Abstract
The 2020 Annual Review Issue, “Preventing Disease in the 21st Century” was selected by the Editors-in-Chief of Pediatric Research to include a variety of disease entities that confront health-care practitioners entrusted to the care of infants and children. In keeping with this mandate, this article reviews the subject of epigenetics, which impacts pediatric research from bench to bedside. Epigenetic mechanisms exert their effects through the interaction of environment, various susceptibility genes, and immunologic development and include: (1) DNA methylation; (2) posttranslational modifications of histone proteins through acetylation and methylation, and (3) RNA-mediated gene silencing by microRNA (miRNA) regulation. The effects of epigenetics during fetal life and early periods of development are first reviewed together with clinical applications of cardiovascular and metabolic disorders in later life. The relationships of epigenetics to the allergic and autoimmune diseases and cancer are next reviewed. A specific focus of the article is directed to the recent recognition that many of these disorders are driven by aberrant immune responses in which immunoregulatory events are often poorly functioning and where through interventive epigenetic measures prevention may be possible by alterations in programming of DNA during fetal and early periods as well as in later life.
Similar content being viewed by others
Introduction
In the 2007 Annual Review Issue of Pediatric Research that was dedicated to “Epigenetics,” Devaskar et al.1 described epigenetics as a science of heritable biological adaptation involving non-Mendelian heritable changes in gene expression that are not mediated by alterations in the DNA sequence. The term epigenetics was coined originally by Conrad Waddington (1905–1975) who defined it as “the branch of biology which studies the causal interactions between genes and their products, which bring the phenotype into being.”2
The topic of the 2020 Annual Review Issue, entitled “Preventing Disease in the 21st Century,” was selected by the Editors-in-Chief of Pediatric Research to include a wide variety of disease entities that not only confront health-care practitioners entrusted to the care of infants and children but also include issues that directly impact management with a focus on prevention. In keeping with this mandate, the present article returns to the subject of epigenetics, which not only has seen a supernova of research contributions since the 2007 report but also continues to impact pediatric research and its translational applications from bench to bedside. Moreover, nowhere is the potential for prevention of disease better seen than in modalities that affect gene expression through epigenetic intervention.
In this article, the effects of environmental stress during fetal development and later life on the development of subsequent cardiovascular and metabolic disorders in adulthood are reviewed together with the relationships of genetics and epigenetics to the allergic and autoimmune diseases and cancer. As a framework for this presentation and for ease of discussion, epigenetics exerts its effects through the interaction of environment, various susceptibility genes, and immunologic development, e.g., age (Fig. 1).3 A specific focus of the article is directed to the recent recognition that many of these disorders are driven by aberrant immune responses in which immunoregulatory events are often poorly functioning and where through interventive epigenetic measures prevention may be possible by alterations in programming of DNA during fetal and early periods as well as later life.
(Reproduced with permission from Bellanti3).
Definition of epigenetics
In the late 1950s, Conrad Hal Waddington, a leading embryologist and geneticist,2 introduced the term epigenetics to define changes in gene function that occur without changes in gene sequence.4 He established a new system to study mechanisms that control gene expression without changing the basic nucleotide sequence of the DNA molecule. The sequencing of the human genome at the turn of this century had led to the hopeful belief that examination of the DNA molecule by candidate gene and genome-wide association studies (GWAS) could ultimately be the basis for an understanding of all complex diseases. While the importance of gene sequencing has unquestionably contributed greatly to our understanding of complex diseases, it was soon recognized that epigenetics played another important role in the control gene expression.5 A major investigative effort subsequently shifted the focus of genetic studies from the structure of the DNA molecule to mechanisms “above the DNA molecule” or “in addition to the DNA molecule” using the new epigenetic concepts for inheritance introduced by Waddington. This investigative shift provided the long sought-after link between genetics and environmental triggers that give rise to clinical disorders ranging from improperly regulated molecular pathways during embryonic and fetal development to a vast number of immunologically mediated diseases occurring in later life that include allergic diseases, the autoimmune disorders, and cancer.5,6
Three mechanisms of epigenetics
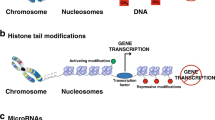
In contrast to the DNA of prokaryotes, which is organized in circular form, the DNA of eukaryotes, is arranged in individual packets known as nucleosomes that are assembled in a linear arrangement to form chromatin (Fig. 2).5,6 The structure of a nucleosome consists of a histone protein encircled by two loops of DNA and when assembled forms chromatin that exists in two molecular configurations: (1) a metabolically active form called euchromatin where the nucleosomes are set apart (an arrangement required for transcription) and (2) a metabolically inactive form, termed heterochromatin, where the nucleosomes are closely packed in a configuration that limits access of the transcription factors to the chromatin.
a Nucleosomes consist of histone proteins (shown in blue) surrounded by two loops of DNA and when assembled as beads on a string form chromatin, the building block of a chromosome. Reversible and site-specific histone modifications occur at multiple sites through acetylation, methylation, and phosphorylation. DNA methylation occurs at 5 position of cytosine residues in a reaction catalyzed by DNA methyltransferases (DNMTs). Together, these modifications provide a unique epigenetic signature that regulates chromatin organization and gene expression. b Schematic of the reversible changes in chromatin organization that influence gene expression: genes are expressed (switched on) when the chromatin is open (active) in the form of euchromatin, and they are inactivated (switched off) when the chromatin is condensed (silent) in the form of heterochromatin. White circles = unmethylated cytosines; red circles = methylated cytosines. (Modified from Rodenhiser and Mann5).
There are three major epigenetic mechanisms that include: (1) DNA methylation, (2) posttranslational modifications of histone proteins through acetylation and methylation, and (3) RNA-mediated gene silencing by microRNA (miRNA). Regulation of DNA methylation, which refers to the addition of a methyl group to the 5-carbon (C5) position of a cytosine by DNMTs and is the best-studied of these epigenetic mechanisms. This DNA modification occurs at a region rich in CpG residues, which are termed CpG islands often found in regulatory regions of the DNA molecule. Gene expression is predominantly influenced by the proximity of these CpG islands to gene promoters as well as their methylation status. These structural modifications not only alter chromatin into a transcriptionally repressive or permissive configuration but also serve as signals to other chromatin-modifying complexes.
Principles of epigenetics: DNA methylation and histone modifications and miRNA regulation
Gene expression is accomplished by activation or inhibition of the DNA and the globular histone proteins in the doublet strands that form chromatin (Fig. 2a). In eukaryotes, genomic DNA is organized into chromatin, which physically restricts accessibility of the information encoded in the genome to regulatory proteins.7 Originally thought of as a simple structural device for compaction of the DNA inside the nucleus, it is now recognized that chromatin is a dynamic and highly regulated structure and that changes in its physical configuration, such as chromatin unwinding, can result in differential accessibility of genomic information during development and growth. This step occurs at DNA synthesis during cell division and RNA synthesis at the time of transcriptional processing.
The functions of DNA methylation can be compared to a switch either in the “open” or “closed” positions.5,6 Genes are inactivated (switched off) when the chromatin is condensed (silent) in the heterochromatin form and are expressed (switched on) when chromatin is open (active) in the euchromatin form (Fig. 2b). These dynamic chromatin states are controlled by reversible epigenetic patterns of DNA methylation and histone modifications carried out by enzymes that include DNA methyltransferases (DNMTs), histone deacetylases (HDACs), histone acetylases, histone methyltransferases, and the methyl-binding domain protein MECP2. Profound and diverse clinical outcomes result from alterations in these normal epigenetic patterns that can deregulate patterns of gene expression. The “closed position” of the switch is associated with increased DNA methylation, genome stability, and decreased gene expression, whereas the “open position” with DNA hypomethylation leads to active transcription and increased gene expression.5,6
RNA-mediated gene silencing by microRNA (miRNA) regulation is accomplished by miRNA, a small non-coding RNA molecule (containing about 22 nucleotides) that functions in RNA silencing and posttranscriptional regulation of gene expression.8 The miRNAs function via base-pairing with complementary sequences within mRNA molecules following which the mRNA molecules are silenced by one or more of the following processes: (1) cleavage of the mRNA strand into two pieces, (2) destabilization of the mRNA through shortening of its poly(A) tail, and (3) less efficient translation of the mRNA into proteins by ribosomes.
Epigenetics and human development: fetal life and early periods of development
During fetal and early periods of development, epigenetics plays a major role in organ development and cell differentiation. It is now generally accepted that environmental encounters during pregnancy alter the programmed fetal development and subsequently cause disorders, such as cardiovascular and metabolic diseases, in adulthood. In a paper by Cutfield et al.,9 the phenotypic and biochemical characteristics during childhood of three low birth weight groups are summarized: children born following in vitro fertilization, small for gestational age, or very premature. Each of these groups is likely to have been exposed to an adverse environment at different developmental stages.10,11,12,13 The triggers and mechanisms leading to programmed changes in growth, development, and metabolism of these groups of children have yet to be identified. Epigenetics has now been proposed as a potential mechanism for these programmed changes through environmentally induced changes in gene expression. Since epigenetics leads to gene regulation and expression that is not dependent on DNA sequence but on chemical modifications of DNA, several lines of evidence suggest that environmental stress in the fetal period alters the epigenetic state of genes, leading to permanent gene dysregulation, which may be associated with disorders that emerge after birth.10,11,12,13 Such stresses include malnutrition, which may be associated with type 2 diabetes, and mental stress, which may be associated with neurodevelopmental disorders. It has also been demonstrated that environmental stress-induced epigenetic alterations can be transmitted to the next generation via disease phenotypes. However, since epigenetic modification is an internal system based on attachment and detachment of chemical residues on a DNA sequence, it is reversible and potentially treatable. In fact, recent studies demonstrated that some drugs and early interventions are effective at preventing epigenetic disorders. Therefore, preventive and preemptive medicine is possible for disorders caused by alterations in programming during fetal and early periods.
A significant challenge in modern clinical genetics involves distinguishing phenotypic overlap characteristics among complex genetic disorders that often lack distinctive characteristic features. This is especially relevant for syndromes involving developmental disabilities (DD) and intellectual disabilities that often share common molecular pathways that are improperly regulated during embryonic and fetal development. Bjornsson et al.14,15 have coined the phrase “Mendelian Disorders of the Epigenetic Machinery” to describe these developmental disorders that are associated with mutations in genes encoding proteins that read, write, and erase these epigenetic modifications to DNA and histone proteins, which regulate chromatin accessibility and transcription.
In a recent report, Sadikovic et al.16 review recent genome-wide mapping approaches that have identified DNA methylation episignatures in patients clinically diagnosed with syndromes manifesting as DD with intellectual impairments. Shown in Table 1 is a list of DNA methylation episignatures identified in several of these developmental disorders, which include gene loci, clinical descriptions of the disorder and number of relevant CpGs, and literature investigative sources.17,18,19,20,21,22,23,24,25,26,27,28,29,30,31,32,33 Although the discovery of these DNA methylation episignatures has provided a nearly perfect performance in molecular diagnosis for these developmental syndromes, the clinical significance remains unanswered. In the Sadikovic et al., report,16 the authors propose that these episignatures be considered as echoes and/or legacies of the initiating mutational events within proteins of the so-called epigenetic machinery and discuss approaches to directly confirm the functional consequences and the implications of these episignatures to patient management and treatment.
Another key aspect of epigenetics and fetal life/health is nutrition.34,35,36 In these studies, the critical role of early-life nutrition on fetal growth and development is highlighted where food intake absence or excess are the two main types of energy malnutrition that predispose to the appearance of diseases in adulthood, according to the hypothesis of “developmental origins of health and disease” (DOHaD).37 Shown in Fig. 3 is a schematic representation of various environmental, maternal, placental, and fetal factors, which can alter different epigenetic processes in the fetus that contribute to DOHaD. Not only has an association between early-life malnutrition and metabolic disorders in later life been established but also emerging evidence suggests that nutrition during this period of life can affect the development of the immune system through epigenetic mechanisms. DNA methylation, covalent modification of histones, and the expression of non-coding RNA, for example, have been shown to play essential roles in the complex interplay between environmental factors and genetics that can lead to inflammatory responses commonly seen in non-alcoholic fatty liver disease and the hepatic manifestations of the metabolic syndrome.34 Thus, current understanding of the mechanisms underlying developmental programming are critically linked to epigenetic modulation of the immune system and environmental factors, such as malnutrition.
(Reproduced with permission from Goyal et al.37).
Epigenetics and human disease: the autoimmune disorders, allergic diseases, and cancer
Epigenetics and autoimmune diseases (ADs)
The autoimmune diseases were not only the first of the clinical disorders to demonstrate an association of epigenetics with clinical disease38 but also the foremost to establish their pathogenesis by the interaction of hereditary (i.e., genetic) and environmental (i.e., epigenetic) factors over time (Fig. 1). The development of ADs and their outcome are now recognized to be triggered by a wide variety of environmental exposures (i.e., cigarette smoking, coffee and alcohol intake, socioeconomic status, chemical agents, vaccines, incidental exposures, infectious agents, and microbiota; Fig. 4).39
Prenatal exposure to these agents is postulated to cause epigenetic changes during intrauterine life thereby effecting early immune programming that may have lasting effects during the child’s life. (Reproduced with modification and with permission from Sabounchi S, Bollyky J, Nadeau K39).
The manner in which systemic lupus erythematosus (SLE) established the contribution of epigenetics in the pathogenesis of autoimmunity was based on studies demonstrating a striking lack of concordance of monozygotic twins to develop SLE. If genetics were the sole controlling pathogenetic mechanism, a concordance of 100% would be expected in the other twin 100% of the time. Instead, a frequency of 50–70% involvement was observed suggesting involvement of an environmentally mediated epigenetic contribution. Moreover, the discordance noted in the early monozygotic twin studies as well as racial and geographic variations in prevalence studies suggested a strong interaction between the environment and the underlying genomic code in SLE.40 The molecular mechanism proposed for this epigenetic modification is through oxidative stress by exposure to environmental agents, such as cigarette smoke, mercury, silica, ultraviolet light, viral infections, and medications that are known to inhibit and/or decrease DNMT1 activity, thus reducing DNA methylation in CD4+ T cells and enhancing autoimmunity.41
Several excellent publications have recently been published summarizing the most recent advances in autoimmunity, which include an update on applications of human epigenetic studies in this field.42,43,44,45 The study of the microbiome remains central to the pathogenesis of autoimmunity and data are being gathered in a growing number of autoimmune conditions, stressing the role of epigenetics and long non-coding RNA. In the cases of specific diseases, such as SLE, rheumatoid arthritis, and psoriatic arthritis, multiple encouraging findings underscore the importance of translational research between basic and clinical domains of science in defining new pathogenetic and therapeutic applications. Cumulatively, the future of autoimmunity appears bright and should be looked upon as one of the fastest growing applications of epigenetics in the field of immunology.
Epigenetics and allergic disease
Several recent studies have been shown to involve epigenetic modifications in allergic disease through similar mechanisms described for the autoimmune disorders Fig. 4.39,46,47 Although still in early infancy, these studies are showing great promise for the application of epigenetics to better diagnostic and therapeutic applications in the allergic diseases. These include better predictive biomarkers that not only identify individuals at risk of developing allergic disease48,49,50,51,52 but also that determine outcomes of immunotherapy and predict responses to other forms of personalized therapy.53 A very recent large-scale international genome-wide meta-analysis conducted prospectively in newborns and cross-sectionally in children identified variation in DNA methylation related to childhood asthma, which may provide additional biomarkers of asthma development and biologic effects that may shed light on disease mechanisms.54
Another emerging investigative area of importance to epigenetics and allergy is the gut microbiome and its modification by diet.55 Several lines of evidence suggest that the gut microbiome may play a critical role in the prevention of allergic disease particularly by commensal bacteria, which have been shown to have important tolerogenic effects on the immune system through the stimulation of T regulatory (Treg) cells.56 In humans, fermentation of a group of carbohydrates, collectively known as resistant starch, are acted upon by these commensal bacteria in the colon to produce products, which have potent Treg-stimulating effects.57 The primary source of these carbohydrates for breastfed infants is human milk in the form of human milk oligosaccharides and plant cell wall carbohydrates found in fiber-rich solid foods in later life. The major products of this fermentation reaction include the short-chain fatty acids, particularly, butyrate, which has been shown to have potent anti-inflammatory effects mediated by Treg induction.58,59 One of the major epigenetic mechanisms by which butyrate exerts its actions is through inhibition of HDACs. Collectively, these studies suggest that modulation of the host epigenome through dietary modification of the gut microbiome may offer innovative therapeutic strategies for the prevention of allergic diseases.60
Epigenetics and cancer
Abnormal DNA methylation in cancer has been the focus of an increasing number of recent studies.61 This is consistent with many of the concepts of DNA methylation described above that control higher-order chromatin structure, which are now being utilized clinically as markers for cancer formation and tumor progression. Aberrant epigenetic DNA methylation patterns in cancer, including DNA methylation, histone modifications, and noncoding expression, have long been reported to play a major role in the genesis of cancer62,63 and to be driven by the accumulation of genetic and epigenetic alterations that result in dysregulation of key oncogenes, tumor-suppressor genes, and DNA repair/housekeeping genes. Although the complete sequential steps required for the development of human cancer are, as yet, unknown, both the activation of oncogenes and the inactivation of tumor-suppressor genes are two critical steps required for tumor initiation and progression (Fig. 5). In the healthy host, the tumor-suppressor region is normally unmethylated and activated in the “open” position while its corresponding DNA oncogene region is methylated and inactivated in the “closed” position. In contrast, during the course of the development of cancer, methylation of the promoter region of the DNA tumor-suppressor region in the “closed” position leads to its inactivation while unmethylation of the corresponding DNA oncogene region in the “open” position leads to its activation.5,6 The cumulative damage resulting from additional multiple epigenetic modifications to other genes are thought to lead to the development of cancer, which is characterized by the increased cellular proliferation, invasiveness, and metastatic potential characteristic of the malignant transformation.
Inactivation of tumor-suppressor genes (by DNA methylation) and inactivation of oncogenes (by DNA hypomethylation) are critical steps in tumor initiation and progression. (Reproduced with modification and with permission from Paska and Hudler62).
There is a dearth of data related to the epigenetics of autoimmune diseases and cancer in the pediatric area but the following are some of the published studies that relate to this area.64,65,66,67,68,69,70,71,72
Conclusions
Epigenetic editing, once thought a futuristic fantasy, has now become a reality. Leveraging cutting-edge and highly specific DNA-binding domains, epigenetic-editing platforms comprise a wide variety of changes, including DNA methylation and hypomethylation and histone posttranslational modifications. Epigenetic editing has the advantage of modifying and presenting DNA sequences and other DNA-binding factors that influence DNA expression without the more complex and more difficult task of gene editing that involves changing the actual DNA sequence itself. Although these approaches have not yet been implemented widely in developmental disorders and the immunological and autoimmune diseases, there is certainly great potential for this in the future, given the extensive existing data linking alterations in epigenetic states, particularly DNA methylation, with virtually every developmental disorder and several autoimmune and allergic disorders. Although several roadblocks remain, the future for epigenetic editing in pediatric research is bright indeed.
References
Devaskar, S. U. & Raychaudhuri, S. Epigenetics-a science of heritable biological adaptation. Pediatr. Res. 61(Pt 2), 1R–4R (2007).
Slack, J. M. Conrad Hal Waddington: the last Renaissance biologist? Nat. Rev. Genet. 3, 889–895 (2002).
Bellanti, J. A. (ed.) Immunology IV: Clinical Applications in Health and Disease (I Care Press, Bethesda, MD, 2012).
Waddington, C. The Study of the Genes (Allen and Unwin, London, 1957).
Rodenhiser, D. & Mann, M. Epigenetics and human disease: translating basic biology into clinical applications. CMAJ 174, 341–348 (2006).
Bellanti, J. A. Genetics/epigenetics/allergy: the gun is loaded … but what pulls the trigger? Allergy Asthma Proc. 40, 76–83 (2019).
Kwon, C. S. & Wagner, D. Unwinding chromatin for development and growth: a few genes at a time. Trends Genet. 23, 403–412 (2007).
Ambros, V. The functions of animal microRNAs. Nature 431, 350–355 (2004).
Cutfield, W. S. et al. Could epigenetics play a role in the developmental origins of health and disease? Pediatr. Res. 61(Pt 2), 68R–75R (2007).
Yeh, T. F. et al. Outcomes at school age after postnatal dexamethasone therapy for lung disease of prematurity. N. Engl. J. Med. 350, 1304–1313 (2004).
Niemann, H. & Wrenzycki, C. Alterations of expression of developmentally important genes in preimplantation bovine embryos by in vitro culture conditions: implications for subsequent development. Theriogenology 53, 21–34 (2000).
Eppig, J. J. & O’Brien, M. J. Comparison of preimplantation developmental competence after mouse oocyte growth and development in vitro and in vivo. Theriogenology 49, 415–422 (1998).
Doherty, A. S. et al. Differential effects of culture on imprinted H19 expression in the preimplantation mouse embryo. Biol. Reprod. 62, 1526–1535 (2000).
Fahrner, J. A. & Bjornsson, H. T. Mendelian disorders of the epigenetic machinery: tipping the balance of chromatin states. Annu. Rev. Genomics Hum. Genet. 15, 269–293 (2014).
Bjornsson, H. T. The Mendelian disorders of the epigenetic machinery. Genome Res. 25, 1473–1481 (2015).
Sadikovic, B., Aref-Eshghi, E., Levy, M. A. & Rodenhiser, D. DNA methylation signatures in mendelian developmental disorders as a diagnostic bridge between genotype and phenotype. Epigenomics 11, 563–575 (2019).
Strong, E. et al. Symmetrical dose-dependent DNA-methylation profiles in children with deletion or duplication of 7q11.23. Am. J. Hum. Genet. 97, 216–227 (2015).
Aref-Eshghi, E. et al. BAFopathies’ DNA methylation epi-signatures demonstrate diagnostic utility and functional continuum of Coffin-Siris and Nicolaides-Baraitser syndromes. Nat. Commun. 9, 4885 (2018).
Schenkel, L. C. et al. Identification of epigenetic signature associated with alpha thalassemia/mental retardation X-linked syndrome. Epigenet. Chromatin 10, 10 (2017).
Aref-Eshghi, E. et al. Genomic DNA methylation signatures enable concurrent diagnosis and clinical genetic variant classification in neurodevelopmental syndromes. Am. J. Hum. Genet. 102, 156–174 (2018). **Describes the identification and use of DNA methylation episignatures in 14 developmental disorders.
Butcher, D. T. et al. CHARGE and Kabuki syndromes: gene-specific DNA methylation signatures identify epigenetic mechanisms linking these clinically overlapping conditions. Am. J. Hum. Genet. 100, 773–788 (2017).
Kernohan, K. D. et al. Identification of a methylation profile for DNMT1-associated autosomal dominant cerebellar ataxia, deafness, and narcolepsy. Clin. Epigenet. 8, 91 (2016).
Schenkel, L. C. et al. Clinical validation of Fragile X syndrome screening by DNA methylation array. J. Mol. Diagn. 18, 834–841 (2016).
Aref-Eshghi, E. et al. Clinical validation of a genome-wide dna methylation assay for molecular diagnosis of imprinting disorders. J. Mol. Diagn. 19, 848–856 (2017).
Prickett, A. R. et al. Genome-wide methylation analysis in Silver-Russell syndrome patients. Hum. Genet. 134, 317–332 (2015).
Wu, D., Gong, C. & Su, C. Genome-wide analysis of differential DNA methylation in Silver-Russell syndrome. Sci. China Life Sci. 60, 692–699 (2017).
Schenkel, L. C. et al. Peripheral blood epi-signature of Claes-Jensen syndrome enables sensitive and specific identification of patients and healthy carriers with pathogenic mutations in KDM5C. Clin. Epigenet. 10, 21 (2018).
Aref-Eshghi, E. et al. The defining DNA methylation signature of Kabuki syndrome enables functional assessment of genetic variants of unknown clinical significance. Epigenetics 12, 923–933 (2017).
Li, Y. et al. An epigenetic signature in peripheral blood associated with the haplotype on 17q21.31, a risk factor for neurodegenerative tauopathy. PLoS Genet. 10, e1004211 (2014).
Choufani, S. et al. NSD1 mutations generate a genome-wide DNA methylation signature. Nat. Commun. 6, 10207 (2015).
Hood, R. L. et al. The defining DNA methylation signature of Floating-Harbor syndrome. Sci. Rep. 6, 38803 (2016).
Bacalini, M. G. et al. Identification of a DNA methylation signature in blood cells from persons with Down Syndrome. Aging 7, 82–96 (2015).
Guastafierro, T. et al. Genome-wide DNA methylation analysis in blood cells from patients with Werner syndrome. Clin. Epigenet. 92, 28–31 (2017).
Campisano, S., La Colla, A., Echarte, S. M. & Chisari, A. N. Interplay between early-life malnutrition, epigenetic modulation of the immune function and liver diseases. Nutr. Res. Rev. 32, 128–145 (2019).
Lesseur, C. & Chen, J. Adverse maternal metabolic intrauterine environment and placental epigenetics: implications for fetal metabolic programming. Curr. Environ. Health Rep. 5, 531–543 (2018).
Tiffon, C. The impact of nutrition and environmental epigenetics on human health and disease. Int. J. Mol. Sci. 19, E3425 (2018).
Goyal, D., Limesand, S. W. & Goyal, R. Epigenetic responses and the developmental origins of health and disease. J. Endocrinol. 242, T105–T119 (2019).
Picascia, A. et al. Epigenetic control of autoimmune diseases: from bench to bedside. Clin. Immunol. 157, 1–15 (2015).
Sabounchi, S., Bollyky, J. & Nadeau, K. Review of Environmental impact on the epigenetic regulation of atopic diseases. Curr. Allergy Asthma Rep. 15, 1–33 (2015).
Teruel, M. & Alarcón-Riquelme, M. E. The genetic basis of systemic lupus erythematosus: what are the risk factors and what have we learned. J. Autoimmun. 74, 161–175 (2016).
Richardson, B. The interaction between environmental triggers and epigenetics in autoimmunity. Clin. Immunol. 192, 1–5 (2018).
Chen, W., Liu, D., Li, Q. Z. & Zhu, H. The function of ncRNAs in rheumatic diseases. Epigenomics 11, 821–833 (2019)
Selmi, C. Autoimmunity in 2018. Clin. Rev. Allergy Immunol. 56, 375–384 (2019).
Selmi, C. & Gershwin, M. E. Sex and autoimmunity: proposed mechanisms of disease onset and severity. Expert Rev. Clin. Immunol. 15, 607–615 (2019).
Charras, A. & Hedrich, C. M. The role of epigenetics in paediatric rheumatic disease. Curr. Opin. Rheumatol. 31, 450–463 (2019).
Potaczek, D. P. et al. Epigenetics and allergy: from basic mechanisms to clinical applications. Epigenomics 9, 539–571 (2017).
North, M. L. & Ellis, A. K. The role of epigenetics in the developmental origins of allergic disease. Ann. Allergy Asthma Immunol. 106, 355–361 (2011).
Tost, J. A translational perspective on epigenetics in allergic diseases. J. Allergy Clin. Immunol. 142, 715–726 (2018).
Peng, C. et al. Epigenome-wide association study of total serum immunoglobulin E in children: a life course. Clin. Epigenet. 10, 55 (2018).
DeVries, A. & Vercelli, D. The neonatal methylome as a gatekeeper in the trajectory to childhood asthma. Epigenomics 9, 585–593 (2017).
DeVries, A. & Vercelli, D. Early predictors of asthma and allergy in children: the role of epigenetics. Curr. Opin. Allergy Clin. Immunol. 15, 435–439 (2015).
DeVries, A. & Vercelli, D. Epigenetics in allergic diseases. Curr. Opin. Pediatr. 27, 719–723 (2015).
Bunning, B. J., DeKruyff, R. H. & Nadeau, K. C. Epigenetic changes during food-specific immunotherapy. Curr. Allergy Asthma Rep. 16, 87 (2016).
Reese, S. E. et al. Epigenome-wide meta-analysis of DNA methylation and childhood asthma. J. Allergy Clin. Immunol. 143, 2062–2074 (2019).
Hughes, R. L. et al. The role of the gut microbiome in predicting response to diet and the development of precision nutrition models-Part I. Overview of current methods. Adv. Nutr. https://doi.org/10.1093/advances/nmz022 (2019).
Walker, M. M., Talley, N. J. & Keely, S. Follow up on atopy and the gastrointestinal tract - a review of a common association 2018. Expert Rev. Gastroenterol. Hepatol. 13, 437–445 (2019).
Cait, A. et al. Reduced genetic potential for butyrate fermentation in the gut microbiome of infants who develop allergic sensitization. J. Allergy Clin. Immunol. https://doi.org/10.1016/j.jaci.2019.06.029 (2019).
Thio, C. L. et al. Regulation of type 2 innate lymphoid cell-dependent airway hyperreactivity by butyrate. J. Allergy Clin. Immunol. 142, 1867–1883 (2018).
Wang, J., Wen, L., Wang, Y. & Chen, F. Therapeutic effect of histone deacetylase inhibitor, sodium butyrate, on allergic rhinitis in vivo. DNA Cell Biol. 35, 203–208 (2016).
Zhu, Z. et al. Short-chain fatty acids as a target for prevention against food allergy by regulatory T cells. JGH Open 3, 190–195 (2019).
Ehrlich, M. DNA hypermethylation in disease: mechanisms and clinical relevance. Epigenetics 8, 1–23 (2019).
Paska, A. V. & Hudler, P. Aberrant methylation patterns in cancer: a clinical view. Biochem. Med. (Zagreb) 25, 161–176 (2015).
Robertson, K. D. DNA methylation and human disease. Nat. Rev. Genet. 6, 597–610 (2005).
Martino, D. et al. Epigenetics in immune development and in allergic and autoimmune diseases. J. Reprod. Immunol. 104-105, 43–48 (2014).
Chavez-Valencia, R. A. et al. The DNA methylation landscape of CD4(+) T cells in oligoarticular juvenile idiopathic arthritis. J. Autoimmun. 86, 29–38 (2018).
Phillips, J. E., Couper, J. J., Penno, M. A. S., Harrison, L. C. & ENDIA Study Group. Type 1 diabetes: a disease of developmental origins. Pediatr. Diabetes 18, 417–421 (2017).
Langie, S. A. et al. DNA methylation and the hygiene hypothesis: connecting respiratory allergy and childhood acute lymphoblastic leukemia. Epigenomics 11, 1519–1537 (2019).
Gentner, M. B. & Leppert, M. L. O. Environmental influences on health and development: nutrition, substance exposure, and adverse childhood experiences. Dev. Med. Child Neurol. 61, 1008–1014 (2019).
McCann, T. S. et al. Biology and targeting of the Jumonji-domain histone demethylase family in childhood neoplasia: a preclinical overview. Expert Opin. Ther. Targets 23, 267–280 (2019).
Mika, A. et al. Epigenetic changes at the Birc5 promoter induced by YM155 in synovial sarcoma. J. Clin. Med. 8, E408 (2019).
Flotho, C. Gene mutations do not operate in a vacuum: the increasing importance of epigenetics in juvenile myelomonocytic leukemia. Epigenetics 14, 236–244 (2019).
Lawless, O. J. et al. In vitro induction of T regulatory cells by a methylated CpG DNA sequence in humans: potential therapeutic applications in allergic and autoimmune diseases. Allergy Asthma Proc. 39, 143–152 (2018).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The author declares no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Bellanti, J.A. Epigenetic studies and pediatric research. Pediatr Res 87, 378–384 (2020). https://doi.org/10.1038/s41390-019-0644-9
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41390-019-0644-9
This article is cited by
-
Epigenetics of pregnancy: looking beyond the DNA code
Journal of Assisted Reproduction and Genetics (2022)