Abstract
Dysfunction of the cortical dorsal visual stream and visuospatial working memory (vsWM) network in individuals with schizophrenia (SZ) likely reflects alterations in both excitatory and inhibitory neurotransmission within nodes responsible for information transfer across the network, including primary visual (V1), visual association (V2), posterior parietal (PPC), and dorsolateral prefrontal (DLPFC) cortices. However, the expression patterns of ionotropic glutamatergic and GABAergic receptor subunits across these regions, and alterations of these patterns in SZ, have not been investigated. We quantified transcript levels of key subunits for excitatory N-methyl-D-aspartate receptors (NMDARs), excitatory alpha-amino-3-hydroxy-5-methyl-4-isoxazole propionic acid receptors (AMPARs), and inhibitory GABAA receptors (GABAARs) in postmortem total gray matter from V1, V2, PPC, and DLPFC of unaffected comparison (UC) and matched SZ subjects. In UC subjects, levels of most AMPAR and NMDAR mRNAs exhibited opposite rostral-to-caudal gradients, with AMPAR GRIA1 and GRIA2 mRNA levels highest in DLPFC and NMDAR GRIN1 and GRIN2A mRNA levels highest in V1. GABRA5 and GABRA1 mRNA levels were highest in DLPFC and V1, respectively. In SZ, most transcript levels were lower relative to UC subjects, with these differences largest in V1, intermediate in V2 and PPC, and smallest in DLPFC. In UC subjects, these distinct patterns of receptor transcript levels across the cortical vsWM network suggest that the balance between excitation and inhibition is achieved in a region-specific manner. In SZ subjects, the large deficits in excitatory and inhibitory receptor transcript levels in caudal sensory regions suggest that abnormalities early in the vsWM pathway might contribute to altered information processing in rostral higher-order regions.
Similar content being viewed by others
Introduction
Visuospatial working memory (vsWM), the ability to transiently maintain and manipulate a limited amount of visuospatial information to guide thought or behavior [1], is mediated by a distributed neural network that includes multiple nodes in the neocortex [2, 3]. In this network, which includes regions of the dorsal visual stream, visual information is conveyed from primary (V1) and association (V2) visual regions in the occipital cortex to higher-order regions in the posterior parietal (PPC) and dorsolateral prefrontal (DLPFC) cortices. Each region appears to contribute to the functional network associated with vsWM performance [4,5,6,7,8,9]. The functional interdependence of these regions is mediated by specific patterns of anatomical connections. For example, excitatory pyramidal neurons furnish axonal projections that convey information between regions, and the activity of these neurons is shaped by inputs from local inhibitory GABA interneurons [10].
Levels of presynaptic markers of GABAergic neurotransmission have been reported to differ across regions of the human vsWM network [11,12,13]. However, little is known about the regional patterns of postsynaptic indices of GABAergic or glutamatergic neurotransmission which depend, in part, on the expression of subunits that form ionotropic N-methyl-D-aspartate receptors (NMDARs), alpha-amino-3-hydroxy-5-methyl-4-isoxazole propionic acid receptors (AMPARs), or GABAA receptors (GABAARs) [14,15,16]. These receptors are each multimeric complexes composed of different types of subunits that influence a variety of receptor properties, including the kinetics of channel opening and closing, ion flow through those channels, and the localization of receptor complexes to synaptic or extra-synaptic sites [17,18,19,20]. For example, AMPARs mediate most fast-spiking excitatory transmission, whereas NMDARs are associated with relatively slower kinetics and higher calcium permeability [19]. Furthermore, subunits within a receptor family regulate specific functions. For example, of the AMPAR subunits, GRIA1 controls the insertion and removal of the receptor complex from the postsynaptic membrane [19] and GRIA2 is calcium-impermeable [19] and is often co-expressed with GRIA1 [21, 22]. Of the NMDAR subunits, GRIN1 is the obligatory subunit that forms a heteromeric complex predominantly with GRIN2A or GRIN2B [23, 24]. Of the GABAAR subunits, GABRA5 and GABRA1 are enriched in GABAARs postsynaptic to somatostatin neuron inputs [25, 26] and parvalbumin basket cell inputs [27], respectively; GABRB2 is heavily colocalized with GABRA1 [28]; and GABRG2, which is present in most synaptic GABAARs [29], is required for postsynaptic receptor clustering [30].
Previous studies have shown that each of these classes of receptors are critical for vsWM. For example, NMDAR antagonism in rhesus monkeys induces temporary vsWM deficits [31], pharmacological potentiation of AMPARs before NMDAR antagonism mitigates the impairments in vsWM [32], and administration of the GABAAR antagonist bicuculline impairs vsWM performance [33]. These experimental findings suggest that disease-related alterations in ionotropic NMDARs, AMPARs, and/or GABAARs in individuals with schizophrenia (SZ) could contribute to impaired vsWM in the disorder [4,5,6,7, 9]. Although postmortem studies cannot directly investigate how these alterations give rise to vsWM deficits observed in schizophrenia, they may provide insight into the underlying neural mechanisms. However, the expression levels of key subunits for these three receptor classes across cortical regions of the vsWM network have not been examined in unaffected comparison (UC) or SZ subjects. Consequently, we sought to answer two questions: (1) In UC subjects, do the levels of key subunits of ionotropic NMDARs, AMPARs, or GABAARs differ across regions of the human vsWM network? (2) Are levels of these transcripts altered in SZ in a manner that is conserved or distinct across cortical regions of the vsWM network?
To address these questions, we quantified the levels of nine key NMDAR, AMPAR, and GABAAR transcripts in total gray matter homogenates from four regions of the vsWM network from UC and SZ subjects. We also used existing data sets to compare the levels of these transcripts in two populations of neurons in UC subjects. We found distinct regional expression patterns that differed by transcript type in UC subjects, suggesting that the balance between excitation and inhibition is achieved in a region-specific manner. In SZ subjects, deficits in excitatory and inhibitory receptor transcript levels were greatest in caudal sensory regions, suggesting that abnormalities early in the vsWM pathway might contribute to altered information processing in rostral higher-order regions.
Materials and methods
Postmortem brain samples
Brain specimens (N = 40) were obtained during routine autopsies conducted at the Allegheny County Medical Examiner’s Office (Pittsburgh, PA) following consent obtained from the next of kin. Consensus DSM-IV diagnoses (or their absence) were made by an independent committee of experienced research clinicians using information obtained from structured interviews with family members and review of medical records, neuropathology exam and toxicology reports [34]. All procedures were approved by the University of Pittsburgh’s Committee for the Oversight of Research and Clinical Training Involving Decedents and Institutional Review Board for Biomedical Research. To control for experimental variance and reduce between-group biological variance, each SZ subject was matched with one UC subject for sex and as closely as possible for age. Subject groups did not differ in mean age, postmortem interval, brain pH, RNA integrity number (Agilent Bioanalyzer, Agilent Technologies, Santa Clara, CA), or tissue storage time at −80 °C (Table 1).
Tissue processing
The right hemisphere of each brain was blocked coronally, immediately frozen, and stored at −80 °C as previously described [34]. Based on their anatomic location and cytoarchitectonic features, four regions (V1, Brodmann area 17; V2, Brodmann area 18; PPC Brodmann area 7; and DLPFC, Brodmann area 46) were identified for sampling [13]. For each cortical region, in locations where tissue sections were cut perpendicular to the pial surface, total gray matter was dissected from cryostat sections (40 μm) in a manner that minimized white matter contamination or RNA degradation [35]. Samples were immediately homogenized in TRIzol reagent (Invitrogen) [36].
Transcript selection and quantitative polymerase chain reaction (qPCR) procedures
Samples from all four regions of both subjects in each pair were processed together throughout the experiment. For each sample, RNA was extracted and purified using the RNAeasy Plus Micro Kit (QIAGEN, Inc., Valencia, CA). Total RNA was converted to complementary DNA using the SuperScript IV VILO Master Mix (ThermoFisher Scientific). Forward and reverse primers were designed to target nine transcripts whose cognate proteins are known to be enriched at synapses involved in ionotropic glutamatergic or GABAergic neurotransmission. Due to the available 384 wells on qPCR plates, we selected nine transcripts of interest to optimize the experimental design for rigorous comparisons across regions and subject groups.
All primers showed 94–107% efficiency, and each amplified product resulted in a single and specific amplicon (Supplementary Table 1). Three normalizer genes (β-actin, glyceraldehyde-3-phosphate dehydrogenase (GAPDH) and cyclophilin-A) were used to normalize the target transcript levels as in prior studies of the same subject cohort [12, 13, 37]. Levels of each transcript were assessed by qPCR using Power SYBR Green fluorescence and the ViiA™7 Real Time qPCR System. Each 384 well plate contained four replicates of 12 transcripts (nine receptor subunits and three normalizers) from four subject pairs. For each transcript, cycle threshold (CT) values were assessed in four wells; any single CT values that increased the SD to ≥0.15 were excluded. The delta CT (dCT) for each target transcript was calculated by subtracting the mean CT of the three normalizer genes from the CT of the gene of interest. Because the dCT represents the log2-transformed expression ratio of each target transcript to the mean of the normalizer genes, the relative expression levels of the target transcripts are reported as the more intuitive expression ratio, or the 2−dCT. Statistical analysis of the mean of the three normalizer genes did not reveal a main effect of diagnosis (F1,40 = 3.2, p = 0.08) or an interaction between diagnosis and region (F3,120 = 0.8, p = 0.48). Across regions within the UC group, normalizers did significantly differ by region (F3,60 = 13.8, p < 0.001), with higher expression levels in V1 relative to the DLPFC, although the magnitude of this difference was very small (1.2%), and much smaller than the smallest regional difference in the transcripts of interest (e.g., GRIN1 mRNA levels were 17.7% higher in V1 than DLPFC). Furthermore, given the different regional patterns of expression in the UC individuals (see Fig. 1), the regional patterns are unlikely to be explained by differences in normalizer levels.
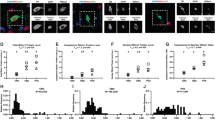
In A–I, regional gradients in levels differ in a transcript-specific manner. In J–L, ratios of subunits all significantly increase from caudal to rostral areas. In each panel, F and p values are from ANCOVA and areas not sharing the same letter are significantly different (p < 0.05) in post hoc analyses. Symbols represent the same subjects in all plots.
Statistics
For all analyses, we used mixed models in R treating observations from each region of each subject as repeated measures to account for the within-subject correlation among the four regions, as done previously [11,12,13]. To assess regional differences in the levels of each transcript in the UC subjects, the model included transcript as the dependent variable, region as a fixed effect, subject as a random term, and age, sex, RIN, and PMI as covariates. F-tests were used to assess the overall regional effect, with Tukey adjusted p values reported to control for multiple comparisons across the nine transcripts. Degrees of freedom were calculated via Satterthwaite’s method [38]. Post hoc comparisons between regions were made using Hotelling’s T2 statistic [39]. Ratios between two transcripts of interest were normalized via Z-score for plotting and interpretation purposes. For each ratio, the mixed model was performed as described above.
For comparisons between UC and SZ subjects, the mixed model included transcript as the dependent variable; diagnosis, region, and diagnosis-by-region interaction as fixed effects; and age, sex, RIN, and PMI as covariates. Control for multiple comparisons across transcripts and posthoc analyses within transcript were conducted as described above. Cohen’s d effect sizes were used to estimate the magnitudes of the differences between regions and diagnoses.
Analysis of RNA sequencing data from pools of neurons
To assess the technical and biological reliability of our qPCR findings, and the extent to which these findings in total gray matter are conserved or different in two neuronal subtypes, we performed a secondary analysis of the same transcripts from a recently published RNA sequencing dataset of layer 3 pyramidal neurons (L3PNs) [40] and from an unpublished RNA sequencing dataset of layer 3 neurons with aggrecan-positive perineuronal nets which are predominately parvalbumin-containing basket cells (L3PVBCs) [41, 42]. Pools of 100 individually dissected L3PNs and 150 individually dissected aggrecan-positive L3PVBCs were collected from V1 and DLPFC of 39 and 40 UC subjects, respectively. All 20 UC subjects assessed via qPCR were included in the cohort used for RNA sequencing. To assess regional differences in the same nine glutamate and GABA receptor subunit transcripts, an analysis of variance (ANOVA) linear regression model was used in LIMMA along with the precision weights obtained during initial voom normalization of these data. To account for the non-independence of samples derived from each brain region from the same subject, the duplicateCorrelations function in LIMMA was implemented [40]. Covariates included in the model were age, sex, PMI, and RIN. Because the nine transcripts included in this study are a very small subset of the genes analyzed by RNA sequencing, we chose a conservative approach for assessing regional differences in the levels of the transcripts assayed. Specifically, statistical significance was determined using the Benjamini-Hochberg procedure with a false discovery rate of 5% across all 16,379 genes assayed.
Results
Expression of receptor subunit transcripts across the vsWM network in UC subjects
For AMPAR transcripts, both GRIA1 and GRIA2 mRNA levels differed significantly across all four areas of the vsWM network, with levels of both transcripts lowest in V1, progressively increasing from caudal to rostral areas, and highest in DLPFC (Fig. 1A, B). In contrast, the NMDAR transcripts GRIN1 (Fig. 1C) and GRIN2A (Fig. 1D) were lowest in DLPFC and highest in V1, whereas GRIN2B mRNA levels were lowest in V1 and highest in DLPFC (Fig. 1E).
For GABAAR transcripts, GABRG2 (Fig. 1F), GABRB2 (Fig. 1G) and GABRA1 (Fig. 1H) mRNA levels all exhibited a significant effect of area, with levels higher in V1 than in DLPFC and intermediate in V2 and PPC. In contrast, GABRA5 mRNA showed the opposite regional pattern with levels lowest in V1, intermediate in V2 and PPC and highest in DLPFC (Fig. 1I).
Because local AMPAR-mediated excitatory postsynaptic currents are required for NMDAR activation [43], we also compared mRNA levels of GRIA1, which is required for the insertion of AMPARs into the plasma membrane, and GRIN1, the obligatory NMDAR subunit, as an index of the AMPAR-mediated influence on NMDAR activation capability in each region. The GRIA1/GRIN1 ratio significantly differed across areas, with the ratio lowest in V1 and progressively higher across V2, PPC, and DLPFC (Fig. 1J). Because the function of NMDA receptors differs based on the relative presence of GRIN2A versus GRIN2B subunits [44], we also compared the ratio of these subunits across areas. The GRIN2B/GRIN2A ratio significantly differed across areas, with the ratio lowest in V1 and progressively higher across V2, PPC and DLPFC (Fig. 1K). Finally, because the relative abundances of somatostatin and parvalbumin interneurons, which preferentially target GABRA5- and GABRA1-containing receptors, respectively, differ substantially across the vsWM network [13, 45, 46], we also calculated the GABRA5/GABRA1 ratio. The GABRA5/GABRA1 ratio significantly differed across areas, with the ratio lowest in V1 and progressively higher across V2, PPC and DLPFC (Fig. 1L).
Replication and cell type specificity of transcript level differences between V1 and DLPFC in UC subjects
To assess the technical and biological reliability of these findings, and the extent to which these gray matter findings are conserved or different in two neuronal subtypes, we performed a secondary analysis of the same transcripts from RNA sequencing datasets of layer 3 pyramidal neurons (L3PNs) and of layer 3 neurons with aggrecan-positive perineuronal nets which are predominately parvalbumin-containing basket cells (L3PVBCs) [41, 42] from V1 and DLPFC of 39 and 40 UC subjects, respectively.
For the AMPA receptor subunits, GRIA1 (Supplementary Fig. S1A) and GRIA2 (Supplementary Fig. S1B), the regional differences in transcript levels were very similar in both L3PNs and L3PVBCs to those found in total gray matter, with levels lowest in V1 and highest in DLPFC. For the NMDAR receptor subunits, the regional differences in L3PNs and L3PVBCs also paralleled those present in total gray matter. GRIN1 (Supplementary Fig. S1C) and GRIN2A (Supplementary Fig. S1D) mRNA levels were higher in V1 than in DLPFC for all three measures, although the difference did not achieve significance for GRIN1 in PVBCs. The pattern of regional differences in levels of GRIN2B mRNA (Supplementary Fig. S1E) were also the same for all three measures, with transcript levels higher in DLPFC than in V1.
In contrast, the regional differences in GABRG2 (Supplementary Fig. S2A), GABRB2 (Supplementary Fig. S2B) and GABRA1 (Supplementary Fig. S2C) mRNA levels present in total gray matter were not detected in either L3PNs or L3PVBCs, suggesting that expression of these transcripts in other cortical layers or cell types might account for the regional differences observed in total gray matter. However, for GABRA5 mRNA (Supplementary Fig. S2D), the presence of higher levels in DLPFC relative to V1 was conserved in L3PNs and L3PVBCs.
The ratios of GRIA1/GRIN1 (Supplementary Fig. S3A), GRIN2B/GRIN2A (Supplementary Fig. S3B), and GABRA5/GABRA1 (Supplementary Fig. S3C) significantly differed across areas in both L3PNs and L3PVBCs with the same patterns present in total gray matter.
Receptor subunit transcript levels across the vsWM network in schizophrenia
For AMPAR subunits, the regional patterns of GRIA1 (Fig. 2A) and GRIA2 (Fig. 2B) mRNA observed in UC subjects were conserved in individuals with SZ. Although mean levels of both transcripts were lower in SZ than UC subjects in all areas, the effect of diagnosis did not achieve statistical significance for either transcript, and the diagnosis-by-area interaction was not significant.
Significant effects of area were observed for all transcripts: GRIA1 (A), GRIA2 (B), GRIN1 (C), GRIN2A (D), and GRIN2B (E). GRIN1 (C) and GRIN2A (D) were significantly lower in at least one area of subjects with schizophrenia. No significant effects were observed in DLPFC for any transcript (A–E). Asterisks (*) indicate a significant omnibus and post hoc result between subject groups within each area.
For all three NMDAR subunits, the effect of area was significant (Fig. 2C–E). For GRIN1 mRNA (Fig. 2C) the effects of diagnosis and of the diagnosis-by-area interaction were both significant, with transcript levels significantly lower in the SZ group in V1 (Cohen’s d = 0.79). Similarly, for GRIN2A mRNA (Fig. 2D) the effects of diagnosis and of the diagnosis-by-area interaction were both significant, with transcript levels significantly lower in the SZ group in V1 (d = 0.62), V2 (d = 1.34) and PPC (d = 1.19). In contrast, GRIN2B mRNA (Fig. 2E) levels did not differ between subject groups in any area.
For all GABAAR subunits, the effect of area was significant (Fig. 3A–D. The regional pattern of GABRG2 mRNA in SZ subjects was the inverse of that present in UC subjects (Fig. 3A); the effects of diagnosis and of the diagnosis-by-area interaction were both significant, with transcript levels significantly lower in the SZ group in V1 (d = 1.38), V2 (d = 1.35), and PPC (d = 1.41) (Fig. 3A). For GABRB2 mRNA (Fig. 3B), the regional pattern present in UC subjects was largely conserved in SZ, and the effects of diagnosis and of the diagnosis-by-area interaction were both significant, with transcript levels significantly lower in the SZ group in V1 (d = 1.21), V2 (d = 1.00), and PPC (d = 1.06). For GABRA1 mRNA (Fig. 3C), unlike the UC subjects, individuals with SZ did not exhibit a prominent difference across areas; the effects of diagnosis and of the diagnosis-by-area interaction were both significant, with transcript levels significantly lower in the SZ group in V1 (d = 1.35), V2 (d = 1.04), and PPC (d = 0.91). For GABRA5 mRNA (Fig. 3D), UC and SZ subjects showed the same regional pattern but no group differences were detected in any area (Fig. 3).
Significant effects of area were observed for all transcripts. Levels of GABRG2 (A), GABRB2 (B), and GABRA1 (C), but not GABRA5 (D), were significantly lower in V1, V2, and PPC in subjects with schizophrenia. No significant effects were observed in DLPFC for any transcript. Asterisks (*) indicate a significant omnibus and post hoc result between subject groups within each area.
The ratios of GRIA1/GRIN1 (Fig. 4A), GRIN2B/GRIN2A (Fig. 4B), and GABRA5/GABRA1 (Fig. 4C) did not significantly differ between groups, with the exception of a higher GABRA5/GABRA1 ratio in PPC in SZ (Fig. 4C).
Discussion
In this study, we found that AMPAR, NMDAR, and GABAAR subunits exhibited different patterns of transcript levels in total gray matter from four regions of the cortical vsWM network in UC subjects: the levels of some transcripts increased from V1 to DLPFC whereas the levels of other transcripts declined. For glutamate receptor subunits, the same patterns were also observed in both L3PNs and L3PVBCs, whereas for GABAAR transcripts, except for GABRA5, the patterns present in total gray matter were not detected in these cell types. Finally, in subjects with SZ transcript levels of excitatory and inhibitory receptor subunits were lower, with the magnitudes of the deficits generally largest in V1 and smallest, if detected, in DLPFC. These findings suggest that (1) in the healthy state, mechanisms for achieving the balance between excitation and inhibition might be region-specific, and (2) in SZ, alterations in early cortical processing of visual information might be important contributors to vsWM impairments.
Receptor transcript levels across regions of the vsWM network in UC subjects
Excitatory subunit transcripts
In UC subjects, levels of AMPAR transcripts in total gray matter decreased from rostral to caudal regions of the vsWM network, whereas NMDAR transcripts exhibited the opposite pattern, except for GRIN2B mRNA which was similar to the AMPAR transcripts pattern. These findings are consistent with (1) a report of increasing levels of GRIA1, GRIA2, and GRIN2B mRNAs, and decreasing levels of GRIN1 and GRIN2A mRNAs, from V1 to DLPFC across the cortical mantle of macaque monkeys [47], (2) the density of receptor proteins across several regions in human postmortem brain [48], and (3) several public transcriptomics datasets containing data from some of these cortical regions in small numbers of brains [14,15,16]. These distinctive regional patterns of AMPAR and NMDAR transcript levels in total gray matter were generally conserved in L3PN and L3PVBC data, suggesting that these patterns reflect differences across regions in levels of the transcript rather than other innate characteristics such as cell type composition or packing density between regions.
Inhibitory subunit transcripts
Levels of GABAAR transcripts in total gray matter differed by subunit. Levels of GABRA5 mRNA were highest in DLPFC and lowest in V1, a pattern that was conserved across total gray matter, L3PNs, and L3PVBCs. GABAARs containing this subunit are frequently located postsynaptic to inputs from the somatostatin subclass of GABA neurons [25, 26]. Given that somatostatin neurons innervate both L3PNs [49,50,51] and PVBCs [52], the conserved pattern of higher GABRA5 transcript levels in the DLPFC versus V1 might reflect the relatively greater presence of somatostatin neurons in DLPFC than in V1 of the primate cortex [13, 45, 46]. Similarly, given that GABAARs containing the GABRA1 subunit are frequently postsynaptic to inputs from PVBCs [27], and that GABRB2 is often colocalized with GABRA1 [28], the relatively greater presence of PV neurons in V1 than DLPFC [13, 45, 46] might account for the finding of higher GABRA1 and GABRB2 transcript levels in total gray matter from V1 than DLPFC. Although this regional pattern of GABRA1 transcript levels was not detected in the present study in L3PNs or PVBCs, both of which are targets of PV neurons [53, 54], such a regional pattern of GABRA1 mRNA in PV neurons was observed in the macaque brain [47].
Altered transcript levels across regions of the cortical vsWM network in schizophrenia
Except for GRIN2A, the regional patterns of the glutamatergic receptor subunits studied here were similar in both SZ and UC subjects. In contrast, the regional patterns of the GABAergic transcripts, except for GABRA5, were substantially different in SZ from those present in UC subjects. Lastly, ratios of GRIA1/GRIN1 and GRIN2B/GRIN2A across regions did not differ by diagnosis, whereas SZ subjects exhibited a significant elevation of GABRA5/GABRA1 in the PPC.
For all transcripts that exhibited an effect of diagnosis, transcript levels were lower in SZ, with the disease effect sizes largest in the more caudal cortical regions and smallest in the DLPFC where they did not achieve statistical significance. Other studies [12, 55,56,57,58,59,60,61,62,63] have evaluated some of the transcripts studied here in the DLPFC, but interpreting those findings in the context of the vsWM network is challenging given that other regions in the network were not examined. The regional pattern of alterations observed here for the GABAAR transcripts is consistent with prior studies in the same subject cohort in which gray matter levels of transcripts (e.g., PV, SST and the 67 KD isoform of glutamic acid decarboxylase (GAD67)) specific to GABA neurons [13, 64] or of proteins (e.g., PV and GAD67) in the axon terminals of PVBCs [11] were lower across the cortical vsWM network in SZ, with the disease effect size larger in V1 than in DLPFC. Even GABA-related transcripts that were upregulated in SZ (e.g., mu opioid receptor) had a larger disease effect size in V1 than in DLPFC [13]. However, it should be noted that studies of the DLPFC in other cohorts reported larger deficits for some transcripts (e.g., GABRA1 [55, 64]) in individuals with SZ than were observed here. Similarly, other indices of excitatory pyramidal neuron markers have been reported to have a larger disease effect in DLPFC than in V1. For example, deficits in dendritic spine density [65] and actin regulators of spine density [37] in SZ were greater in DLPFC than in V1.
However, alterations of excitatory drive within any region engaged during visual processing tasks may have a substantial impact on SZ pathology [66,67,68]. Indeed, a recent study utilizing a V1 orientation selectivity model suggests that reduced recurrent excitation may be a primary pathology resulting in a secondary and potentially compensatory reduction of recurrent inhibition within early sensory regions [69].
Most transcripts showed this pattern of greatest alteration in caudal relative to rostral regions in SZ. For example, GRIN2A mRNA levels were lower in V1, V2, and PPC (but not DLPFC), while GRIN2B mRNA levels were not altered in any studied regions in SZ. Although speculative, GRIN2A, but not GRIN2B, mRNA levels are highly dependent on levels of synaptic activity in vitro [70]. Thus, lower levels of synaptic signaling, due to upstream deficits in excitatory signaling reflected in fewer dendritic spines [71], could contribute to lower expression of GRIN2A mRNA. Alternatively, GRIN2A has been identified as an ultra-rare but highly penetrant genetic risk loci for SZ [72], but such genetic risk is unlikely to explain the high proportion of individuals with SZ who exhibited lower levels of GRIN2A reported here. Lastly, our study does not assess the possibility of altered synaptic compartment localization of the proteins of these receptor subunits in SZ. Therefore, although the mRNA levels of GRIN2B are unaltered here, abnormalities of its cognate protein could still play a role in SZ.
No significant effect of diagnosis was observed for the ratios of GRIA1/GRIN1 and GRIN2B/GRIN2A which may reflect similar alterations in both components of each ratio in SZ. Interestingly, GABRA5/GABRA1 ratio was higher in the PPC in SZ, suggesting both the levels and the balance of inhibitory strength at the distal dendrites versus the perisomatic region of pyramidal cells might be altered in this region.
Understanding the functional impact of the relative magnitudes of the disease effect size across regions for different measures might be informed by the idea of a cascade model of pathophysiology in SZ in which alterations in early sensory processing of visual information contribute in a bottom-up fashion to inaccurate higher-level information processing and thus cognitive impairments such as vsWM [73]. These early visual processing deficits are thought to be particularly prominent in the magnocellular thalamic inputs to the dorsal visual pathway that subserves vsWM, other higher-order visuospatial processes [74], and processes involved in cognitive control [75, 76]. Indeed, the generation of occipital fixation-related potentials (FRP) are thought to reflect “priming” of the visual cortex to process incoming information, a measure which is reduced in subjects with SZ [77]. Although FRPs originate from visual sensory cortex, the source of the inputs for stimulus-evoked FRPs remains unknown, and could include top–down projections from frontoparietal cortex, or bottom-up projections from subcortical regions [78]. Thus, the presence of large alterations in glutamate and GABAAR subunit transcript levels, and in markers of GABA neurons [11, 13] in V1 and V2, in combination with prominent alterations of L3PNs, a key neuronal element in the cortical vsWM circuit [79] in the DLPFC [37, 65, 68] suggest that vsWM impairments in SZ may arise from alterations both in the transmission of accurate visual information between lower and higher-order cortical areas and from alterations in the processing of that information in the intrinsic circuitry of the DLPFC.
Data availability
Individual subject data are shown in the figures, and both deidentified subject data and R code for the analysis will be provided upon request.
References
Baddeley A. Working memory. Science. 1992;255:556–9.
Eriksson J, Vogel EK, Lansner A, Bergström F, Nyberg L. Neurocognitive architecture of working memory. Neuron. 2015;88:33–46.
Christophel TB, Klink PC, Spitzer B, Roelfsema PR, Haynes JD. The distributed nature of working memory. Trends Cogn Sci. 2017;21:111–24.
Henseler I, Falkai P, Gruber O. A systematic fMRI investigation of the brain systems subserving different working memory components in schizophrenia. Eur J Neurosci. 2009;30:693–702.
Henseler I, Falkai P, Gruber O. Disturbed functional connectivity within brain networks subserving domain-specific subcomponents of working memory in schizophrenia: relation to performance and clinical symptoms. J Psychiatr Res. 2010;44:364–72.
Meyer-Lindenberg A, Poline JB, Kohn PD, Holt JL, Egan MF, Weinberger DR, et al. Evidence for abnormal cortical functional connectivity during working memory in schizophrenia. Am J Psychiatry. 2001;158:1809–17.
Schlösser R, Gesierich T, Kaufmann B, Vucurevic G, Hunsche S, Gawehn J, et al. Altered effective connectivity during working memory performance in schizophrenia: a study with fMRI and structural equation modeling. Neuroimage. 2003;19:751–63.
Müller NG, Knight RT. The functional neuroanatomy of working memory: contributions of human brain lesion studies. Neuroscience. 2006;139:51–8.
Repovš G, Barch DM. Working memory related brain network connectivity in individuals with schizophrenia and their siblings. Front Hum Neurosci. 2012;6:137.
Constantinidis C, Williams GV, Goldman-Rakic PS. A role for inhibition in shaping the temporal flow of information in prefrontal cortex. Nat Neurosci. 2002;5:175–80.
Fish KN, Rocco BR, DeDionisio AM, Dienel SJ, Sweet RA, Lewis DA. Altered parvalbumin basket cell terminals in the cortical visuospatial working memory network in schizophrenia. Biol Psychiatry. 2021;90:47–57.
Hoftman GD, Dienel SJ, Bazmi HH, Zhang Y, Chen K, Lewis DA. Altered gradients of glutamate and gamma-aminobutyric acid transcripts in the cortical visuospatial working memory network in schizophrenia. Biol Psychiatry. 2018;83:670–79.
Tsubomoto M, Kawabata R, Zhu X, Minabe Y, Chen K, Lewis DA, et al. Expression of transcripts selective for GABA neuron subpopulations across the cortical visuospatial working memory network in the healthy state and schizophrenia. Cereb Cortex. 2019;29:3540–50.
Akbarian S, Liu C, Knowles JA, Vaccarino FM, Farnham PJ, Crawford GE, et al. The PsychENCODE project. Nat Neurosci. 2015;18:1707–12.
Hawrylycz MJ, Lein ES, Guillozet-Bongaarts AL, Shen EH, Ng L, Miller JA, et al. An anatomically comprehensive atlas of the adult human brain transcriptome. Nature. 2012;489:391–99.
Kang HJ, Kawasawa YI, Cheng F, Zhu Y, Xu X, Li M, et al. Spatio-temporal transcriptome of the human brain. Nature. 2011;478:483–9.
Sieghart W. Structure and pharmacology of gamma-aminobutyric acidA receptor subtypes. Pharmacol Rev. 1995;47:181–234.
Isaac JT, Ashby MC, McBain CJ. The role of the GluR2 subunit in AMPA receptor function and synaptic plasticity. Neuron. 2007;54:859–71.
Traynelis SF, Wollmuth LP, McBain CJ, Menniti FS, Vance KM, Ogden KK, et al. Glutamate receptor ion channels: structure, regulation, and function. Pharmacol Rev. 2010;62:405–96.
Paoletti P, Bellone C, Zhou Q. NMDA receptor subunit diversity: impact on receptor properties, synaptic plasticity and disease. Nat Rev Neurosci. 2013;14:383–400.
Lu W, Shi Y, Jackson AC, Bjorgan K, During MJ, Sprengel R, et al. Subunit composition of synaptic AMPA receptors revealed by a single-cell genetic approach. Neuron. 2009;62:254–68.
Mayer ML, Armstrong N. Structure and function of glutamate receptor ion channels. Annu Rev Physiol. 2004;66:161–81.
Monyer H, Sprengel R, Schoepfer R, Herb A, Higuchi M, Lomeli H, et al. Heteromeric NMDA receptors: molecular and functional distinction of subtypes. Science. 1992;256:1217–21.
Monyer H, Burnashev N, Laurie DJ, Sakmann B, Seeburg PH. Developmental and regional expression in the rat brain and functional properties of four NMDA receptors. Neuron. 1994;12:529–40.
Ali AB, Thomson AM. Synaptic alpha 5 subunit-containing GABAA receptors mediate IPSPs elicited by dendrite-preferring cells in rat neocortex. Cereb Cortex. 2008;18:1260–71.
Silberberg G, Markram H. Disynaptic inhibition between neocortical pyramidal cells mediated by Martinotti cells. Neuron. 2007;53:735–46.
Klausberger T, Roberts JD, Somogyi P. Cell type- and input-specific differences in the number and subtypes of synaptic GABA(A) receptors in the hippocampus. J Neurosci. 2002;22:2513–21.
Fritschy JM, Mohler H. GABAA-receptor heterogeneity in the adult rat brain: differential regional and cellular distribution of seven major subunits. J Comp Neurol. 1995;359:154–94.
Angelotti TP, Macdonald RL. Assembly of GABAA receptor subunits: alpha 1 beta 1 and alpha 1 beta 1 gamma 2S subunits produce unique ion channels with dissimilar single-channel properties. J Neurosci. 1993;13:1429–40.
Alldred MJ, Mulder-Rosi J, Lingenfelter SE, Chen G, Lüscher B. Distinct gamma2 subunit domains mediate clustering and synaptic function of postsynaptic GABAA receptors and gephyrin. J Neurosci. 2005;25:594–603.
Roussy M, Luna R, Duong L, Corrigan B, Gulli RA, Nogueira R, et al. Ketamine disrupts naturalistic coding of working memory in primate lateral prefrontal cortex networks. Mol Psychiatry. 2021;26:6688–703.
Ranganathan M, DeMartinis N, Huguenel B, Gaudreault F, Bednar MM, Shaffer CL, et al. Attenuation of ketamine-induced impairment in verbal learning and memory in healthy volunteers by the AMPA receptor potentiator PF-04958242. Mol Psychiatry. 2017;22:1633–40.
Rao SG, Williams GV, Goldman-Rakic PS. Destruction and creation of spatial tuning by disinhibition: GABA(A) blockade of prefrontal cortical neurons engaged by working memory. J Neurosci. 2000;20:485–94.
Glausier JR, Kelly MA, Salem S, Chen K, Lewis DA. Proxy measures of premortem cognitive aptitude in postmortem subjects with schizophrenia. Psychol Med. 2020;50:507–14.
Chung DW, Chung Y, Bazmi HH, Lewis DA. Altered ErbB4 splicing and cortical parvalbumin interneuron dysfunction in schizophrenia and mood disorders. Neuropsychopharmacology. 2018;43:2478–86.
Chomczynski P. A reagent for the single-step simultaneous isolation of RNA, DNA and proteins from cell and tissue samples. Biotechniques. 1993;15:532–4.
Kimoto S, Hashimoto T, Berry KJ, Tsubomoto M, Yamaguchi Y, Enwright JF, et al. Expression of actin- and oxidative phosphorylation-related transcripts across the cortical visuospatial working memory network in unaffected comparison and schizophrenia subjects. Neuropsychopharmacology. 2022;47:2061–70.
Kuznetsova A, Brockhoff PB, Rune HB. {lmerTest} Package: tests in linear mixed effects models. J Stat Softw. 2017;82:1–26.
Hotelling H. The generalization of student’s ratio. Ann Math Stat. 1931;2:360–78.
Enwright Iii JF, Arion D, MacDonald WA, Elbakri R, Pan Y, Vyas G, et al. Differential gene expression in layer 3 pyramidal neurons across 3 regions of the human cortical visual spatial working memory network. Cereb Cortex. 2022;32:5216–29.
Enwright Iii JF, Huo Z, Arion D, Corradi JP, Tseng G, Lewis DA. Transcriptome alterations of prefrontal cortical parvalbumin neurons in schizophrenia. Mol Psychiatry. 2018;23:1606–13.
Enwright JF, Sanapala S, Foglio A, Berry R, Fish KN, Lewis DA. Reduced labeling of parvalbumin neurons and perineuronal nets in the dorsolateral prefrontal cortex of subjects with schizophrenia. Neuropsychopharmacology. 2016;41:2206–14.
Di Maio V, Ventriglia F, Santillo S. AMPA/NMDA cooperativity and integration during a single synaptic event. J Comput Neurosci. 2016;41:127–42.
Sheng M, Cummings J, Roldan LA, Jan YN, Jan LY. Changing subunit composition of heteromeric NMDA receptors during development of rat cortex. Nature. 1994;368:144–47.
Torres-Gomez S, Blonde JD, Mendoza-Halliday D, Kuebler E, Everest M, Wang XJ, et al. Changes in the proportion of inhibitory interneuron types from sensory to executive areas of the primate neocortex: implications for the origins of working memory representations. Cereb Cortex. 2020;30:4544–62.
Dienel SJ, Ciesielski AJ, Bazmi HH, Profozich EA, Fish KN, Lewis DA. Distinct laminar and cellular patterns of GABA neuron transcript expression in monkey prefrontal and visual cortices. Cereb Cortex. 2021;31:2345–63.
Chen A, Sun Y, Lei Y, Li C, Liao S, Meng J, et al. Single-cell spatial transcriptome reveals cell-type organization in the macaque cortex. Cell. 2023;186:3726–43.e24.
Zilles K, Palomero-Gallagher N. Multiple transmitter receptors in regions and layers of the human cerebral cortex. Front Neuroanat. 2017;11:78.
Horn ME, Nicoll RA. Somatostatin and parvalbumin inhibitory synapses onto hippocampal pyramidal neurons are regulated by distinct mechanisms. Proc Natl Acad Sci USA. 2018;115:589–94.
de Lima AD, Morrison JH. Ultrastructural analysis of somatostatin-immunoreactive neurons and synapses in the temporal and occipital cortex of the macaque monkey. J Comp Neurol. 1989;283:212–27.
Melchitzky DS, Lewis DA. Dendritic-targeting GABA neurons in monkey prefrontal cortex: comparison of somatostatin- and calretinin-immunoreactive axon terminals. Synapse. 2008;62:456–65.
Xu H, Jeong H-Y, Tremblay R, Rudy B. Neocortical somatostatin-expressing GABAergic interneurons disinhibit the thalamorecipient layer 4. Neuron. 2013;77:155–67.
Williams SM, Goldman-Rakic PS, Leranth C. The synaptology of parvalbumin-immunoreactive neurons in the primate prefrontal cortex. J Comp Neurol. 1992;320:353–69.
Freund TF, Katona I. Perisomatic inhibition. Neuron. 2007;56:33–42.
Hashimoto T, Arion D, Unger T, Maldonado-Aviles JG, Morris HM, Volk DW, et al. Alterations in GABA-related transcriptome in the dorsolateral prefrontal cortex of subjects with schizophrenia. Mol Psychiatry. 2008;13:147–61.
Beneyto M, Abbott A, Hashimoto T, Lewis DA. Lamina-specific alterations in cortical GABAA receptor subunit expression in schizophrenia. Cereb Cortex. 2011;21:999–1011.
Duncan CE, Webster MJ, Rothmond DA, Bahn S, Elashoff M, Shannon Weickert C, et al. receptor alpha-subunit expression in normal postnatal human development and schizophrenia. J Psychiatr Res. 2010;44:673–81.
Beneyto M, Meador-Woodruff JH. Lamina-specific abnormalities of AMPA receptor trafficking and signaling molecule transcripts in the prefrontal cortex in schizophrenia. Synapse. 2006;60:585–98.
Dracheva S, McGurk SR, Haroutunian V. mRNA expression of AMPA receptors and AMPA receptor binding proteins in the cerebral cortex of elderly schizophrenics. J Neurosci Res. 2005;79:868–78.
Weickert CS, Fung SJ, Catts VS, Schofield PR, Allen KM, Moore LT, et al. Molecular evidence of N-methyl-D-aspartate receptor hypofunction in schizophrenia. Mol Psychiatry. 2013;18:1185–92.
Akbarian S, Sucher NJ, Bradley D, Tafazzoli A, Trinh D, Hetrick WP, et al. Selective alterations in gene expression for NMDA receptor subunits in prefrontal cortex of schizophrenics. J Neurosci. 1996;16:19–30.
Beneyto M, Meador-Woodruff JH. Lamina-specific abnormalities of NMDA receptor-associated postsynaptic protein transcripts in the prefrontal cortex in schizophrenia and bipolar disorder. Neuropsychopharmacology. 2008;33:2175–86.
Sokolov BP. Expression of NMDAR1, GluR1, GluR7, and KA1 glutamate receptor mRNAs is decreased in frontal cortex of “neuroleptic-free” schizophrenics: evidence on reversible up-regulation by typical neuroleptics. J Neurochem. 1998;71:2454–64.
Hashimoto T, Bazmi HH, Mirnics K, Wu Q, Sampson AR, Lewis DA. Conserved regional patterns of GABA-related transcript expression in the neocortex of subjects with schizophrenia. Am J Psychiatry. 2008;165:479–89.
Glantz LA, Lewis DA. Decreased dendritic spine density on prefrontal cortical pyramidal neurons in schizophrenia. Arch Gen Psychiatry. 2000;57:65–73.
Glausier JR, Enwright JF 3rd, Lewis DA. Diagnosis- and cell type-specific mitochondrial functional pathway signatures in schizophrenia and bipolar disorder. Am J Psychiatry. 2020;177:1140–50.
Schoonover KE, Dienel SJ, Lewis DA. Prefrontal cortical alterations of glutamate and GABA neurotransmission in schizophrenia: Insights for rational biomarker development. Biomark Neuropsychiatry. 2020;3:100015.
Schoonover KE, Miller NE, Fish KN, Lewis DA. Scaling of smaller pyramidal neuron size and lower energy production in schizophrenia. Neurobiol Dis. 2024;191:106394.
Qian N, Lipkin RM, Kaszowska A, Silipo G, Dias EC, Butler PD, et al. Computational modeling of excitatory/inhibitory balance impairments in schizophrenia. Schizophr Res. 2022;249:47–55.
Hoffmann H, Gremme T, Hatt H, Gottmann K. Synaptic activity-dependent developmental regulation of NMDA receptor subunit expression in cultured neocortical neurons. J Neurochem. 2000;75:1590–9.
Glausier JR, Lewis DA. Dendritic spine pathology in schizophrenia. Neuroscience. 2013;251:90–107.
Singh T, Poterba T, Curtis D, Akil H, Al Eissa M, Barchas JD, et al. Rare coding variants in ten genes confer substantial risk for schizophrenia. Nature. 2022;604:509–16.
Javitt DC. When doors of perception close: bottom-up models of disrupted cognition in schizophrenia. Annu Rev Clin Psychol. 2009;5:249–75.
Butler PD, Zemon V, Schechter I, Saperstein AM, Hoptman MJ, Lim KO, et al. Early-stage visual processing and cortical amplification deficits in schizophrenia. Arch Gen Psychiatry. 2005;62:495–504.
Dias EC, Butler PD, Hoptman MJ, Javitt DC. Early sensory contributions to contextual encoding deficits in schizophrenia. Arch Gen Psychiatry. 2011;68:654–64.
Dias EC, Bickel S, Epstein ML, Sehatpour P, Javitt DC. Abnormal task modulation of oscillatory neural activity in schizophrenia. Front Psychol. 2013;4:540.
Dias EC, Van Voorhis AC, Braga F, Todd J, Lopez-Calderon J, Martinez A, et al. Impaired fixation-related theta modulation predicts reduced visual span and guided search deficits in schizophrenia. Cereb Cortex. 2020;30:2823–33.
Rajkai C, Lakatos P, Chen CM, Pincze Z, Karmos G, Schroeder CE. Transient cortical excitation at the onset of visual fixation. Cereb Cortex. 2008;18:200–9.
Arnsten AF. Prefrontal cortical network connections: key site of vulnerability in stress and schizophrenia. Int J Dev Neurosci. 2011;29:215–23.
Acknowledgements
Brain tissue for this study was provided by the University of Pittsburgh Brain Tissue Donation Program. We thank Kelly Rogers for tissue sample preparation. Most importantly, we thank all the family members who donated tissue.
Funding
This work was supported by the National Institute of Mental Health (grant numbers NIMH043784 to DAL, NIMH122943 to KES).
Author information
Authors and Affiliations
Contributions
Authors Schoonover, Dienel, Bazmi, Enwright, and Lewis were responsible for experimental design and data analysis. Author Bazmi was responsible for qPCR primer design and experimental execution. All authors were involved in manuscript preparation and editing.
Corresponding author
Ethics declarations
Competing interests
DL currently receives research funding from Merck for work unrelated to these studies. All other authors report no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Schoonover, K.E., Dienel, S.J., Holly Bazmi, H. et al. Altered excitatory and inhibitory ionotropic receptor subunit expression in the cortical visuospatial working memory network in schizophrenia. Neuropsychopharmacol. (2024). https://doi.org/10.1038/s41386-024-01854-x
Received:
Revised:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41386-024-01854-x