Abstract
Depression in older adults with cognitive impairment increases progression to dementia. Microbiota is associated with current mood and cognition, but the extent to which it predicts future symptoms is unknown. In this work, we identified microbial features that reflect current and predict future cognitive and depressive symptoms. Clinical assessments and stool samples were collected from 268 participants with varying cognitive and depressive symptoms. Seventy participants underwent 2-year follow-up. Microbial community diversity, structure, and composition were assessed using high-resolution 16 S rRNA marker gene sequencing. We implemented linear regression to characterize the relationship between microbiome composition, current cognitive impairment, and depressive symptoms. We leveraged elastic net regression to discover features that reflect current or future cognitive function and depressive symptoms. Greater microbial community diversity associated with lower current cognition in the whole sample, and greater depression in participants not on antidepressants. Poor current cognitive function associated with lower relative abundance of Bifidobacterium, while greater GABA degradation associated with greater current depression severity. Future cognitive decline associated with lower cognitive function, lower relative abundance of Intestinibacter, lower glutamate degradation, and higher baseline histamine synthesis. Future increase in depressive symptoms associated with higher baseline depression and anxiety, lower cognitive function, diabetes, lower relative abundance of Bacteroidota, and lower glutamate degradation. Our results suggest cognitive dysfunction and depression are unique states with an overall biological effect detectable through gut microbiota. The microbiome may present a noninvasive readout and prognostic tool for cognitive and psychiatric states.
Similar content being viewed by others
Introduction
Over 50 million people live with dementia [1, 2], burdened with symptoms such as memory loss and inability to engage in complex cognitive functions [3,4,5,6,7]. Characterized as a progressive neurodegenerative condition, dementia advances from preclinical stages to mild cognitive impairment (MCI) to major neurocognitive impairment, such as Alzheimer’s Disease (AD) [8, 9]. Cognitive symptoms are often comorbid with behavioral symptoms: 30–50% of those with cognitive decline experience late-life depression [10]. Cognitive decline comorbid with depression, or even sub-diagnostic depressive symptoms, decreases quality of life and increases likelihood of progression to dementia [11,12,13,14,15,16,17]. Prevalence and incidence of dementia and depressive symptoms are on the rise [18, 19], with the number of impacted individuals expected to double by 2050 [20, 21]. Successful prevention or management of depressive and cognitive symptoms may improve health outcomes in late life [11, 22, 23], highlighting the urgent need to identify novel treatment and prognostic approaches for depressive symptoms and dementia [24].
Recent technological advances have enabled systems-level analysis to identify novel biomarkers for diagnostics and monitoring of treatment response in a variety of psychiatric diseases. One such novel readout emerging from these studies is the microbiome [8, 25,26,27,28,29,30,31,32]. Microbial community diversity, or alpha diversity, is the most common metric reported in the gut microbiome literature and represents a reliable indicator of overall health status [33]. Alterations in alpha diversity in either direction (i.e., increase or decrease) associate with non-comorbid depression and AD [31, 34,35,36,37,38]. Further, many studies identified taxa that may play a role in depression [28, 31, 36, 38,39,40,41,42,43,44,45,46,47] or AD [34, 35], while a smaller number explored functional microbiota changes in depression [45, 48], often reporting conflicting results. Nevertheless, the debate continues over the utility of the microbiome and microbial community diversity metrics as a readout of varying degrees of psychiatric disease or cognitive function.
Studies comparing gut microbiome profiles in co-occurring cognitive impairment and depressive symptoms in late life are currently lacking. This is a significant knowledge gap not only due to the high comorbidity and complex interactions between cognitive function and mood but also due to their pernicious impact and frequently detrimental outcomes [15, 24, 49, 50]. Previous studies often reported conflicting results, further highlighting the need for more research in this area. Other issues include small sample sizes, categorical approaches to psychiatric diagnosis and cognitive impairment, and a lack of consideration for important covariates (i.e., age, anxiety, antidepressant use, years of education, Body Mass Index [BMI], diabetes, hypertension) [51].
This paper examines the novel hypotheses that microbial community diversity, composition, and function may reflect current and predict future cognitive function and depressive symptoms in late life. We studied a large community clinic sample of individuals with cognitive impairment and depressive symptoms by taking a dimensional approach to both cognitive function and depressive symptoms. This approach enabled us to determine whether gut microbiome community diversity associated with current depression severity (and its dependence on antidepressant use), cognitive function, and whether these sets of factors moderated one another given the synergistic effect of comorbid cognitive decline and depressive symptoms. Finally, we identify behavioral and microbial features that reflect current or predict future cognitive function and depressive symptoms using predictive machine learning approaches.
Methods
Participants and study design
Participants were recruited for the Biobank Innovations for chronic Cerebrovascular disease With ALZheimer’s disease Study [52] (BICWALZS), led by the Korea Disease Control and Prevention Agency for the Korea Biobank Project. BICWALZS is an ongoing biobank platform study conducted at five universities’ memory clinics and a community geriatric mental health center to coordinate and oversee research on cognitive decline and dementia. Participants were voluntarily recruited if they visited a participating neurology or memory clinic. Some of the participants were followed annually up to 4 years. Written informed consent was obtained from all participants and caregivers. Those with current/history of a severe neurological or medical condition that would interfere with the study (e.g., Parkinson’s disease, cerebral infarction, organ failure) were excluded from the study. BICWALZS is registered in the Korean National Clinical Trial Registry (KCT0003391) and approved by Institutional Review Board (AJIRB-BMR-SUR-16-362).
At the time of this analysis, BICWALZS had recruited 713 participants from 6 sites. We included participants who provided a stool sample at baseline, N = 292, and a subset of those (N = 70/292) who completed cognitive and depressive assessment at 2-year follow-up (mean follow-up duration 23.53 ± 1.78 months). All participants were Korean. All participants in this study were recruited from two sites: a memory clinic affiliated with Ajou University Hospital and from Suwon Community Geriatric Mental Health Center.
Assessments
All participants received comprehensive psychiatric and neuropsychological evaluations described elsewhere [52, 53]. Current diagnosis of major or minor depressive disorder was determined by a psychiatrist. Diagnosis of subjective cognitive decline (SCD) was established if no impairment was detected on the Clinical Dementia Rating (CDR) [54] and Seoul Neuropsychological Screening Battery (SNSB) [55]. MCI was diagnosed based on 0.5 CDR score and expanded Mayo Criteria on mild cognitive impairment [56]. AD was diagnosed using National Institute on Aging-Alzheimer’s Association core clinical probable AD criteria [57]. Vascular dementia was diagnosed using major vascular neurocognitive disorder criteria [58].
Current depressive symptoms were evaluated using the Korean-Version of the Montgomery-Asberg Depression Rating Scale [59] (MADRS) and Korean version of the Short form of Geriatric Depression Scale (SGDS-K) [60]. Anxiety symptoms were evaluated using the South Korean version of Beck’s Anxiety Inventory (KBAI) [61, 62]. General cognitive function was evaluated using the Mini Mental Status Examination (MMSE) [63]. Additional questionnaires included The Mini Nutritional Assessment (MNA) [64], International Physical Activity Questionnaire (IPAQ) [65], lifetime alcohol consumption (average of weekly standard drinks multiplied by years of drinking), and cigarette smoking (average of packs smoked per day multiplied by years of smoking). Participants noted their history of Diabetes Mellitus, hypertension, myocardial infarction, and cardiac ischemia. Participants completed the same measures at their 2-year follow-up.
Microbiome data collection and preprocessing
Stool samples were collected at the Ajou University Hospital biobank the day before clinical assessment using a sterilized stool container and stored at −20 °C until further processing (Fig. 1A). We used Illumina MiSeq platform to amplify the V3 and V4 regions of the 16 S rRNA marker gene. V3 and V4 Illumina adapters and dual barcode sequences were used to generate paired end reads of 300 bases of length in each direction.
A Stool samples were collected and pre-processed to create an amplicon sequence variant table. B Multiple Alpha Diversity Indices were calculated to quantify gut microbiota community richness and evenness. C–F We ran four linear regression models to examine whether gut microbiome community diversity was associated with current depression severity (and its dependence on antidepressant use), cognitive function, and whether these sets of factors moderated one-another given the deleterious effect of comorbid cognitive decline and depressive symptoms. Before interpreting any of the models, we used visual inspection to ensure normally distributed model residuals. The model containing the Shannon Index as the dependent variable had the residual distribution most representative of a normal distribution, which did not violate the normality assumption. G Distribution of cognitive symptoms, alpha diversity, and depressive symptoms. H Higher microbial community diversity (alpha diversity) was associated with and greater depression severity in those participants who were not currently on antidepressants. I Higher microbial community diversity was associated with lower cognitive function in the whole sample. This figure was created with BioRender.com.
Demultiplexed sequences were pre-processed with QIIME2 [66] (version 2022.2). First, we trimmed the primers from demultiplexed sequences. The average number of reads was 25,040 ± 6425. A trained researcher visually inspected the results to determine the read quality. Next, we denoised the data using DADA2 [67] (version 1.18), to remove chimeric sequences (sequences formed from two or more biological sequences joined together) and produce an amplicon sequence variant table. The data was truncated to minimize inclusion of poor-quality bases, while maximizing the overlap between the forward and backward reads. Taxonomy was assigned using the Silva database [68] (version 138.1). Only samples with over 10,000 reads after pre-processing were used for the subsequent analyses.
Statistical analysis
Prior to statistical analysis, we excluded 24 participants: 15 participants were excluded for having a psychiatric disorder other than depression as their primary diagnosis (anxiety disorder: N = 3, sleep disorder: N = 1, alcohol use: N = 5, psychotic disorder: N = 3, bipolar disorder: N = 1, other: N = 2). Additional 6 participants were excluded for incomplete data, 1 for low-quality stool sample, and 2 for experimenter error. The final sample consisted of N = 268 participants at baseline. This cohort had 17 participants with SCD, 189 participants with MCI, 40 participants with AD, and 22 participants with another dementia (subcortical vascular dementia, AD with small vessel disease, or AD with vascular factors). Clinically, 62 participants had no psychiatric diagnosis, whereas 124 and 82 participants had a primary diagnosis of major and minor depressive disorder, respectively.
We used alpha diversity (Fig. 1B) to examine the association between high-scale gut microbiota composition, depressive symptoms, antidepressant use, and cognitive function. Alpha diversity summarizes the number and distribution of species within a community, allowing for community comparison. Alpha diversity was calculated using R library phyloseq on minimally filtered, untrimmed data to produce four indices of alpha diversity, including Fisher, Simson, Chao1, and Shannon Index. Each diversity index was used as the dependent variable in four multiple linear regression models built in R using the lm() function. All models had the same independent variables: general cognitive function (MMSE), depressive symptoms (MADRS), current antidepressant use [45] (yes or no), and an interaction factor between depressive symptoms and antidepressant use. Finally, all four models had the same covariates: participants’ age [45], sex [45], BMI [45], nutrition [69] (MNA), anxiety [70] (KBAI), exercise [71] (IPAQ), drinking [72], smoking [72], the site of testing, and presence or absence of specific medical conditions, including hypertension [73], myocardial infarction [74], cardiac ischemia [75], and diabetes mellitus [76]. The VIF function from the car package was used to assess the variance inflation factor and confirm absence of multicollinearity by ensuring that VIF < 5. Regression coefficients were standardized using lm.beta package. Processing scripts are available from the corresponding authors on reasonable request.
We used visual inspection to ensure normally distributed model residuals (Fig. 1C–F). The model containing the Shannon Index as the dependent variable had the residual distribution most representative of a normal distribution, which did not violate the normality assumption. To test for the presence of a three-way interaction between cognitive function, depressive symptoms, and antidepressant use, we built another model with Shannon Index as the dependent variable, cognitive status, depressive symptoms, antidepressant use and their interaction as the independent variable, while controlling for the same variables as previously.
Exploratory analyses were conducted to identify the predictive potential of microbial populations and microbial products in predicting cognitive functioning and depressive symptoms at baseline and 2-year follow-up. Taxonomic analyses were done on centered log-ratio transformed data agglomerated on phylum or genus level, where any unidentified taxa and any taxa occurring in less than 65% of the participants were removed (this excluded 11/15 taxa, 305/325 genus in the N = 268 sample, and 303/325 genus in the N = 70 sample). For microbial product analysis, we used PICRUSt2 [77] for functional inference in the form of Kyoto Encyclopedia of Genes and Genomes (KEGG) orthologs (KOs) [78]. We used Gut-Brain Modules (GBMs) using the Gomixer [45] library in R to extract modules corresponding to neuroactive compounds. The GBMs were restricted to a subset which may impact cognitive function [79] or depression severity [45] (Supplementary Table 1). The resulting modules were centered log-ratio transformed and any modules occurring in less than 50% of the participants were removed (this excluded 4/45 cognitive modules, and 0/19 depression modules).
We built six cross-validated elastic net models on 268 datapoints using eNetXplorer package in R to discover features predictive of baseline cognitive function and depressive symptoms. All models contained clinical and demographic controls, including participants’ age [45], sex [45], anxiety symptoms [80], years of education [81], BMI [45], use of antidepressants [45], hypertension, myocardial infarction, cardiac ischemia, and diabetes mellitus. Further, the models contained centered log-ratio transformed microbiota on phylum, genus, or functional level, and either depressive symptoms or cognitive functioning – whichever one was not the dependent variable. We used five-fold cross-validation and ran 250 permutations per model. We optimized over 50 values of lambda, and 11 values of alpha ranging from 0 to 1.
We used the same framework on 70 datapoints to predict future cognitive function and depressive symptoms at the 2-year follow-up using baseline measures. All models contained baseline information on participants’ age, sex, anxiety symptoms, years of education, BMI, use of antidepressants, cognitive function, depressive symptoms, time in months between baseline and follow-up, and log-ratio transformed microbiota phylum/genus/GBMs. As MADRS was not collected at the 2-year follow-up, we used the SGDS-K as an indicator of depressive symptoms. Both MADRS and SGDS-K are reliable scales [82, 83] and their baseline correlation in this sample was r(68) = 0.72, p < 0.0001.
Results
Participants’ baseline clinical and demographic information is reported in Table 1. At the two-year follow-up, most participants exhibited a decrease in both cognitive function and depressive symptoms (Table 2). On average, participants experienced a 0.75 (min = −16, max = +17, SD = 4.59) reduction in MMSE and 0.41(min = −14, max = +11, SD = 3.89) reduction in GDS. Comparison of participants who agreed to versus declined to provide a stool sample is in Supplementary Table 2.
We found that both cognitive function and depressive symptoms serve as significant predictors of microbiome composition, primarily alpha diversity, (r = 0.11, F(13, 254) = 2.52, p = 0.003, Fig. 1G–I, Table 3). Higher microbial community diversity (alpha diversity) associated with lower cognitive function in the whole sample (Fig. 1I) and greater depression severity in those participants who were not currently on antidepressants (Fig. 1H). Greater alpha diversity was also associated with lower lifetime alcohol consumption. There was no three-way-interaction between microbial community diversity, MADRS, MMSE, and antidepressant use (Supplementary Table 3).
We leveraged machine learning to determine if microbial community metrics predict current MMSE and MADRS. Since these are data driven, exploratory analyses aimed at feature identification as opposed to hypothesis testing, we discussed all variables below p < 0.1 threshold. This is common when interpreting machine learning models as the goal is feature identification and the models are a combination of a complex set of weights including both significant and non-significant features especially common in microbiome literature [45].
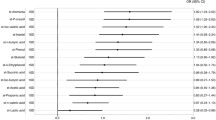
Taxonomic annotation at the phylum level could accurately predict current MMSE with out-of-bag correlation coefficient r(268) = 0.42, permuted p-value < 0.0001, alpha=0.06, lambda = 0.0081; and with genus data with out-of-bag correlation coefficient r(268) = 0.38, permuted p-value < 0.0001, alpha=0, lambda=0.3496; and with GBM data with out-of-bag correlation coefficient r(268) = 0.35, permuted p-value < 0.0001, alpha=0, lambda=1.8980. These results are shown in Table 4, Fig. 2A, and Supplementary Table 4. In all models, lower MMSE (worse cognitive function) was associated with lower education and higher depressive symptoms. In the Phylum model, lower anxiety, no antidepressant use, lower BMI, and greater relative abundance of Bacteroidota associated with lower cognitive functioning. In the Genus model, lower Bifidobacterium abundance were associated with lower cognitive functioning. On functional level, lower propionate degradation associated with lower cognitive function.
A Lower current cognitive function is associated with lower education, no antidepressant use, higher relative levels of Bacteroidota, lower Bifidobacterium, lower BMI, and lower anxiety. Higher current depression is associated with antidepressant use, lower education, diabetes, higher anxiety, and higher GABA degradation. B Lower cognition after 2 years is associated with lower baseline cognition, lower baseline abundance of Intestinibacter, absence of hypertension, lower baseline glutamate degradation, and higher baseline histamine synthesis. Higher depression after 2 years is associated with higher baseline anxiety and depression, diabetes, lower baseline cognition, lower baseline Bacteroidota, and lower baseline glutamate degradation. This figure was created with BioRender.com.
We could predict current MADRS accurately with the phylum data with out-of-bag correlation coefficient r(268) = 0.67, permuted p-value < 0.0001, alpha=0.3, lambda=1.81; with the genus data with out-of-bag correlation coefficient r(268) = 0.66, permuted p-value < 0.0001, alpha=0.1, lambda=7.93; and with GBM data with out-of-bag correlation coefficient r(268) = 0.67, permuted p-value < 0.0001, alpha=0.1, lambda=7.93. Higher MADRS (greater depression severity) was associated with higher anxiety symptoms, antidepressant use, and lower cognitive functioning across all models (Table 5, Fig. 2A, Supplementary Table 5). On Phylum level, presence of Diabetes Mellitus associated with higher depressive symptoms. On Genus level, lower education associated with greater depression symptoms. On functional level, higher microbial gamma-amino butyric acid (GABA) degradation capability associated with higher depression severity.
As our results suggest that key aspects of the microbiome, such as taxonomic annotation and alpha diversity, predict current MMSE and MADRS, we next examined if microbial features could predict cognitive and depressive outcomes at the two-year follow up visit.
We could predict future MMSE using the phylum data with out-of-bag correlation coefficient r(70) = 0.67, permuted p-value < 0.0001, alpha=0.2, lambda=0.6706; with genus data with out-of-bag correlation coefficient r(70) = 0.64, permuted p-value < 0.0001, alpha=0.1, lambda=2.1455; and with GBM data with out-of-bag correlation coefficient r(70) = 0.62, permuted p-value < 0.0001, alpha=0.1, lambda=2.5892 (Table 6, Fig. 2B, Supplementary Table 6). Across all models, lower MMSE at 2-year follow-up (indicating cognitive decline) was associated with lower baseline cognitive function. Cognitive decline was also associated absence of hypertension in the phylum model, lower Intestinibacter in the genus model, and lower Glutamate degradation and greater histamine synthesis potential in the GBM model.
We could predict future SGDS-K using the phylum data with out-of-bag correlation coefficient r(70) = 0.56, permuted p-value < 0.0001, alpha=0.0, lambda=0.3020; with genus data with out-of-bag correlation coefficient r(70) = 0.54, permuted p-value < 0.0001, alpha=0.1, lambda=6.7140; and with GBM data with out-of-bag correlation coefficient r(70) = 0.52, permuted p-value < 0.0001, alpha=0.0, lambda=5.0645 (Table 7, Fig. 2B, Supplementary Table 7). In all three models, higher SGDS-K at 2-year follow-up (higher depression) was associated with higher baseline depression. Higher 2-year SGDS-K was also associated with lower Bacteroidota in the Phylum model and higher anxiety in the Genus model. In the GBM model, higher SGDS-K was additionally associated with greater anxiety, presence of diabetes mellitus, lower cognitive function, and lower Glutamate Degradation potential by the gut microbiota.
Discussion
Our analysis of a large, transdiagnostic sample of older adults illustrates novel associations between gut microbiota, cognitive function, and depressive symptoms. Greater cognitive impairment and greater depression severity (in those not on antidepressants) was associated with greater gut microbiota diversity. Our data did not support an association between the interaction of three factors and alpha diversity. In taxonomic analyses, higher abundance of Bacteroidota phylum and lower abundance of Bifidobacterium genus was associated with worse cognitive function at baseline. Moreover, greater gut microbial GABA degradation associated with higher baseline depression severity using an analysis of microbial metabolic pathways. Finally, we found that baseline gut microbiota predicts cognitive function and depressive symptoms at 2-year follow-up. Worse cognitive function at 2-year follow up was associated with lower baseline cognitive function and glutamate degradation, lower relative abundance of Intestinibacter, and higher baseline histamine synthesis. Worse depressive symptoms at 2-year follow up was associated with higher baseline depression and anxiety, diabetes, lower cognitive function, lower relative abundance of Bacteroidota and lower baseline glutamate degradation potential.
Alpha diversity is associated with cognitive and depressive symptoms
Based on the previous literature showing associations between cognitive function and alpha diversity, we first determined whether cognitive function in an aging population was associated with gut microbiota. Lower cognitive function was associated with greater alpha diversity. These results are intriguing, given a recent meta-analysis demonstrating lower alpha diversity in those with AD compared to healthy controls [35]. It is essential to note that alpha diversity is a relative measure – high diversity is not implicitly a better or worse outcome for a community [84]. There could be multiple reasons for the differences in findings, including different patient demographics and categorical versus continuous approach to cognitive function. Nevertheless, both studies support the idea of gut dysbiosis in cognitive impairment and point toward an urgent need for a more thorough understanding of the gut-brain axis in cognitive impairment.
Based on associations between mid-life depression and gut microbiota, we investigated whether depressive symptoms in late life associated with alpha diversity. Alpha diversity was significantly associated with depressive symptoms in a way moderated by antidepressant use – greater depression severity associated with greater diversity in those not using antidepressants. These findings align with human and animal studies that implicate gut microbiota as an antidepressant response mediator [85,86,87,88,89,90,91]. Our findings do not align with a recent meta-analysis that did not detect alpha diversity alterations in midlife depression [92]. This may be because most previous studies lack control for covariates such as BMI and antidepressant use [30, 92, 93].
Finally, we examined whether the potential synergistic effect between comorbid cognitive decline and depressive symptoms may be reflected in the gut microbiota community composition [94]. We did not find a significant three-way interaction between cognitive function, depressive symptoms, and antidepressant use. At this time, we cannot conclude if we failed to detect the synergistic effect because it does not exist, or because it cannot be detected with the resolution offered by 16 S marker gene sequencing.
Gut microbiome reflects current cognitive function and depressive symptoms
Next, we determined which demographic, clinical, and gut microbiome features reflect current cognitive status. We first confirmed known predictors of cognitive function, including education [81], depressive symptoms [95], anxiety [96], antidepressant use [97], and BMI [98]. We further demonstrated that a higher abundance of Bacteroidota and lower abundance of Bifidobacterium predict lower cognitive functioning. Bacteroidota has previously been found to associate with cognitive function in animal models [99, 100], Parkinson’s Disease [101, 102], and AD without depression [103]. We replicate and extend these findings, supporting the hypothesis that decreased abundance of Bacteroidota may be protective against dementia, potentially by reducing the amyloid load through immune-mediated pathways [104,105,106]. In contrast, Bifidobacterium is a beneficial gut genus with significant health benefits, as it suppresses inflammation and ameliorates amyloid accumulation [107, 108]. Supplementing Bifidobacterium improves cognitive function in animal models and people with varying levels of cognitive impairment [107,108,109,110,111,112,113].
Similarly, we identified features that reflect current depressive symptoms. Depression severity was associated with cognitive function [95], antidepressant use [114], anxiety symptoms [115], education level, and diabetes mellitus [116]. Higher microbial GABA degradation also associated with higher depression severity. GABA, a major inhibitory neurotransmitter, has been implicated across psychiatric disorders primarily for its role in inhibitory tuning [117]. Reduced GABAergic function is a molecular hallmark of depression [118]. Those with depression exhibit higher GABA degradation and lower GABA biosynthesis [51]. A reduction in GABAergic function plays a crucial role in cognitive function, influencing symptoms that manifest across depression and aging [119]. Microbial-derived GABA impacts GABA levels across the body and is associated with changes in behavior and functional connectivity [120]. Taken together, these findings may also point towards unique interactions between antidepressants and microbiome in psychiatric aging and drug degradation [85, 121].
Gut microbiome predicts future cognitive function and depressive symptoms
Having established that gut microbiota is associated with current cognitive function, we next wanted to determine whether gut microbiota dynamics have predictive value for future cognitive function. Lower future cognitive function was associated with lower baseline cognitive function – a known predictor of future cognitive function [122, 123]. Surprisingly, lower future cognitive function was also associated with absence of hypertension. While some have reported a positive association between late life and hypertension and cognitive function [124], others argue that such relationship is minimal [125] or negative [126]. On taxonomic level, cognitive decline was associated with decreased relative abundance of Intestinibacter – a hallmark of gut dysbiosis suggesting inflammation-driven cognitive decline and increased biological aging [127,128,129,130,131,132,133]. Functionally, cognitive decline was associated with lower baseline glutamate degradation and higher histamine synthesis potential. The association between lower glutamate degradation potential and future cognitive decline points to the role of the gut-brain axis in glutamate excitotoxicity leading to neurodegeneration [134]. The association between increased bacterial histamine synthesis potential and future cognitive decline supports the hypothesis of neuroinflammatory-induced neurodegeneration in dementia [135].
Similarly, we confirmed features predictive of increased future depression, including higher baseline depression and anxiety, lower baseline cognitive function, and presence of diabetes mellitus. Future increase in depressive symptoms was also associated with lower relative abundance of Bacteroidota at baseline, a finding previously reported in mid-life depression [136]. These findings point to the complex interactions between depression, anxiety, aging, and cognition, further emphasizing the need to treat late-life psychiatric symptoms [137, 138]. Interestingly, higher baseline Bacteroidota associated with lower baseline cognitive function, while lower baseline Bacteroidota also associated with future depression increase, emphasizing Bacteroidota is a large phylum with a variety of species that differently impact host health. Finally, increased future depression was associated with lower baseline glutamate degradation potential. In a recent investigation, modifications in microbial metabolites preceding the metabolic processes of glutamate and GABA have been established as having a direct correlation with depression [139]. In this study, we validate and broaden the pivotal significance of microbial GABA and glutamate metabolism by elucidating that variations in both metabolic pathways are intricately linked to the manifestation of present and prospective depressive symptoms, respectively.
This study has several limitations. Our sampling was non-probabilistic, and our sample was predominately female. Our sample is inherently biased to include people with subjective or diagnosed cognitive decline. It is unclear to what extent our findings generalize to non-Korean individuals. Mood and cognition changes at the two-year-follow-up were not statistically significant, indicating that a longer follow-up period may be required. Our microbiome data was collected using 16 S rRNA marker gene sequencing and thus is not able to capture changes to gene content or microbial function. Our analysis focused on the most frequent taxa; therefore, our findings may not extend to rare ones. Extensive longitudinal studies with diverse participants sampled using shotgun sequencing are required to further elucidate the relationships between gut microbiota, cognitive impairment, and depressive symptoms in late life.
This is the first longitudinal, transdiagnostic study that investigated the current and future impacts of the gut microbiome on cognitive decline and depressive symptoms in a large sample of older adults. As such, it represents an essential step forward at the intersection of psychiatry, aging, and the microbiome. Our results suggest that the gut microbiome contributes to cognitive function and depressive symptoms across stages of cognitive impairment, whereby GABA-degrading microbiota species may be of particular interest. Further, the microbiome may predict future cognitive decline and depressive symptoms, potentially offering a biomarker for identifying people who may experience cognitive or mood decline. Such models would be of great benefit for treatment personalization which may alter disease progression and increase quality of life among the elderly.
Data availability
16 S amplicon data has been archived in NCBI SRA PRJNA 953406. Clinical data and processing scripts are available from the corresponding authors on reasonable request.
References
Factsheet W. The top 10 causes of death. Geneva: World Health Organization; 2020.
Organization WH. Global status report on the public health response to dementia. 2021.
Cooper C, Li R, Lyketsos C, Livingston G. Treatment for mild cognitive impairment: systematic review. Br J Psychiatry. 2013;203:255–64.
ongsiriyanyong S, Limpawattana P. Mild cognitive impairment in clinical practice: a review article. Am J Alzheimer’s Dis Other Dement. 2018;33:500–7.
Langa KM, Levine DA. The diagnosis and management of mild cognitive impairment: a clinical review. Jama. 2014;312:2551–61.
Mariani E, Monastero R, Mecocci P. Mild cognitive impairment: a systematic review. J Alzheimer’s Dis. 2007;12:23–35.
Sachdev PS, Lipnicki DM, Kochan NA, Crawford JD, Thalamuthu A, Andrews G, et al. The prevalence of mild cognitive impairment in diverse geographical and ethnocultural regions: the COSMIC collaboration. PloS one. 2015;10:e0142388.
Jiang C, Li G, Huang P, Liu Z, Zhao B. The gut microbiota and Alzheimer’s disease. J Alzheimer’s Dis. 2017;58:1–15.
Sperling RA, Aisen PS, Beckett LA, Bennett DA, Craft S, Fagan AM, et al. Toward defining the preclinical stages of Alzheimer’s disease: Recommendations from the National Institute on Aging-Alzheimer’s Association workgroups on diagnostic guidelines for Alzheimer’s disease. Alzheimer’s Dement. 2011;7:280–92.
Zubenko GS, Zubenko WN, McPherson S, Spoor E, Marin DB, Farlow MR, et al. A collaborative study of the emergence and clinical features of the major depressive syndrome of Alzheimer’s disease. Am J Psychiatry. 2003;160:857–66.
Brendel M, Pogarell O, Xiong G, Delker A, Bartenstein P, Rominger A. Depressive symptoms accelerate cognitive decline in amyloid-positive MCI patients. Eur J Nucl Med Mol imaging. 2015;42:716–24.
Defrancesco M, Marksteiner J, Deisenhammer E, Hinterhuber H, Weiss EM. Association of mild cognitive impairment (MCI) and depression. Neuropsychiatrie. 2009;23:144–50.
Ismail Z, Elbayoumi H, Fischer CE, Hogan DB, Millikin CP, Schweizer T, et al. Prevalence of depression in patients with mild cognitive impairment: a systematic review and meta-analysis. JAMA psychiatry. 2017;74:58–67.
Ma L. Depression, anxiety, and apathy in mild cognitive impairment: current perspectives. Front Aging Neurosci. 2020;12:9.
Richard E, Reitz C, Honig LH, Schupf N, Tang MX, Manly JJ, et al. Late-life depression, mild cognitive impairment, and dementia. JAMA Neurol. 2013;70:383–9.
Alexopoulos GS. Depression in the elderly. lancet. 2005;365:1961–70.
Ezzati A, Katz MJ, Derby CA, Zimmerman ME, Lipton RB. Depressive symptoms predict incident dementia in a community sample of older adults: results from the Einstein aging study. J Geriatr psychiatry Neurol. 2019;32:97–103.
Nations U. Department of Economic and Social Affairs, Population Division (2019). World Population Ageing 2019: Highlights (ST/ESA/SER. A/430).
Zivin K, Wharton T, Rostant O. The economic, public health, and caregiver burden of late-life depression. Psychiatr Clin. 2013;36:631–49.
Roseborough A, Ramirez J, Black SE, Edwards JD. Associations between amyloid β and white matter hyperintensities: a systematic review. Alzheimer’s Dement. 2017;13:1154–67.
Rasmussen J, Langerman H. Alzheimer’s disease–why we need early diagnosis. Degenerative Neurol Neuromuscul Dis. 2019;9:123.
Chan JY, Yiu KK, Kwok TC, Wong SY, Tsoi KK. Depression and antidepressants as potential risk factors in dementia: a systematic review and meta-analysis of 18 longitudinal studies. J Am Med Dir Assoc. 2019;20:279–86. e1.
Dafsari FS, Jessen F. Depression—an underrecognized target for prevention of dementia in Alzheimer’s disease. Transl Psychiatry. 2020;10:1–13.
Wang S-M, Han K-d, Kim N-Y, Um YH, Kang D-W, Na H-R, et al. Late-life depression, subjective cognitive decline, and their additive risk in incidence of dementia: a nationwide longitudinal study. Plos one. 2021;16:e0254639.
Guo M, Peng J, Huang X, Xiao L, Huang F, Zuo Z. Gut microbiome features of Chinese patients newly diagnosed with Alzheimer’s disease or mild cognitive impairment. J Alzheimer’s Dis. 2021;80:299–310.
Liu P, Wu L, Peng G, Han Y, Tang R, Ge J, et al. Altered microbiomes distinguish Alzheimer’s disease from amnestic mild cognitive impairment and health in a Chinese cohort. Brain, Behav, Immun. 2019;80:633–43.
Saji N, Murotani K, Hisada T, Tsuduki T, Sugimoto T, Kimura A, et al. The relationship between the gut microbiome and mild cognitive impairment in patients without dementia: a cross-sectional study conducted in Japan. Sci Rep. 2019;9:1–10.
Aizawa E, Tsuji H, Asahara T, Takahashi T, Teraishi T, Yoshida S, et al. Possible association of Bifidobacterium and Lactobacillus in the gut microbiota of patients with major depressive disorder. J Affect Disord. 2016;202:254–7.
Averina OV, Zorkina YA, Yunes RA, Kovtun AS, Ushakova VM, Morozova AY, et al. Bacterial metabolites of human gut microbiota correlating with depression. Int J Mol Sci. 2020;21:9234.
Cheung SG, Goldenthal AR, Uhlemann A-C, Mann JJ, Miller JM, Sublette ME. Systematic review of gut microbiota and major depression. Frontiers in psychiatry. 2019;10:34.
Jiang H, Ling Z, Zhang Y, Mao H, Ma Z, Yin Y, et al. Altered fecal microbiota composition in patients with major depressive disorder. Brain, Behav, Immun. 2015;48:186–94.
Meyer K, Lulla A, Debroy K, Shikany JM, Yaffe K, Meirelles O, et al. Association of the Gut Microbiota With Cognitive Function in Midlife. JAMA Netw Open. 2022;5:e2143941-e.
Hagerty SL, Hutchison KE, Lowry CA, Bryan AD. An empirically derived method for measuring human gut microbiome alpha diversity: Demonstrated utility in predicting health-related outcomes among a human clinical sample. PLoS One. 2020;15:e0229204.
Murray ER, Kemp M, Nguyen TT. The Microbiota–Gut–Brain Axis in Alzheimer’s Disease: A Review of Taxonomic Alterations and Potential Avenues for Interventions. Arch Clin Neuropsychol. 2022;37:595–607.
Hung C-C, Chang C-C, Huang C-W, Nouchi R, Cheng C-H. Gut microbiota in patients with Alzheimer’s disease spectrum: a systematic review and meta-analysis. Aging (Albany NY). 2022;14:477.
Huang Y, Shi X, Li Z, Shen Y, Shi X, Wang L, et al. Possible association of Firmicutes in the gut microbiota of patients with major depressive disorder. Neuropsychiatr Dis Treat. 2018;14:3329.
Lai W-T, Deng W-F, Xu S-X, Zhao J, Xu D, Liu Y-H, et al. Shotgun metagenomics reveals both taxonomic and tryptophan pathway differences of gut microbiota in major depressive disorder patients. Psychol Med. 2021;51:90–101.
Mason BL, Li Q, Minhajuddin A, Czysz AH, Coughlin LA, Hussain SK, et al. Reduced anti-inflammatory gut microbiota are associated with depression and anhedonia. J Affect Disord. 2020;266:394–401.
Chahwan B, Kwan S, Isik A, van Hemert S, Burke C, Roberts L. Gut feelings: A randomised, triple-blind, placebo-controlled trial of probiotics for depressive symptoms. J Affect Disord. 2019;253:317–26.
Chen J-J, Zheng P, Liu Y-Y, Zhong X-G, Wang H-Y, Guo Y-J, et al. Sex differences in gut microbiota in patients with major depressive disorder. Neuropsychiatr Dis Treat. 2018;14:647.
Chen Z, Li J, Gui S, Zhou C, Chen J, Yang C, et al. Comparative metaproteomics analysis shows altered fecal microbiota signatures in patients with major depressive disorder. Neuroreport. 2018;29:417–25.
Chung Y-CE, Chen H-C, Chou H-CL, Chen I-M, Lee M-S, Chuang L-C, et al. Exploration of microbiota targets for major depressive disorder and mood related traits. J Psychiatr Res. 2019;111:74–82.
Kelly JR, Borre Y, O’Brien C, Patterson E, El Aidy S, Deane J, et al. Transferring the blues: depression-associated gut microbiota induces neurobehavioural changes in the rat. J Psychiatr Res. 2016;82:109–18.
Naseribafrouei A, Hestad K, Avershina E, Sekelja M, Linløkken A, Wilson R, et al. Correlation between the human fecal microbiota and depression. Neurogastroenterol Motil. 2014;26:1155–62.
Valles-Colomer M, Falony G, Darzi Y, Tigchelaar EF, Wang J, Tito RY, et al. The neuroactive potential of the human gut microbiota in quality of life and depression. Nat Microbiol. 2019;4:623–32.
Vinberg M, Ottesen N, Meluken I, Sørensen N, Pedersen O, Kessing L, et al. Remitted affective disorders and high familial risk of affective disorders associate with aberrant intestinal microbiota. Acta Psychiatr Scandinavica. 2019;139:174–84.
Zheng P, Zeng B, Zhou C, Liu M, Fang Z, Xu X, et al. Gut microbiome remodeling induces depressive-like behaviors through a pathway mediated by the host’s metabolism. Mol psychiatry. 2016;21:786–96.
Donoso F, Egerton S, Bastiaanssen TF, Fitzgerald P, Gite S, Fouhy F, et al. Polyphenols selectively reverse early-life stress-induced behavioural, neurochemical and microbiota changes in the rat. Psychoneuroendocrinology. 2020;116:104673.
Butters MA, Young JB, Lopez O, Aizenstein HJ, Mulsant BH, Reynolds CF III, et al. Pathways linking late-life depression to persistent cognitive impairment and dementia. Dialogues Clin Neurosci. 2022;10:345–57.
Köhler S, Thomas AJ, Barnett NA, O’Brien JT. The pattern and course of cognitive impairment in late-life depression. Psychol Med. 2010;40:591–602.
Stevens BR, Roesch L, Thiago P, Russell JT, Pepine CJ, Holbert RC, et al. Depression phenotype identified by using single nucleotide exact amplicon sequence variants of the human gut microbiome. Mol Psychiatry. 2021;26:4277–87.
Roh HW, Kim N-R, Lee D-g, Cheong J-Y, Seo SW, Choi SH, et al. Baseline Clinical and Biomarker Characteristics of Biobank Innovations for Chronic Cerebrovascular Disease With Alzheimer’s Disease Study: BICWALZS. Psychiatry Investig. 2022;19:100.
Karim HT, Aizenstein HJ, Mizuno A, Ly M, Andreescu C, Wu M, et al. Independent replication of advanced brain age in mild cognitive impairment and dementia: detection of future cognitive dysfunction. Mol Psychiatry. 2022;27:5235–43.
Morris JC. The clinical dementia rating (CDR): current version and scoring rules. Neurology. 1993;43:2412–4.
Ahn H-J, Chin J, Park A, Lee BH, Suh MK, Seo SW, et al. Seoul Neuropsychological Screening Battery-dementia version (SNSB-D): a useful tool for assessing and monitoring cognitive impairments in dementia patients. J Korean Med Sci. 2010;25:1071–6.
Winblad B, Palmer K, Kivipelto M, Jelic V, Fratiglioni L, Wahlund LO, et al. Mild cognitive impairment–beyond controversies, towards a consensus: report of the International Working Group on Mild Cognitive Impairment. J Intern Med. 2004;256:240–6.
McKhann GM, Knopman DS, Chertkow H, Hyman BT, Jack CR Jr, Kawas CH, et al. The diagnosis of dementia due to Alzheimer’s disease: Recommendations from the National Institute on Aging-Alzheimer’s Association workgroups on diagnostic guidelines for Alzheimer’s disease. Alzheimer’s Dement. 2011;7:263–9.
American Psychiatric Association D, Association AP. Diagnostic and statistical manual of mental disorders: DSM-5. Washington, DC: American psychiatric association; 2013.
Ahn Y-M, Lee K-Y, Yi J-S, Kang M-H, Kim D-H, Kim J-L, et al. A validation study of the Korean-version of the Montgomery-Asberg depression rating scale. J Korean Neuropsychiatr Assoc. 2005;44:466–476.
Kim JM, Prince MJ, Shin IS, Yoon JS. Validity of Korean form of Geriatric Depression Scale (KGDS) among cognitively impaired Korean elderly and development of a 15‐item short version (KGDS‐15). Int J Methods Psychiatr Res. 2001;10:204–10.
Yook S, Kim Z. A clinical study on the Korean version of Beck Anxiety Inventory: comparative study of patient and non-patient. Korean J Clin Psychol. 1997;16:185–97.
Beck AT, Epstein N, Brown G, Steer RA. An inventory for measuring clinical anxiety: psychometric properties. J Consulting Clin Psychol. 1988;56:893.
Folstein MF, Robins LN, Helzer JE. The mini-mental state examination. Arch Gen psychiatry. 1983;40:812–812.
Guigoz Y, Vellas B, Garry PJ. Assessing the nutritional status of the elderly: The Mini Nutritional Assessment as part of the geriatric evaluation. Nutr Rev. 1996;54:S59.
Booth M. Assessment of physical activity: an international perspective. Res Q Exerc sport. 2000;71:114–20.
Hall M, Beiko RG. 16S rRNA gene analysis with QIIME2. Microbiome analysis: Springer; 2018. p. 113-29.
Callahan BJ, McMurdie PJ, Rosen MJ, Han AW, Johnson AJA, Holmes SP. DADA2: High-resolution sample inference from Illumina amplicon data. Nat methods. 2016;13:581–3.
Quast C, Pruesse E, Yilmaz P, Gerken J, Schweer T, Yarza P, et al. The SILVA ribosomal RNA gene database project: improved data processing and web-based tools. Nucleic acids Res. 2012;41:D590–D6.
Kim D, Hofstaedter CE, Zhao C, Mattei L, Tanes C, Clarke E, et al. Optimizing methods and dodging pitfalls in microbiome research. Microbiome. 2017;5:1–14.
Foster JA, Neufeld K-AM. Gut–brain axis: how the microbiome influences anxiety and depression. Trends Neurosci. 2013;36:305–12.
Mailing LJ, Allen JM, Buford TW, Fields CJ, Woods JA. Exercise and the gut microbiome: a review of the evidence, potential mechanisms, and implications for human health. Exerc sport Sci Rev. 2019;47:75–85.
Capurso G, Lahner E. The interaction between smoking, alcohol and the gut microbiome. Best Pract Res Clin Gastroenterol. 2017;31:579–88.
Avery EG, Bartolomaeus H, Maifeld A, Marko L, Wiig H, Wilck N, et al. The gut microbiome in hypertension: recent advances and future perspectives. Circ Res. 2021;128:934–50.
Vahed SZ, Barzegari A, Zuluaga M, Letourneur D, Pavon-Djavid G. Myocardial infarction and gut microbiota: an incidental connection. Pharmacol Res. 2018;129:308–17.
Sun L, Jia H, Li J, Yu M, Yang Y, Tian D, et al. Cecal gut microbiota and metabolites might contribute to the severity of acute myocardial ischemia by impacting the intestinal permeability, oxidative stress, and energy metabolism. Front Microbiol. 2019;10:1745.
Barlow GM, Yu A, Mathur R. Role of the gut microbiome in obesity and diabetes mellitus. Nutr Clin Pract. 2015;30:787–97.
Douglas GM, Maffei VJ, Zaneveld JR, Yurgel SN, Brown JR, Taylor CM, et al. PICRUSt2 for prediction of metagenome functions. Nat Biotechnol. 2020;38:685–8.
Kanehisa M, Goto S, Sato Y, Furumichi M, Tanabe M. KEGG for integration and interpretation of large-scale molecular data sets. Nucleic acids Res. 2012;40:D109–D14.
Connell E, Le Gall G, Pontifex MG, Sami S, Cryan JF, Clarke G, et al. Microbial-derived metabolites as a risk factor of age-related cognitive decline and dementia. Mol Neurodegener. 2022;17:1–26.
Tiller JW. Depression and anxiety. Med J Aust. 2013;199:S28–S31.
Lövdén M, Fratiglioni L, Glymour MM, Lindenberger U, Tucker-Drob EM. Education and cognitive functioning across the life span. Psychol Sci Public Interest. 2020;21:6–41.
Müller-Thomsen T, Arlt S, Mann U, Maß R, Ganzer S. Detecting depression in Alzheimer’s disease: evaluation of four different scales. Arch Clin Neuropsychol. 2005;20:271–6.
Levin JB, Aebi ME, Smyth KA, Tatsuoka C, Sams J, Scheidemantel T, et al. Comparing patient-reported outcomes measure information system depression scale with legacy depression measures in a community sample of older adults with varying levels of cognitive functioning. Am J Geriatr Psychiatry. 2015;23:1134–43.
Shade A. Diversity is the question, not the answer. ISME J. 2017;11:1–6.
Cussotto S, Strain CR, Fouhy F, Strain RG, Peterson VL, Clarke G, et al. Differential effects of psychotropic drugs on microbiome composition and gastrointestinal function. Psychopharmacology. 2019;236:1671–85.
Getachew B, Aubee JI, Schottenfeld RS, Csoka AB, Thompson KM, Tizabi Y. Ketamine interactions with gut-microbiota in rats: relevance to its antidepressant and anti-inflammatory properties. BMC Microbiol. 2018;18:1–10.
Kurokawa S, Tomizawa Y, Miyaho K, Ishii D, Takamiya A, Ishii C, et al. Fecal microbial and metabolomic change during treatment course for depression: an observational study. J Psychiatr Res. 2021;140:45–52.
Lukić I, Getselter D, Ziv O, Oron O, Reuveni E, Koren O, et al. Antidepressants affect gut microbiota and Ruminococcus flavefaciens is able to abolish their effects on depressive-like behavior. Transl psychiatry. 2019;9:1–16.
Macedo D, Chaves Filho AJM, de Sousa CNS, Quevedo J, Barichello T, Júnior HVN, et al. Antidepressants, antimicrobials or both? Gut microbiota dysbiosis in depression and possible implications of the antimicrobial effects of antidepressant drugs for antidepressant effectiveness. J Affect Disord. 2017;208:22–32.
Sun L, Zhang H, Cao Y, Wang C, Zhao C, Wang H, et al. Fluoxetine ameliorates dysbiosis in a depression model induced by chronic unpredicted mild stress in mice. Int J Med Sci. 2019;16:1260.
Zhang W, Qu W, Wang H, Yan H. Antidepressants fluoxetine and amitriptyline induce alterations in intestinal microbiota and gut microbiome function in rats exposed to chronic unpredictable mild stress. Transl psychiatry. 2021;11:1–16.
Sanada K, Nakajima S, Kurokawa S, Barceló-Soler A, Ikuse D, Hirata A, et al. Gut microbiota and major depressive disorder: A systematic review and meta-analysis. J Affect Disord. 2020;266:1–13.
Inserra A, Rogers GB, Licinio J, Wong ML. The microbiota‐inflammasome hypothesis of major depression. Bioessays. 2018;40:1800027.
Leszek J, Trypka E, Koutsouraki E, Michmizos D, Yarla NS, Tarasov VV, et al. Late-life depression and Alzheimer disease: a potential synergy of the underlying mechanisms. Curr Med Chem. 2018;25:5389–94.
Wang S, Blazer DG. Depression and cognition in the elderly. Annu Rev Clin Psychol. 2015;11:331–60.
Santabárbara J, Lipnicki DM, Olaya B, Villagrasa B, Bueno-Notivol J, Nuez L, et al. Does anxiety increase the risk of all-cause dementia? An updated meta-analysis of prospective cohort studies. J Clin Med. 2020;9:1791.
Korten NC, Penninx BW, Kok RM, Stek ML, Voshaar RCO, Deeg DJ, et al. Heterogeneity of late-life depression: relationship with cognitive functioning. Int Psychogeriatr. 2014;26:953–63.
Qizilbash N, Gregson J, Johnson ME, Pearce N, Douglas I, Wing K, et al. BMI and risk of dementia in two million people over two decades: a retrospective cohort study. lancet Diabetes Endocrinol. 2015;3:431–6.
Shi H, Wang Q, Zheng M, Hao S, Lum JS, Chen X, et al. Supplement of microbiota-accessible carbohydrates prevents neuroinflammation and cognitive decline by improving the gut microbiota-brain axis in diet-induced obese mice. J Neuroinflamm. 2020;17:1–21.
Shi H, Ge X, Ma X, Zheng M, Cui X, Pan W, et al. A fiber-deprived diet causes cognitive impairment and hippocampal microglia-mediated synaptic loss through the gut microbiota and metabolites. Microbiome. 2021;9:1–20.
Grant H, Anderton R, Gasson N, Lawrence BJ. The gut microbiome and cognition in Parkinson’s disease: a systematic review. Nutri Neurosci. 2022;10:932–41.
Keshavarzian A, Green SJ, Engen PA, Voigt RM, Naqib A, Forsyth CB, et al. Colonic bacterial composition in Parkinson’s disease. Mov Disord. 2015;30:1351–60.
Vogt NM, Kerby RL, Dill-McFarland KA, Harding SJ, Merluzzi AP, Johnson SC, et al. Gut microbiome alterations in Alzheimer’s disease. Sci Rep. 2017;7:1–11.
Wasén C, Vincentini J, Gauthier C, Lopes J, Butovsky O, Cox L, et al. The gut microbiota affects amyloid deposition in Alzheimer’s disease models by suppressing immune-mediated Aβ clearance. Research Square:rs.3.rs-2110304/v1 [Preprint]. 2022 [cited 2022 Oct 10]: [29 p.]. Available from: https://doi.org/10.21203/rs.3.rs-2110304/v1.
Dai C-L, Liu F, Iqbal K, Gong C-X. Gut Microbiota and Immunotherapy for Alzheimer’s Disease. Int J Mol Sci. 2022;23:15230.
Wu S, Liu X, Jiang R, Yan X, Ling Z. Roles and mechanisms of gut microbiota in patients with Alzheimer’s disease. Front Aging Neurosci. 2021;13:650047.
Sharma M, Wasan A, Sharma RK. Recent developments in probiotics: An emphasis on Bifidobacterium. Food Biosci. 2021;41:100993.
Xiao J, Katsumata N, Bernier F, Ohno K, Yamauchi Y, Odamaki T, et al. Probiotic Bifidobacterium breve in improving cognitive functions of older adults with suspected mild cognitive impairment: a randomized, double-blind, placebo-controlled trial. J Alzheimer’s Dis. 2020;77:139–47.
Kobayashi Y, Kinoshita T, Matsumoto A, Yoshino K, Saito I, Xiao J-Z. Bifidobacterium breve A1 supplementation improved cognitive decline in older adults with mild cognitive impairment: an open-label, single-arm study. J Prev Alzheimer’s Dis. 2019;6:70–5.
Kobayashi Y, Kuhara T, Oki M, Xiao J-Z. Effects of Bifidobacterium breve A1 on the cognitive function of older adults with memory complaints: a randomised, double-blind, placebo-controlled trial. Beneficial Microbes. 2019;10:511–20.
Lee D-Y, Shin Y-J, Kim J-K, Jang H-M, Joo M-K, Kim D-H. Alleviation of cognitive impairment by gut microbiota lipopolysaccharide production-suppressing Lactobacillus plantarum and Bifidobacterium longum in mice. Food Funct. 2021;12:10750–63.
Lee H-J, Lee K-E, Kim J-K, Kim D-H. Suppression of gut dysbiosis by Bifidobacterium longum alleviates cognitive decline in 5XFAD transgenic and aged mice. Sci Rep. 2019;9:1–12.
Zhu G, Zhao J, Zhang H, Chen W, Wang G. Administration of bifidobacterium breve improves the brain function of aβ1-42-treated mice via the modulation of the gut microbiome. Nutrients. 2021;13:1602.
Gerlach AR, Karim HT, Peciña M, Ajilore O, Taylor W, Butters MA, et al. MRI predictors of pharmacotherapy response in major depressive disorder. NeuroImage: Clin. 2022;36:103157.
Andreescu C. The “Late-Life” Snag in Late-Life Anxious Depression. Am J Geriatr Psychiatry. 2021;29:348–51.
Bădescu S, Tătaru C, Kobylinska L, Georgescu E, Zahiu D, Zăgrean A, et al. The association between diabetes mellitus and depression. J Med life. 2016;9:120.
Duman RS, Sanacora G, Krystal JH. Altered connectivity in depression: GABA and glutamate neurotransmitter deficits and reversal by novel treatments. Neuron. 2019;102:75–90.
Newton DF, Fee C, Nikolova YS, Sibille E. Altered GABAergic function, cortical microcircuitry, and information processing in depression. Neurobiology of depression: Elsevier; 2019. p. 315-29.
Prévot T, Sibille E. Altered GABA-mediated information processing and cognitive dysfunctions in depression and other brain disorders. Mol psychiatry. 2021;26:151–67.
Strandwitz P, Kim KH, Terekhova D, Liu JK, Sharma A, Levering J, et al. GABA-modulating bacteria of the human gut microbiota. Nat Microbiol. 2019;4:396–403.
Weersma RK, Zhernakova A, Fu J. Interaction between drugs and the gut microbiome. Gut. 2020;69:1510–9.
Beltran JF, Wahba BM, Hose N, Shasha D, Kline RP. Initiative AsDN. Inexpensive, non-invasive biomarkers predict Alzheimer transition using machine learning analysis of the Alzheimer’s Disease Neuroimaging (ADNI) database. PloS one. 2020;15:e0235663.
Bierman E, Comijs H, Jonker C, Beekman A. Symptoms of anxiety and depression in the course of cognitive decline. Dement Geriatr Cogn Disord. 2007;24:213–9.
Harrison SL, Stephan BC, Siervo M, Granic A, Davies K, Wesnes KA, et al. Is there an association between metabolic syndrome and cognitive function in very old adults? The Newcastle 85+ Study. J Am Geriatr Soc. 2015;63:667–75.
Moll AC, Woodard JL. Hypertension and cognition are minimally associated in late life. Hypertens Res. 2022;45:1622–31.
Dregan A, Stewart R, Gulliford MC. Cardiovascular risk factors and cognitive decline in adults aged 50 and over: a population-based cohort study. Age Ageing. 2013;42:338–45.
Sheng C, Lin L, Lin H, Wang X, Han Y, Liu S-L. Altered gut microbiota in adults with subjective cognitive decline: the SILCODE study. J Alzheimer’s Dis. 2021;82:513–26.
Angoorani P, Ejtahed H-S, Siadat SD, Sharifi F, Larijani B. Is There Any Link between Cognitive Impairment and Gut Microbiota? A Systematic Review. Gerontology. 2022;68:1201–13.
Bonnechère B, Karamujić‐Čomić H, Radjabzadeh D, Ahmad S, Ikram MA, Hankemeier T, et al. The role of the gut microbiome in cognitive function and Alzheimer’s disease: Biomarkers (non‐neuroimaging)/Novel biomarkers. Alzheimer’s Dement. 2020;16:e043197.
Grigor’eva IN. Gallstone disease, obesity and the Firmicutes/Bacteroidetes ratio as a possible biomarker of gut dysbiosis. J Personalized Med. 2020;11:13.
Noh JY, Wu C-S, DeLuca JA, Devaraj S, Jayaraman A, Alaniz RC, et al. Novel Role of Ghrelin Receptor in Gut Dysbiosis and Experimental Colitis in Aging. Int J Mol Sci. 2022;23:2219.
Kim SJ, Kim S-E, Kim A, Kang S, Park M-Y, Sung M-K. Dietary fat intake and age modulate the composition of the gut microbiota and colonic inflammation in C57BL/6J mice. BMC Microbiol. 2019;19:1–11.
Deleemans JM, Chleilat F, Reimer RA, Baydoun M, Piedalue K-A, Lowry DE, et al. The Chemo-Gut Pilot Study: Associations between Gut Microbiota, Gastrointestinal Symptoms, and Psychosocial Health Outcomes in a Cross-Sectional Sample of Young Adult Cancer Survivors. Curr Oncol. 2022;29:2973–94.
Chang C-H, Lin C-H, Lane H-Y. d-glutamate and Gut Microbiota in Alzheimer’s Disease. Int J Mol Sci. 2020;21:2676.
Westfall S, Lomis N, Kahouli I, Dia SY, Singh SP. Prakash S. Microbiome, probiotics and neurodegenerative diseases: deciphering the gut brain axis. Cell Mol Life Sci. 2017;74:3769–87.
Zhong Q, Chen J-J, Wang Y, Shao W-H, Zhou C-J, Xie P. Differential gut microbiota compositions related with the severity of major depressive disorder. Front Cell Infec Microbiol. 2022;12:907239.
Wolkowitz OM, Epel ES, Reus VI, Mellon SH. Depression gets old fast: do stress and depression accelerate cell aging? Depress Anxiety. 2010;27:327–38.
Kolobaric A, Karim HT, Banihashemi L, Mizuno A, Aizenstein HJ, Andreescu C. Are all anxieties created equal? Stress-related networks and anxiety phenotypes in old age. Am J Geriatr Psychiatry. 2022;30:801–12.
Mayneris-Perxachs J, Castells-Nobau A, Arnoriaga-Rodríguez M, Martin M, de la Vega-Correa L, Zapata C, et al. Microbiota alterations in proline metabolism impact depression. Cell Metab. 2022;34:681–701. e10.
Acknowledgements
We thank the staff of the BICWALZS and Suwon Geriatric Mental Health Center for their involvement in data acquisition.
Funding
This study was conducted with biospecimens and data from the consortium of the Biobank Innovations for Chronic cerebrovascular disease With ALZheimer’s disease Study (BICWALZS), which was funded by the Korea Disease Control and Prevention Agency for the Korea Biobank Project (#6637-303). This work was supported by the National Research Foundation of Korea (NRF) funded by the Ministry of Science and ICT (NRF-2019R1A5A2026045). This research was also supported by a grant of the Korea Health Technology R&D Project through the Korea Health Industry Development Institute (KHIDI), funded by the Ministry of Health & Welfare, Republic of Korea (HR21C1003). Role of the Funder/Sponsor: The funding source had no role in the design and conduct of the study; collection, management, analysis, and interpretation of the data; preparation, review, or approval of the manuscript; and decision to submit the manuscript for publication.
Author information
Authors and Affiliations
Contributions
Conceptualization/Funding acquisition – CHH, HWR, JYC, SJS; Data generation – CHH, HWR, YKK, TSS, CSK, COK, SYY, SWH, SJS; Data Curation – AK, SJS; Formal Analysis – AK, CA, EJ, HJA, HTK, SJS; Original Draft – AK, CA, EJ, HTK; and Reviewing and Editing – AK, EJ, CA, HTK, SJS.
Corresponding authors
Ethics declarations
Competing interests
AK serves as a consultant for Radicle Science. CA, EJ, CHH, HWR, JYC, YKK, TSS, CSK, COK, SYY, SWH, HJA, HTK and SJS report no biomedical financial interests or potential conflicts of interest.
Ethics approval and consent to participate
Written informed consent was obtained from all participants and caregivers. BICWALZS is registered in the Korean National Clinical Trial Registry (KCT0003391) and approved by Institutional Review Board (AJIRB-BMR-SUR-16-362).
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Kolobaric, A., Andreescu, C., Jašarević, E. et al. Gut microbiome predicts cognitive function and depressive symptoms in late life. Mol Psychiatry (2024). https://doi.org/10.1038/s41380-024-02551-3
Received:
Revised:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41380-024-02551-3