation [16]. However, whether LXRα and ABCA1 mediate the effects of celastrol on lipophagy and EMT in ccRCC remains elusive. In the present study, we investigated the role of celastrol in EMT process of ccRCC and reported that celastrol promoted lipophagy via the LXRα/ABCA1 pathway, thereby inhibiting tumor growth of ccRCC.
Materials and methods
Drugs and chemicals
Celastrol (No. C0869), atorvastatin (No. PZ0001), β-cyclodextrin (β-CD; No. C4767), and 3-methyladenine (3-MA; No. M9281) were purchased from Sigma-Aldrich (St. Louis, MO, USA). Oxidized low-density lipoprotein (Ox-LDL; No. YB-002-1) was purchased from Yiyuan Biotechnology (Guangzhou, China). T0901317 (No. ab142808) was purchased from Abcam (Cambridge, MA, USA). GSK2033 (No. HY-108688) was purchased from MedChemExpress LLC (Monmouth Junction, NJ, USA).
Human ccRCC clinical specimens
A total of 4 fresh frozen ccRCC patient samples acquired at the Second XiangYa Hospital of Central South University between 2017 and 2019 were obtained. The samples included both cancer tissue and adjacent normal tissue (1 cm away from the margin of the tumor tissues). No included patient had a history of adjuvant therapy (chemotherapy or radiotherapy), and informed consent was obtained prior to surgery. The use of these samples was ratified by the Declaration of Helsinki and the Ethics Committee of Hunan University of Chinese Medicine.
Cell culture
Human ccRCC cell lines (786-O, A498, SN12C, and OS-RC-2) and a renal epithelial cell line (HK-2) were obtained from the Cell Bank of the Chinese Academy of Sciences (Shanghai, China). ccRCC cells were cultured in RPMI-1640 medium (Gibco, Grand Island, NY, USA) supplemented with 10% fetal bovine serum (FBS; Gibco, Grand Island, NY, USA) and 1% penicillin/streptomycin (Gibco, Grand Island, NY, USA). Dulbecco’s modified Eagle’s medium (DMEM; Gibco, Grand Island, NY, USA) containing 10% FBS and 1% streptomycin-penicillin was used for HK-2 cell culture. All cells were maintained at 37 °C in a humidified atmosphere with 5% CO2.
Small interfering RNAs (siRNAs) and transfection
Three double-stranded siRNAs targeting ABCA1 and a negative control siRNA were designed and synthesized by Sangon Biotech (Shanghai, China) and transfected into 786-O cells using Lipofectamine™ 2000 (Invitrogen, Carlsbad, CA, USA) following the manufacturer’s guidelines.
Immunoblotting analysis
Cell lysates were extracted in radioimmunoprecipitation assay (RIPA; Cwbiotech, China) buffer containing protease inhibitor cocktail (Cwbiotech, China). Protein concentrations were determined with a BCA™ Protein Assay Kit according to the protocol provided by the manufacturer (Cwbiotech, China). Proteins (20 μL) loaded in each lane were subjected to sodium dodecyl sulfate-polyacrylamide gel electrophoresis and were then transferred to a polyvinylidene difluoride (PVDF; Merck Millipore, Billerica, MA, USA) membrane. After blocking with 5% skim milk for 2 h, the PVDF membranes were incubated first with a primary antibody overnight at 4 °C and then with a secondary antibody for 1.5 h. Protein bands were detected using an imaging system (Tanon, China).
Quantitative real-time PCR (qRT-PCR)
Total RNA was extracted using TRIzol reagent (No. 15596018, Invitrogen, Carlsbad, California, USA) and was then treated with RNase-free DNase I (No. 10977015 Invitrogen, Carlsbad, California, USA). Total RNA (0.5 μg) was reverse transcribed using an RNA LA PCR Kit (Thermo Fisher Scientific, Waltham, MA, USA), and qPCR was performed in an ABI 7500 Real-Time fluorescence quantitative PCR instrument (ABI, Waltham, MA, USA). GAPDH was used as the internal control.
Cell Counting kit-8 assay (CCK-8)
Cell viability was evaluated using the CCK-8 kit (Beyotime, China) according to the manufacturer’s instructions. Cells were seeded in 96-well flat bottom microtiter plates at a density of 5 × 103 cells per well. The absorbance was measured on a microplate reader (Synergy HT, Bio-Tek, Biotek Winooski, Vermont, USA) at 450 nm.
Wound healing assay
A wound was created in the center of the cell monolayer with a sterile plastic pipette tip after treatment for 24 h. The migration ability of ccRCC cells was assessed by comparing several marked points along the wounded area in the 0, 6, 12, and 24 h micrographs. The percentage of healed wound area was calculated by dividing the healed area after 0, 6, 12, or 24 h by the initial wound area at time zero.
Transwell invasion assay
The transwell invasion assay was performed using transwell inserts containing a polycarbonate membrane (Corning Incorporated, Corning, NY, USA) coated with Matrigel (BD, Franklin Lakes, NJ, USA). A total of 2 × 105 cells were placed in the upper chamber with RPMI-1640 containing 1% FBS, while the lower chamber was filled with RPMI-1640 containing 10% FBS. The cells that passed through the membrane were fixed with methanol and stained with hematoxylin (Solarbio, China). The invaded area was measured in 5 randomly selected fields using Image Pro Plus 6.0 software (Media Cybernetics, USA).
Oil Red O staining
Oil Red O staining was used to evaluate the cholesterol content. Fresh tumor tissues harvested from mice after sacrifice were immediately fixed with 4% paraformaldehyde (PFA) and were then mounted in OCT compound (Tissue-Tek, Torrance, CA, USA) and frozen at −20 °C. Cryosections (8 µm thick) were then cut and mounted on glass slides for Oil Red O staining (Solarbio, China). In brief, frozen sections were placed in 60% isopropanol for 5 min and were then stained with Oil Red O solution for 20 min. Sections were then rinsed with PBS and counterstained with hematoxylin. Images were acquired with an inverted phase contrast microscope (Olympus DP73, TH4-200, Tokyo, Japan). The cholesterol content was quantified with Image Pro Plus 6.0 software (Media Cybernetics, USA).
To evaluate the cholesterol content in 786-O cells, cells were first fixed with 4% PFA and then stained with Oil Red O solution for 20 min. Hematoxylin was used as a counterstaining dye, and cells were imaged with an inverted phase contrast microscope (Olympus DP73, TH4-200, Tokyo, Japan).
DiI-ox-LDL staining assay
786-O cells were subjected to the same conditions as described above prior to treatment with 5 µg/mL DiI-ox-LDL for 4 h. Slides were stained with 4’,6’-diamidino-2-phenylindole dihydrochloride (DAPI). After incubation, accumulation of DiI-ox-LDL in cells was determined by measuring the fluorescence intensity with a fluorescence microscope, and images were acquired with an inverted phase contrast microscope (Olympus DP73, TH4-200, Tokyo, Japan). The fluorescence intensity was quantified in at least 3 random fields per slide.
Lipid analysis by high-performance liquid chromatography (HPLC)
Cells were placed in 0.9% NaCl solution on ice and homogenized by sonication for 10 s. Subsequently, an equal volume of freshly prepared cold (−20 °C) potassium hydroxide in ethanol (150 g/L) was added to the supernatant of each cell lysate. The mixture was vortexed until a clear solution was obtained. Then, an equal volume of solvent (hexane/isopropanol of 3:2, v/v) was added, and the mixture was first vortexed for 5 min and then centrifuged at 800 × g for 5 min. The organic phases were collected and dried in a SpeedVac. Each organic phase was then dissolved in 100 μL of isopropanol/acetonitrile (20:80, v/v) and placed in an ultrasonic water bath at 25 °C for 5 min. Finally, the samples were subjected to HPLC analysis (Agilent 1100; Agilent Technologies, Santa Clara, CA, USA). HPLC was performed using a Hypersil C18 column with isopropanol/acetonitrile (20:80, v/v) as the eluent at a flow rate of 1 mL/min.
BODIPY-cholesterol assay
We examined cholesterol efflux using BODIPY-cholesterol (No. GC42964, Glpbio, Montclair, CA, USA). Cells were cultured in serum-free medium containing 0.1 mL of labeling medium for 1 h. Cells were washed twice with MEM-HEPES and were then cultured in serum-free medium containing treatment factors for 24 h. After centrifugation at 12,000 × g for 10 min, the liquid supernatant was collected, and the fluorescence intensity value, which represented the total cholesterol (TC) efflux, was recorded using a fluorescence photometer (excitation 482 nm, emission 515 nm).
Autophagic flux analysis
786-O cells were transiently transfected with monomeric red fluorescent protein (mRFP)-green fluorescent protein (GFP)-LC3 adenovirus to examine the formation of fluorescent puncta indicating autophagosomes. Cells were cultured in 24-well plates and transfected with 0.8 μg/well mRFP-EGFP-LC3 adenovirus using LipofectamineTM 2000 (Invitrogen, Carlsbad, California, USA). After transfection, cells were treated with or without 1.0 μM celastrol for 24 h and were then incubated with DAPI for 15 min. Image acquisition was performed using a confocal laser scanning microscope (Leica, Wetzlar, Germany).
Transmission electron microscopy (TEM)
786-O cells were fixed with 1.6% glutaraldehyde prior to postfixation in osmium tetroxide and uranyl acetate en bloc staining. Samples were processed and embedded in Spurr’s epoxy resin, thin sectioned, and counterstained with lead citrate. Digital images were acquired with a Hitachi HT7700 TEM (Hitachi, Tokyo, Japan).
Subcutaneous xenograft experiment
Animal experiments were approved by the Animal Care and Use Committee at Hunan University of Chinese Medicine. The two main defects of BALB/c nude mice are failure of hair growth and hypoplasia of the thymic epithelium due to developmental failure of the thymic anlage. BALB/c nude mice also have a poor response to thymus-dependent antigens because of a defect in helper T-cell activity. Male BALB/c nude mice (Slac, SCXK(Xiang)2014-0002, 4 weeks old) were injected subcutaneously with 5 × 106 786-O cells. Cells were resuspended in ice-cold PBS in a final per-injection volume of 200 μL. Tumor volumes were recorded at the indicated time points by measuring the tumors with calipers and calculating the volumes with the formula L×W2/2 (L is the length of the longer axis and W is the length of the shorter axis). BALB/c nude mice (n = 4) were randomly assigned to the control, high-fat diet (HFD), celastrol (0.25, 0.5, 1.0 mg/kg, i.p.) or atorvastatin (1.5 mg/kg, i.g.) group. Mice in the control group were fed a normal diet. The HFD and treatment groups were fed an HFD (1.0% cholesterol, 0.4% hyocholic acid, 10.0% lard, and 87.6% common feed). After 4 weeks of treatment with the corresponding medicine, tumors were harvested for further analyses.
Plasma biochemical analysis
Blood was collected after 4 weeks of treatment with the corresponding medicine into heparinized tubes. Biochemical parameters, including plasma TC, TG, LDL, and very low density lipoprotein (VLDL) levels, were measured using an autoanalyzer (Beckman Coulter, Miami, FL, USA).
Histology and immunohistochemistry analysis
Tissues were fixed with 4% PFA and processed for histologic examination by steps including embedding in paraffin, sectioning, and staining with hematoxylin and eosin (H&E). The samples were then observed under an optical microscope (40×) (Olympus, Tokyo, Japan).
Four-micrometer sections from selected paraffin blocks of each specimen were used for immunohistochemical analysis. Sections were first incubated overnight at 4 °С with each primary antibody and then incubated for 1 h at room temperature with secondary antibodies. Proteins were visualized using EnVision Detection Systems (Dako Japan, Tokyo, Japan). Immunochemical results were evaluated by an experienced pathologist with a semiquantitative approach assigning an H-score (or “Histoscore”) to the tumor sample. The percentage of tumor cells was determined for each different nuclear staining intensity (0/+/++/+++), and the sum of the individual H-scores for each intensity level was then calculated using the following equation: H-score = 1 × (% of cells with an intensity of 1 + ) + 2 × (% of cells with an intensity of 2 + ) + 3 × (% of cells with an intensity of 3 + ).
Statistical analysis
All values are expressed as means ± SDs. All experiments were performed at least three times. Single comparisons and multiple comparisons were performed by Student’s t test and one-way ANOVA, respectively. P < 0.05 was considered to indicate a significant difference. GraphPad Prism (Version 6.0; La Jolla, CA, USA) was used for statistical analysis.
Results
LXRα and ABCA1 expression was decreased in ccRCC
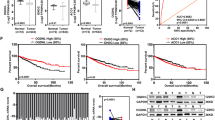
The mRNA expression levels of LXRα and ABCA1 in primary ccRCC tumors and normal tissues were obtained from the TCGA_KIRC and GTEx_KIRC datasets. Bioinformatic analysis indicated that the mRNA expression levels of both LXRα and ABCA1 in ccRCC tissues were lower than those in adjacent normal tissues (Fig. 1a). To verify LXRα and ABCA1 expression levels in ccRCC tissues and matched adjacent normal tissues, we extracted protein for immunoblotting. Our results indicated that the protein expression levels of LXRα and ABCA1 were decreased in ccRCC tissues (Fig. 1b, c). We further examined the expression of LXRα and ABCA1 in a panel of ccRCC cell lines (786-O, A498, SN12C, and OS-RC-2) relative to HK-2 cells. The results showed that compared with those in HK-2 cells, the mRNA and protein levels of LXRα and ABCA1 in ccRCC cells were reduced (Fig. 1d, e). These findings revealed that the expression of LXRα and ABCA1 was decreased in ccRCC tissues and cell lines; these molecules could be potential biomarkers for ccRCC diagnosis and treatment.
a The mRNA levels of LXRα and ABCA1 in ccRCC samples were obtained from the Cancer Genome Atlas (TCGA)_KIRC and Genotype-Tissue Expression (GTEx)_KIRC datasets. b Protein expression levels and (c) statistical analysis of LXRα and ABCA1 in ccRCC tissues and adjacent normal tissues. *P < 0.05, **P < 0.01 vs. control. d, e The mRNA and protein expression levels of LXRα and ABCA1 in HK-2 and ccRCC cell lines (786-O, A498, SN12C, and OS-RC-2) were measured by qRT-PCR and Western blotting, respectively. Glyceraldehyde-3-phosphate dehydrogenase (GAPDH) was used as the internal control. *P < 0.05, **P < 0.01, ***P < 0.001 vs. control.
Celastrol inhibited the invasion and migration of ccRCC cells
To investigate the anti-proliferative activity and cytotoxicity of celastrol in ccRCC cells, 786-O and SN12C cells were treated with celastrol (0, 0.25, 0.5, 1.0, 1.5, and 2.0 μM) for 12 h, 24 h and 48 h. Celastrol inhibited the growth of ccRCC cells in time- and dose-dependent manners (Fig. 2a, Supplementary Fig. S1a). We found that celastrol dose-dependently inhibited cell migration and invasion (Fig. 2b–d, Supplementary Fig. S2b-d), and this result was also confirmed by the increased E-cadherin (E-cad) and decreased vimentin (Vim) and matrix metalloproteinase 2 (MMP-2) expression levels in 786-O and SN12C cells (Fig. 2e, Supplementary Fig. S1e). Based on these findings, 1.0 μM celastrol was selected for subsequent experiments.
a 786-O cells were pretreated with celastrol (0, 0.25, 0.5, 1.0, 1.5, and 2.0 μM) for different times (12, 24, and 48 h). Cell viability was measured by a CCK-8 assay in cells treated at various concentrations and different times. 24 h: ***P < 0.001 vs. control. 48 h: ###P < 0.001 vs. control. b Representative images of the wound area at 0, 6, 12, and 24 h after wounding and (c) quantification of the effect of celastrol (0, 0.25, 0.5, and 1.0 μM) or atorvastatin (ATV, 5.0 μM) on 786-O cell motility. Scale bars: 50 μm. 24 h: *P < 0.05, **P < 0.01 vs. control. 48 h: ##P < 0.01, ###P < 0.001 vs. control. d The invasive ability of 786-O cells treated with celastrol for 24 h was evaluated by a transwell assay. Scale bars: 50 μm. **P < 0.01, ***P < 0.001 vs. control. e 786-O cells were treated with celastrol for 24 h. The expression levels of EMT-associated proteins (E-cad, Vim, MMP-2) were analyzed by Western blotting. GAPDH was used as the internal control. *P < 0.05, **P < 0.01 vs. control.
Celastrol inhibited the invasion of ccRCC cells by reducing lipid accumulation
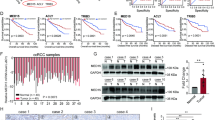
Accumulation of lipids is crucial for cancer cells to support their highly proliferative growth [22]. Next, we examined alterations in lipid metabolism by using Oil Red O staining and HPLC. The results showed that celastrol inhibited lipid accumulation (Fig. 3a) and reduced the TC, TG and CE levels in 786-O cells (Fig. 3b). Furthermore, we found that celastrol upregulated the protein and mRNA expression of LXRα and ABCA1 (Fig. 3c, Supplementary Fig. S2a).
a Representative images of 786-O cells treated with celastrol (0, 0.25, 0.5, 1.0 μM) and ATV stained with Oil Red O and counterstained with hematoxylin to visualize nuclei. Scale bar: 100 μm. b The TC, FC, and CE contents in 786-O cells treated with celastrol (0, 0.25, 0.5, 1.0 μM) were determined by HPLC. *P < 0.05, **P < 0.01, ***P < 0.001 vs. control. c The protein levels of LXRα and ABCA1 were analyzed by Western blotting. 786-O cells were pretreated with celastrol (0, 0.25, 0.5, 1.0 μM) for 24h. GAPDH was used as the internal control. *P < 0.05, **P < 0.01, ***P < 0.001 vs. control. (d) Oil Red O staining of 786-O cells after treatment with celastrol (1.0 μM) and ox-LDL (100 μg/mL) for 24 h. *P < 0.05 vs. control. ##P < 0.01 vs. ox-LDL. e DiI-ox-LDL staining in 786-O cells was assessed in the presence of celastrol and ox-LDL, and the DiI fluorescence intensity was quantified with Image Pro Plus. *P < 0.05 vs. control. #P < 0.05 vs. ox-LDL. f The protein expression levels of ABCA1, LXRα, E-cad, Vim and MMP-2 were evaluated by Western blot analysis. 786-O cells were pretreated with celastrol and ox-LDL. GAPDH was used as the internal control. *P < 0.05, **P < 0.01, ***P < 0.001 vs. control. #P < 0.05, ##P < 0.01 vs. ox-LDL.
To verify whether celastrol inhibited the invasion of ccRCC cells by regulating lipid metabolism, 786-O cells were treated with celastrol (1.0 μM) or β-CD (10 μM) for 24 h. The results showed that both celastrol and β-CD reduced lipid accumulation (Supplementary Fig. S2b). In addition, both celastrol and β-CD upregulated the protein expression of LXRα, ABCA1 and E-cad and downregulated the expression of Vim and MMP-2 (Supplementary Fig. S2c). Conversely, we used ox-LDL (100 μg/mL) to establish a lipid-loaded model. Celastrol significantly abolished lipid accumulation and reduced the DiI-derived fluorescence intensity induced by ox-LDL (Fig. 3d, e). Moreover, ox-LDL reduced the levels of LXRα, ABCA1, and E-cad and elevated the levels of Vim and MMP-2, whereas celastrol produced the opposite effects (Fig. 3f). Taken together, these results indicated that celastrol inhibited the invasion of ccRCC cells by reducing lipid accumulation.
Celastrol triggered autophagy to reduce lipid storage
It has been reported that autophagy is a potential cell survival mechanism in ccRCC cells [23]. We then determined whether celastrol could contribute to the induction of autophagy. Western blot analysis revealed that celastrol significantly induced LC3-I to LC3-II conversion (a marker for autophagy) and reduced the level of p62 (an autophagic degradation protein) and the p-mTOR/mTOR ratio (indicating inhibition of autophagy) (Fig. 4a). When 786-O cells were exposed to ox-LDL for 12 h, they responded with an increased LC3-II/I ratio along with a decreased p62 level and p-mTOR/mTOR ratio (Fig. 4b). However, after 786-O cells were exposed to ox-LDL for 24 h, the level of LC3-I to LC3-II conversion was decreased, while the p62 level was increased (Fig. 4b). 3-Methyladenine (3-MA), an autophagy inhibitor, significantly increases lipid storage. As expected, 3-MA (2 mM) promoted lipid accumulation, and this promotion was inhibited by celastrol (Fig. 4c, d). 3-MA also increased the TC, FC, and CE contents, and this effect was reversed by celastrol (Supplementary Fig. S3a). Subsequently, we monitored autophagic flux using mRFP-GFP-LC3 adenovirus. Because GFP is sensitive to acidic conditions, GFP fluorescence is quenched when autophagosomes fuse with lysosomes, and only red fluorescence can be detected [24]. The simultaneous appearance of GFP and mRFP fluorescence indicated that the autophagosomes were not bound to lysosomes. Ox-LDL and 3-MA significantly impaired the formation of autophagosomes (yellow puncta in merged images). Celastrol enhanced autophagosome formation (Fig. 4e). Next, cells were incubated with rapamycin (Rap, a general inducer of autophagy). Rap restored autophagic flux after ox-LDL treatment by enhancing autophagosome formation, as indicated by the increase in yellow puncta (Fig. 4e). In addition, we observed increases in the ABCA1 and E-cad levels and the LC3-II/I ratio, as well as reductions in the Vim and p62 levels and the p-mTOR/mTOR ratio following exposure to celastrol and 3-MA (Supplementary Fig. S3b). Therefore, these results showed that the induction of autophagy by celastrol contributed to reducing lipid storage.
The expression levels of autophagy-related proteins in 786-O cells treated with celastrol (a) and ox-LDL (b) were measured by Western blot analysis. α-Tubulin was used as the internal control. *P < 0.05, **P < 0.01 vs. control. (c) Photomicrographs of Oil Red O-stained 786-O cells with 3-MA (2 mM) and celastrol treatment. Scale bar: 50 μm. *P < 0.05, **P < 0.01 vs. control. d A DiI-ox-LDL staining assay was performed to evaluate the intracellular lipid content. Scale bar: 50 μm. *P < 0.05, **P < 0.01 vs. control, ###P < 0.001 vs. ox-LDL. e Autophagic flux in 786-O cells was evaluated by mRFP-GFP-LC3 double immunofluorescence labeling. The mean number of yellow puncta, indicating autophagosomes, and the mean number of red puncta, indicating autolysosomes, are plotted. Scale bar: 25 μm. mRFP: *P < 0.05 vs. control. GFP/mRFP: #P < 0.05, ##P < 0.01 vs. control.
Celastrol induced lipophagy through activation of LXRα signaling
An LXRα agonist (T0901317) and an LXRα antagonist (GSK2033) were used to determine whether celastrol induces lipophagy via LXRα signaling. Although treatment with T0901317 (500 nM) alone had no effect, its combination with celastrol resulted in a reduction in lipid storage (Supplementary Fig. S4a). We further measured the levels of downstream targets of LXRα, such as ABCA1, ABCG1 and SREBP-1c. Interestingly, celastrol dramatically upregulated LXRα, ABCA1 and ABCG1 but had little effect on SREBP-1c (Fig. 5a). The LXRα activator maximally increased the levels of LXRα, ABCA1, E-cad, and LC3-II/I but reduced the levels of Vim, MMP-2, and p62, consistent with the effects of celastrol (Fig. 5a). In addition, inhibition of LXRα with GSK2033 (50 nM) was inefficient at reducing lipid accumulation, whereas celastrol significantly reduced lipid accumulation (Supplementary Fig. S3b). Relative to control treatment, GSK2033 obviously downregulated the expression of LXRα, ABCA1, and E-cad and upregulated the expression of Vim and MMP-2, while celastrol abolished these effects (Fig. 5b). However, the expression of ABCG1 and sterol regulatory element-binding protein 1c (SREBP-1c) was not affected by treatment with GSK2033 and celastrol (Fig. 5b). To better understand the lipophagy process, we performed TEM to clarify that autophagosomes enveloped LDs and found double-membrane vesicles analogous to autophagosomes around LDs and degradative structures enriched in LDs. LDs were easily identified as round, light-density structures, not enclosed with a bilayer lipid membrane. Our results showed that both celastrol and T0901317 induced autophagy very strongly, as indicated by the formation of autophagosomes and autophagolysosomes. Similar to T0901317, celastrol resulted in packed degradative structures enriched in LDs (Fig. 5c). Conversely, GSK2033 resulted in the formation of a decreased number of autophagolysosomes, autophagosomes and lipophagic vesicle-like structures, while these alterations were significantly reversed by celastrol (Fig. 5d). Therefore, celastrol triggered lipophagy by activating LXRα signaling.
a The expression of EMT-associated proteins, autophagy-related proteins, and lipid metabolism proteins in 786-O cells treated with celastrol and T0901317 (500 nM) was evaluated by Western blot analysis. *P < 0.05, **P < 0.01, ***P < 0.001 vs. control. (b) Western blot analysis was used to evaluate the expression of EMT-associated proteins, autophagy-related proteins, and lipid metabolism proteins in 786-O cells treated with celastrol and GSK2033 (50 nM). GAPDH was used as the internal control. *P < 0.05, **P < 0.01, ***P < 0.001 vs. control. #P < 0.05, ##P < 0.01 vs. GSK2033. c 786-O cells were treated with and without celastrol or T0901317 and then processed for TEM. Scale bar: 2 μm. d Transmission electron micrographs of 786-O cells treated with celastrol or GSK2033. Scale bar: 2 μm. AP autophagosomes, red arrows; AL autophagolysosomes, yellow arrows; LD lipid droplets and degradative structures enriched in lipid droplets, green arrows.
ABCA1 was required for celastrol-promoted cholesterol efflux
Previous studies also confirmed that autophagy increases cholesterol efflux in macrophage-derived foam cells via lysosomal acid lipase [10]. We then tested the hypothesis that ABCA1 was sufficient for cholesterol efflux. We silenced ABCA1 expression using specific siRNAs. The knockdown efficiency of siABCA1 was confirmed by Western blot analysis. The results showed a stronger reduction in ABCA1-Homo 6779 expression in three cell lines (Supplementary Fig. S5a). Transient transfection with si-NC did not alter ABCA1 expression (Supplementary Fig. S5a). To investigate whether ABCA1 triggered cholesterol efflux, we examined lipid deposition and measured cholesterol efflux. Oil Red O staining indicated that knockdown of ABCA1 significantly increased lipid storage in 786-O cells, an effect that was reversed by celastrol (Fig. 6a). We utilized DiI-labeled ox-LDL to trace ox-LDL uptake and found that silencing ABCA1 enhanced red fluorescence, whereas celastrol attenuated this effect (Fig. 6b). BODIPY 493/503 is commonly used to fluorescently label neutral lipids. The results showed that knockdown of ABCA1 promoted lipid storage, while celastrol reduced intracellular lipid accumulation (Fig. 6c). Furthermore, knockdown of ABCA1 dramatically suppressed cholesterol efflux, whereas celastrol produced the opposite effect (Fig. 6c). Consistent with the Oil Red O staining and BODIPY-cholesterol assay results, celastrol decreased the alterations in TC, FC, and CE levels caused by silencing of ABCA1 (Supplementary Fig. S5b). These results suggested that celastrol promoted cholesterol efflux from 786-O cells via ABCA1. Furthermore, we found that celastrol reversed the ABCA1 silencing-induced inhibition of autophagy and EMT, based on the increases in the E-cad level, LC3-II/I ratio, and p-mTOR/mTOR ratio and the reductions in Vim, MMP-2, and p62 levels (Supplementary Fig. S5c). Electron microscopy was used to further elucidate the mechanism of LD degradation by autophagic vesicles. Knockdown of ABCA1 significantly decreased autophagosome and autophagolysosome formation and reduced the number of degradative structures enriched in LDs (Fig. 6d). In contrast, induction of autophagy by celastrol increased autophagosome and autolysosome formation, as well as lipolysis (Fig. 6d). Thus, celastrol reduced lipid accumulation through activation of autophagy, and cholesterol efflux was mediated by autophagy predominantly in an ABCA1-dependent manner.
a Photomicrographs of Oil Red O-stained 786-O cells with ABCA1 knockdown. Scale bar: 50 μm. b The evaluation of lipid accumulation by DiI-ox-LDL internalization assays. Scale bar: 50 μm. *P < 0.05 vs. control, #P < 0.05 vs. siABCA1. c The BODIPY fluorescence intensity that remained in 786-O cells was observed using fluorescence microscopy. Scale bar: 20μm. The BODIPY fluorescence intensity in the supernatants was measured by fluorescence photometry. *P < 0.05, **P < 0.01 vs. control, #P < 0.05, ###P < 0.001 vs. siABCA1. d TEM was used to detect lipophagy of 786-O cells transfected with siABCA1 and further treated with celastrol. AP: autophagosomes, red arrows; AL autophagolysosomes, yellow arrows; LD lipid droplets and degradative structures enriched in lipid droplets, green arrows. Scale bar: 5 μm. *P < 0.05, **P < 0.01 vs. control, #P < 0.05, ##P < 0.01 vs. siABCA1.
Celastrol inhibited HFD-induced tumor growth in vivo
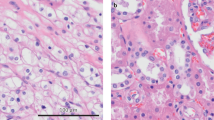
We then determined the effect of celastrol in vivo. An HFD-induced xenograft model of ccRCC was established by injection of 786-O cells. As shown in Fig. 7a, the celastrol (1.0 mg/kg) groups showed significant inhibition of tumor growth compared to the HFD group. In agreement with recent results [16], administration of celastrol significantly reduced body weight (Supplementary Fig. S6a). To further evaluate tumor formation, we performed H&E staining of tumor tissues. In the HFD group, we observed certain tumor characteristics, such as significantly increased cell density, distinct nuclear atypia, and common mitotic figures (Fig. 7b). These results suggested the successful establishment of the model. Interestingly, we observed greater lipid droplet formation in the HFD group than in the control group, and this effect was clearly reduced by celastrol treatment (Fig. 7c, d). Biochemical analysis revealed that celastrol also reduced plasma TC, TG, LDL and VLDL concentrations in nude mice fed a HFD (Fig. 7e). Immunohistochemical results demonstrated that celastrol-treated tumor tissues exhibited significantly increased levels of ABCA1 and E-cad, as well as a slightly increased level of LC3; however, the levels of Vim, MMP-2, and p62 were decreased (Fig. 7f). Celastrol also upregulated the protein expression of ABCA1, LXRα, and E-cad, increased the LC3-II/I ratio, and downregulated the expression of Vim, MMP-2, and p62 (Supplementary Fig. S6b). Therefore, celastrol alleviated HFD-promoted lipid deposition and exhibited potential antitumor activity in vivo via a mechanism similar to that observed in vitro.
a Nude mice were subcutaneously injected with 786-O cells. Tumors were weighed after celastrol treatment for 4 weeks. #P < 0.05 vs. the HFD group. b Histological evaluation of xenografts was performed by H&E staining. (c and d) Representative images of lipid deposition areas, which were quantified with Oil Red O staining and Image Pro Plus 6.0 software. **P < 0.01 vs. the control group, #P < 0.05, ##P < 0.01 vs. the HFD group. e The biochemical analysis of plasma TC, TG, LDL, and VLDL concentrations. **P < 0.01 vs. the control group, #P < 0.05, ##P < 0.01, ###P < 0.001 vs. the HFD group. f LXRα, ABCA1, E-cad, Vim, MMP-2, LC3-II/I, and p62 expression in xenografts was analyzed by immunohistochemistry. Scale bars: 20 µm. Bar graph showing the quantification of immunostaining-positive areas as determined by using Image Pro Plus 6.0 software. *P < 0.05, **P < 0.01 vs. the control group, #P < 0.05, ##P < 0.01, ###P < 0.001 vs. the HFD group.
Discussion
Abnormal lipid metabolism is closely associated with tumorigenesis. Specifically, it contributes to invasion and metastasis [25, 26]. Although ccRCC can be characterized as a “lipid metabolic disease,” it remains incompletely understood, because lipid metabolic targets are therapeutically tractable [27]. Accumulating evidence has shown that LXRα plays distinct roles in cancer. LXRα promotes RCC cell metastasis by regulating the NOD-like receptor protein 3 inflammasome [28]. In contrast, LXRα suppresses the proliferation of human oral cancer cells by promoting cholesterol efflux through upregulation of ABCA1 expression [29]. Moreover, the anticancer activity of ABCA1 is compromised following inhibition of ABCA1 gene expression by oncogenic mutations or cancer-specific ABCA1 loss-of-function mutations [30]. According to our bioinformatic analysis, clinical sample analysis and in vitro results, the expression levels of LXRα and ABCA1 are decreased in ccRCC. We concluded that LXRα and ABCA1 might play an important role in the development of ccRCC. Despite these observations, little evidence exists about the effect of LXRα/ABCA1 on lipophagy during ccRCC progression.
Previous studies have shown that celastrol inhibits the growth of gastric carcinoma and nasopharyngeal carcinoma cells via suppression of inflammation and induction of cell cycle arrest and apoptosis [18, 19]. Our study provides evidence that celastrol inhibits migration, invasion, and tumor growth in ccRCC. EMT, during which cancer cells acquire migratory and invasive phenotypes by activating several specific signaling pathways and their downstream transcription factors, has long been considered to play an essential role in cancer metastasis [31, 32]. Loss of epithelial markers such as E-cad and gain of mesenchymal markers such as Vim and MMP-2 are the most common hallmarks of EMT [33]. We showed that celastrol obviously suppressed EMT progression to inhibit the migration and invasion of ccRCC cells. A recent study reported that celastrol not only inhibited adipocyte differentiation (as assessed by lipid accumulation and the TC content) but also increased lipolysis (as assessed by glycerol release and free fatty acid release) in 3T3-L1 adipocytes [34]. Consistent with these results, our study demonstrated that celastrol inhibited intracellular lipid accumulation. ox-LDL, an important marker for atherosclerosis, has been assumed to be involved in cancer progression since multiple studies reported elevated levels of serum ox-LDL in colon, breast, and ovarian cancers [35]. Our study demonstrated that celastrol inhibited lipid storage in cells with lipid loading induced by ox-LDL. Interestingly, this reduction in intracellular lipid accumulation significantly suppressed EMT following upregulation of LXRα and ABCA1 expression. As mentioned previously, celastrol has been reported to inhibit the xenograft growth in models of ovarian cancer, prostate cancer, glioma, and other cancers [36,37,38]. Our in vivo findings also confirmed that celastrol inhibited tumor growth in HFD-promoted mice. Thus, celastrol might inhibit EMT by reducing lipid accumulation. ccRCC cells possess high metabolic plasticity in their use of metabolic pathways, which is also dependent on the dynamically changing tumor microenvironment. Normal tissues also depend on the same metabolic pathways for survival and proliferation. These characteristics constitute a major challenge for the therapeutic targeting of lipid metabolism.
As a modulator of pathogenesis, autophagy has been widely studied as a promising, novel therapeutic target in multiple diseases [39]. In general, autophagic flux can be assessed by simultaneously analyzing the levels of LC3-II and LC3-I, which label autophagosome membranes, and measuring the level of p62, a protein cleared through autophagic degradation. The combination of an increased LC3-II/I ratio and a decreased p62 level is consistent with an overall increase in autophagic flux. In the present study, we found that celastrol triggered autophagy, increased the synthesis of autophagy-related membranes and partially promoted autophagic flux. Therefore, celastrol might inhibit the function of lysosomes to some extent. Moreover, an inverse relationship existed between abnormal lipid accumulation and autophagy. Interestingly, when we used 3-MA to limit the formation of autophagosomes, intracellular lipids accumulated significantly. Celastrol reversed the effects of 3-MA on autophagy and lipid accumulation. We further revealed that celastrol-triggered autophagy inhibited EMT. Although moderate lipid exposure was shown to decrease autophagic flux, the fusogenic ability of the autophagic/lysosomal compartments was reinforced under exposure to Rap. The significant decreases in the expression levels of LXRα and its target gene ABCA1 induced by 3-MA showed that autophagy was associated with LXRα and ABCA1. It was also clear that the effects of celastrol on autophagy were LXRα-dependent, as inhibition of LXRα attenuated autophagy marker expression induced by celastrol.
A previous study demonstrated a close relationship between autophagy and lipid metabolism [10]. Fusion of autophagosomes with lysosomes during the autophagic process is a vital step that leads to degradation of LDs. Activation of LXRα by an LXRα agonist and/or celastrol triggered autophagy to promote LD degradation. Loss of autophagy mediated by an LXRα antagonist attenuated LD degradation. Although the mechanism of LD sequestration/degradation through autophagy needs clarification in future studies, our data indicated that autophagy might contribute to LD degradation. In addition, the autophagic degradation of organelles, including LDs, releases free fatty acids and cholesterol, which need to be handled appropriately to prevent lipotoxicity. Lipophagy has been reported in various tissues and cells, such as 3T3-L1 adipocytes, macrophages, and glial cells [17, 40, 41]. Despite these findings, much remains to be elucidated about lipophagy. First, the mechanism and extent of selectivity by which LDs are degraded by autophagy are unclear. Unlike those for other autophagic pathways targeting specific organelles, the ligands and receptors for lipophagy have not yet been defined [42]. Second, engulfment of LDs by the isolation membrane has not been definitively observed. LC3, the most reliable marker of the isolation membrane/autophagosome, also exists in LDs and may be involved in LD formation [43, 44].
On the one hand, the LXRα agonist T0901317 is a potent inducer of ABC transporters [45]. ABCA1 and ABCG1 are major transporters that have crucial roles in mediating cholesterol efflux [46, 47]. On the other hand, LXRα activates lipid synthesis by inducing SREBP-1c [48]. Intriguingly, we found that activation of LXRα by celastrol increased ABCA1 expression but did not significantly alter the expression of ABCG1 and SREBP-1c, implying that cholesterol efflux might be predominantly mediated by ABCA1. ABCA1, as a cell cholesterol exporter, can mediate the transport of cholesterol across cellular membranes, providing an efficient pathway for cells to unload excess cholesterol [49]. To initiate reverse cholesterol transport, phospholipids and cholesterol are exported by ABCA1 and subsequently loaded onto lipid-free apolipoprotein A-I (ApoA-I) to form nascent high-density lipoprotein particles. ABCA1-mediated lipid export to ApoA-I is the rate-limiting step in high-density lipoprotein biogenesis [38]. When induced by cellular cholesterol loading, ABCA1 constitutively generates exovesiculated membrane domains, even in the absence of apolipoproteins [50]. Our previous study reported that the high amount of cholesterol accumulation in atherosclerotic plaques could be attributed to inhibition of ABCA1 expression [51]. Here, we observed that celastrol reduced lipid accumulation and promoted ABCA1-mediated cholesterol efflux in 786-O cells. Moreover, we found that celastrol triggered autophagy, enhanced ABCA1 expression, and blocked the EMT process, while knockdown of ABCA1 led to the opposite effects. ABCA1 has been implicated in the initiation of autophagy-centric LD degradation by mediating the docking of autophagosomes and lysosomes to LDs during lipophagy. Here, we clearly established that autophagy triggered by celastrol can mediate cholesterol efflux, which is primarily ABCA1-dependent. The interconnections between LXRα/ABCA1 and EMT were also evident in ccRCC cells: (I) treatment with the LXRα agonist T0901317, an agent that directly activates LXRα, inhibited EMT; (II) suppression of LXRα with GSK2033 obviously promoted EMT; and (III) knockdown of ABCA1 was sufficient to induce EMT despite the presence of celastrol.
In conclusion, the present study suggested that the anticancer effects of celastrol in ccRCC were caused by the triggering of autophagy and degradation of LDs via the activation of LXRα signaling, which then promoted ABCA1-mediated cholesterol efflux and impaired EMT progression, ultimately inhibiting the migration and invasion of ccRCC cells (Fig. 8). In addition, this study identified a critical function of lipophagy that could have important implications for cancers with disorders of lipid metabolism. Taken together, these results indicate that therapeutic strategies to enhance autophagy may constitute a potential approach to prevent lipid metabolism diseases and related pathologies. In-depth and rigorous molecular analyses of traditional Chinese medicines may reveal new strategies for the prevention or treatment of cancer. Clinical studies are required to evaluate the efficacy and safety of traditional medicines. Integration of traditional medicine into conventional treatment regimens may be an alternative approach to ccRCC therapy in the future. Therefore, collectively, these findings provide insight into the anticancer activities of celastrol associated with lipophagy.
References
Miller KD, Nogueira L, Mariotto AB, Rowland JH, Yabroff KR, Alfano CM, et al. Cancer treatment and survivorship statistics, 2019. CA Cancer J Clin. 2019;69:363–85.
Siegel RL, Miller KD, Jemal A. Cancer statistics, 2017. CA Cancer J Clin. 2017;67:7–30.
Choueiri TK. Clinical treatment decisions for advanced renal cell cancer. J Natl Compr Canc Netw. 2013;11:694–7.
Long J, Zhang CJ, Zhu N, Du K, Yin YF, Tan X, et al. Lipid metabolism and carcinogenesis, cancer development. Am J Cancer Res. 2018;8:778–91.
Wettersten HI, Aboud OA, Lara PN Jr, Weiss RH. Metabolic reprogramming in clear cell renal cell carcinoma. Nat Rev Nephrol. 2017;13:410–9.
Sanchez DJ, Simon MC. Genetic and metabolic hallmarks of clear cell renal cell carcinoma. Biochim Biophys Acta Rev Cancer. 2018;1870:23–31.
Cheng C, Geng F, Cheng X, Guo D. Lipid metabolism reprogramming and its potential targets in cancer. Cancer Commun. 2018;38:27.
Beloribi-Djefaflia S, Vasseur S, Guillaumond F. Lipid metabolic reprogramming in cancer cells. Oncogenesis. 2016;5:e189.
Qin W, Li C, Zheng W, Guo Q, Zhang Y, Kang M, et al. Inhibition of autophagy promotes metastasis and glycolysis by inducing ROS in gastric cancer cells. Oncotarget. 2015;6:39839–54.
Singh R, Kaushik S, Wang Y, Xiang Y, Novak I, Komatsu M, et al. Autophagy regulates lipid metabolism. Nature. 2009;458:1131–5.
Villa GR, Hulce JJ, Zanca C, Bi J, Ikegami S, Cahill GL, et al. An LXR-cholesterol axis creates a metabolic co-dependency for brain cancers. Cancer Cell. 2016;30:683–93.
Carpenter KJ, Valfort AC, Steinauer N, Chatterjee A, Abuirqeba S, Majidi S, et al. LXR-inverse agonism stimulates immune-mediated tumor destruction by enhancing CD8 T-cell activity in triple negative breast cancer. Sci Rep. 2019;9:19530.
Tsui KH, Chung LC, Feng TH, Lee TY, Chang PL, Chen WT, et al. Divergent effect of liver X receptor agonists on prostate-specific antigen expression is dependent on androgen receptor in prostate carcinoma cells. Prostate. 2015;75:603–15.
Li T, Hu SM, Pang XY, Wang JF, Yin JY, Li FH, et al. The marine-derived furanone reduces intracellular lipid accumulation in vitro by targeting LXRα and PPARα. J Cell Mol Med. 2020;24:3384–98.
Tavazoie MF, Pollack I, Tanqueco R, Ostendorf BN, Reis BS, Gonsalves FC, et al. LXR/ApoE activation restricts innate immune suppression in cancer. Cell. 2018;172:825–40.
Liu J, Lee J, Salazar Hernandez MA, Mazitschek R, Ozcan U. Treatment of obesity with celastrol. Cell. 2015;161:999–1011.
Ouimet M, Franklin V, Mak E, Liao X, Tabas I, Marcel YL. Autophagy regulates cholesterol efflux from macrophage foam cells via lysosomal acid lipase. Cell Metab. 2011;13:655–67.
Guo D, Zhang W, Yang H, Bi J, Xie Y, Cheng B, et al. Celastrol induces necroptosis and ameliorates inflammation via targeting biglycan in human gastric carcinoma. Int J Mol Sci. 2019;20:5716.
Hsieh MJ, Wang CW, Lin JT, Chuang YC, Hsi YT, Lo YS, et al. Celastrol, a plant-derived triterpene, induces cisplatin-resistance nasopharyngeal carcinoma cancer cell apoptosis though ERK1/2 and p38 MAPK signaling pathway. Phytomedicine. 2019;58:152805.
Chang W, He W, Li PP, Song SS, Yuan PF, Lu JT, et al. Protective effects of Celastrol on diethylnitrosamine-induced hepatocellular carcinoma in rats and its mechanisms. Eur J Pharmacol. 2016;784:173–80.
Feng X, Guan D, Auen T, Choi JW, Salazar Hernández MA, Lee J, et al. IL1R1 is required for celastrol’s leptin-sensitization and antiobesity effects. Nat Med. 2019;25:575–82.
Zhao M, Bu Y, Feng J, Zhang H, Chen Y, Yang G, et al. SPIN1 triggers abnormal lipid metabolism and enhances tumor growth in liver cancer. Cancer Lett. 2020;470:54–63.
Cao Q, Bai P. Role of autophagy in renal cancer. J Cancer. 2019;10:2501–9.
Klionsky DJ, Abeliovich H, Agostinis P, Agrawal DK, Aliev G, Askew DS, et al. Guidelines for the use and interpretation of assays for monitoring autophagy in higher eukaryotes. Autophagy. 2008;4:151–75.
Santos CR, Schulze A. Lipid metabolism in cancer. FEBS J. 2012;279:2610–23.
Luo X, Cheng C, Tan Z, Li N, Tang M, Yang L, et al. Emerging roles of lipid metabolism in cancer metastasis. Mol Cancer. 2017;16:76. https://doi.org/10.1186/s12943-017-0646-3.
Neumann CKA, Silver DJ, Venkateshwari V, Zhang R, Traughber CA, Przybycin C, et al. MBOAT7-driven phosphatidylinositol remodeling promotes the progression of clear cell renal carcinoma. Mol Metab. 2020;34:136–45.
Wang K, Xu T, Ruan H, Xiao H, Liu J, Song Z, et al. LXRα promotes cell metastasis by regulating the NLRP3 inflammasome in renal cell carcinoma. Cell Death Dis. 2019;10:159.
Liang X, Cao Y, Xiang S, Xiang Z. LXRα-mediated downregulation of EGFR suppress colorectal cancer cell proliferation. J Cell Biochem. 2019;120:17391–404.
Smith B, Land H. Anticancer activity of the cholesterol exporter ABCA1 gene. Cell Rep. 2012;2:580–90.
Nieto MA, Huang RY, Jackson RA, Thiery JP. EMT: 2016. Cell. 2016;166:21–45.
Lambert AW, Pattabiraman DR, Weinberg RA. Emerging biological principles of metastasis. Cell. 2017;168:670–91.
Serrano-Gomez SJ, Maziveyi M, Alahari SK. Regulation of epithelial-mesenchymal transition through epigenetic and post-translational modifications. Mol Cancer. 2016;15:18.
Choi SK, Park S, Jang S, Cho HH, Lee S, You S, et al. Cascade regulation of PPARγ(2) and C/EBPα signaling pathways by celastrol impairs adipocyte differentiation and stimulates lipolysis in 3T3-L1 adipocytes. Metabolism. 2016;65:646–54.
González-Chavarría I, Fernandez E, Gutierrez N, González-Horta EE, Sandoval F, Cifuentes P, et al. LOX-1 activation by oxLDL triggers an epithelial mesenchymal transition and promotes tumorigenic potential in prostate cancer cells. Cancer Lett. 2018;414:34–43.
Xu LN, Zhao N, Chen JY, Ye PP, Nan XW, Zhou HH, et al. Celastrol inhibits the growth of ovarian cancer cells in vitro and in vivo. Front Oncol. 2019;9:2. https://doi.org/10.3389/fonc.2019.00002.
Pang X, Yi Z, Zhang J, Lu B, Sung B, Qu W, et al. Celastrol suppresses angiogenesis-mediated tumor growth through inhibition of AKT/mammalian target of rapamycin pathway. Cancer Res. 2010;70:1951–9.
Liu X, Zhao P, Wang X, Wang L, Zhu Y, Song Y, et al. Celastrol mediates autophagy and apoptosis via the ROS/JNK and Akt/mTOR signaling pathways in glioma cells. J Exp Clin Cancer Res. 2019;38:184.
Tan YQ, Zhang J, Zhou G. Autophagy and its implication in human oral diseases. Autophagy. 2017;13:225–36.
Lizaso A, Tan KT, Lee YH. β-adrenergic receptor-stimulated lipolysis requires the RAB7-mediated autolysosomal lipid degradation. Autophagy. 2013;9:1228–43.
Martinez-Vicente M, Talloczy Z, Wong E, Tang G, Koga H, Kaushik S, et al. Cargo recognition failure is responsible for inefficient autophagy in Huntington’s disease. Nat Neurosci. 2010;13:567–76.
Texada MJ, Malita A, Christensen CF, Dall KB, Faergeman NJ, Nagy S, et al. Autophagy-mediated cholesterol trafficking controls steroid production. Dev Cell. 2019;48:659–71.
Shibata M, Yoshimura K, Furuya N, Koike M, Ueno T, Komatsu M, et al. The MAP1-LC3 conjugation system is involved in lipid droplet formation. Biochem Biophys Res Commun. 2009;382:419–23.
Shibata M, Yoshimura K, Tamura H, Ueno T, Nishimura T, Inoue T, et al. LC3, a microtubule-associated protein1A/B light chain3, is involved in cytoplasmic lipid droplet formation. Biochem Biophys Res Commun. 2010;393:274–9.
He P, Smith A, Gelissen IC, Ammit AJ. The effect of statins and the synthetic LXR agonist T0901317 on expression of ABCA1 transporter protein in human lung epithelial cell lines in vitro. Pharmacol Rep. 2019;71:1219–26.
Price NL, Rotllan N, Zhang X, Canfrán-Duque A, Nottoli T, Suarez Y, et al. Specific disruption of Abca1 targeting largely mimics the effects of miR-33 knockout on macrophage cholesterol efflux and atherosclerotic plaque development. Circ Res. 2019;124:874–80.
Frambach S, de Haas R, Smeitink JAM, Rongen GA, Russel FGM, Schirris TJJ. Brothers in arms: ABCA1- and ABCG1-mediated cholesterol efflux as promising targets in cardiovascular disease treatment. Pharmacol Rev. 2020;72:152–90.
Hsieh J, Koseki M, Molusky MM, Yakushiji E, Ichi I, Westerterp M, et al. TTC39B deficiency stabilizes LXR reducing both atherosclerosis and steatohepatitis. Nature. 2016;535:303–7.
Liu Y, Tang C. Regulation of ABCA1 functions by signaling pathways. Biochim Biophys Acta. 2012;1821:522–9.
Qian H, Zhao X, Cao P, Lei J, Yan N, Gong X. Structure of the human lipid exporter ABCA1. Cell. 2017;169:1228–39.
Zhang CJ, Zhu N, Liu Z, Shi Z, Long J, Zu XY, et al. Wnt5a/Ror2 pathway contributes to the regulation of cholesterol homeostasis and inflammatory response in atherosclerosis. Biochim Biophys Acta Mol Cell Biol Lipids. 2020;1865:158547.
Acknowledgements
This work was supported by grants from the National Natural Science Foundation of China (No. 81973668, No. 81774130 and No. 81270359); the Natural Science Foundation of Hunan Province for Distinguished Young Scholars (No. 2018JJ1018); and the First-Class Discipline of Pharmaceutical Science of Hunan.
Author information
Authors and Affiliations
Contributions
LQ designed the research; CJZ and JL performed the research and analyzed the data; CJZ and YXW performed the in vivo experiments; NZ and HTW provided clinical samples; CJZ wrote the paper; and BYL and DFL revised the paper.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Rights and permissions
About this article
Cite this article
Zhang, Cj., Zhu, N., Long, J. et al. Celastrol induces lipophagy via the LXRα/ABCA1 pathway in clear cell renal cell carcinoma. Acta Pharmacol Sin 42, 1472–1485 (2021). https://doi.org/10.1038/s41401-020-00572-6
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41401-020-00572-6
Keywords
This article is cited by
-
Selective autophagy in cancer: mechanisms, therapeutic implications, and future perspectives
Molecular Cancer (2024)
-
CCDC25 suppresses clear cell renal cell carcinoma progression by LATS1/YAP-mediated regulation of the hippo pathway
Cancer Cell International (2024)
-
A novel autophagy-related long non-coding RNAs prognostic risk score for clear cell renal cell carcinoma
BMC Urology (2022)
-
The regulation, function, and role of lipophagy, a form of selective autophagy, in metabolic disorders
Cell Death & Disease (2022)
-
A novel therapeutic strategy for atherosclerosis: autophagy-dependent cholesterol efflux
Journal of Physiology and Biochemistry (2022)