Abstract
Lysosomes, since their discovery, have been primarily known for degrading cellular macromolecules. However, in recent studies, they have begun to emerge as crucial regulators of cell homeostasis. They are at the crossroads of catabolic and anabolic pathways and are intricately involved in cellular trafficking, nutrient signaling, energy metabolism, and immune regulation. Their involvement in such essential cellular functions has renewed clinical interest in targeting the lysosome as a novel way to treat disease, particularly cancer. Acute myeloid leukemia (AML) is an aggressive blood cancer with a low survival probability, particularly in older patients. The genomic landscape of AML has been extensively characterized but few targeted therapies (with the exception of differentiation therapy) can achieve a long-term cure. Therefore, there is an unmet need for less intensive and more tolerable therapeutic interventions. In this review, we will give an overview on the myriad of functions performed by lysosomes and their importance in malignant disease. Furthermore, we will discuss their relevance in hematopoietic cells and different ways to potentially target them in AML.
Similar content being viewed by others
Introduction
History and renewed interest
Lysosomes were first discovered by Christian de Duve in 1955 while working on a specific enzyme, glucose-6-phospatase, a target of insulin in hepatic tissue. Glucose-6-phosphatase was found to be encased in what was described as “sac-like particles” [1]. Several more pH sensitive hydrolases were then discovered in the same fraction of the cell and within a few years, the role of lysosomes in the digestion of extra- and intra-cellular material was established.
In the 1990s, the discovery of autophagy [2], generated a new appreciation for lysosomes as a degradative organelle critical for cellular recycling and homeostasis. However, as the final step in autophagic flux, lysosomes still lacked prestige due to their relegation to garbage disposal units. More recent studies have now elevated the lysosome to center stage with a new understanding of their role as a sensor of cell stress and coordinator of the response to a diverse range of environmental cues including nutrient, growth factor, and immune-related signaling [3, 4]. With that, clinical interest in the organelle for therapeutic intervention has also re-emerged [5].
In this review, we will focus on the key signaling functions of lysosomes, their potential role in the pathophysiology of malignant disease and consider why lysosomes might represent a good therapeutic target for acute myeloid leukemia (AML).
Overview of lysosome function, biogenesis, and interaction with other organelles
Lysosomes are acidic organelles with an internal pH of 4.5 to 5.5, maintained by the presence of the vacuolar-type H+ ATPase (V-ATPase) on the lysosomal membrane [6]. This pH range allows luminal hydrolytic enzymes, which degrade macromolecules, to function optimally. Lysosomes are heterogeneous in morphology, distribution, and function depending on the species and cell type. They can rapidly change their distribution, number, size, and activity to meet cellular needs. The formation of primary lysosomes requires fusion between endosomes and Golgi-derived vesicles containing hydrolytic enzymes tagged for lysosomal delivery [7].
The outer lysosomal membrane is rich in transmembrane proteins, predominantly lysosome-associated membrane proteins (LAMPs). Five types of LAMPs have been identified so far: LAMP-1/CD107a, LAMP-2/CD107b, LAMP-3/DC-LAMP, LAMP-4/Macrosialin/CD68 and LAMP-5/BAD-LAMP. LAMP-1 and LAMP-2 account for almost 80% of the lysosomal membrane proteins. Other key lysosomal membrane proteins (LMPs) include, ion channels and various receptors for specific cargo, tethering/fusion or signaling, such as the chloride channel protein 7 (CLC7), Niemann-Pick C1 protein (NPC1), synaptotagmin (SYT7), and the Vacuolar (V)-ATPase proton (H+ ) transporter [8].
Lysosomal genes are transcriptionally co-regulated in a network of genes referred to as the CLEAR network (Coordinated Lysosomal Expression and Regulation)—enabling a rapid response to metabolic demand. This gene network is regulated by transcription factors belonging to the microphthalmia/transcription factor E (MiT/TFE) family, including TFEB, TFE3, and MITF. They bind to a 10-bp palindromic motif sequence in the promoter sequence of lysosomal genes referred to as the CLEAR element. TFEB has also been shown to co-regulate the expression of several autophagy genes [9].
Lysosomes are the terminal compartment for the delivery of cargo from clathrin- and caveolin-dependent and -independent endocytosis, autophagosomes, phagosomes, and chaperone-mediated autophagy (CMA) (Fig. 1). Interaction and fusion with other organelles are regulated by specific GTPases, Soluble N-éthylmaleimide-sensitive-factor Attachment protein REceptor (SNARE) complexes and Ca2 + release from the lumen of the lysosome [10]. The primary role of lysosomal fusion is considered to be degradation of internal or external cargo into products that can be exported back to the cytoplasm for metabolic reuse, or in the case of professional antigen-presenting cells (APCs), presented as peptides by major histocompatibility complex (MHC) molecules to instigate an immune response [11]. However, fusion events, cargo components or contact with other ligands or organelles can also trigger signaling events [12].
Lysosomes are the terminal organelle for endocytic and autophagic pathways. External cargos are delivered to lysosomes via clathrin- and caveolin-dependent pathways or receptor-independent pathways such as phagocytosis. Late endosomes can either fuse with lysosomes, or they can release their contents outside the cell via extracellular vesicles known as exosomes. Cytoplasmic cargos are delivered to lysosomes for degradation by autophagosomes (macroautophagy), with the help of heat shock protein (HSP) A8 chaperone complex (CMA) or simply by invagination of the lysosomal membrane itself followed by engulfment (microautophagy). Created with BioRender.com.
Signaling events arising from the intra-luminal compartment
The luminal compartment of lysosomes is the hydrolytic engine of the organelle, containing more than 60 acidic hydrolytic enzymes including proteases, nucleases, lipases, glycosidase phosphatases, and sulfatases. Deficiencies in any of these enzymes can have a major impact on cellular homeostasis. The lysosome is also a reservoir for important ions and metabolites including Ca2 + /Fe2 + /Zn2 + , H+ /Na+/K+, Cl-, and ATP. Intra-luminal levels of amino acids such as arginine and leucine are also regulated by lysosomal receptors and linked to external signaling [13].
Degradative enzymes and lysosome-dependent cell death
Cargo digestion is a key function of lysosomal enzymes. Several classes of enzymes will however also influence events outside the lysosome. Cathepsins are lysosomal proteases classed into, serine (A and G), aspartic (D and E), and cysteine proteases (B, C, F, H, K L, O, S, V, W, and X). The release of these proteases into the extracellular space has been associated with degradation of the extracellular matrix, cell migration, and invasion of cancer cells [14]. In addition, the release of proteases following lysosomal membrane leakage has been associated with cell death [15]. Several forms of lysosomal cell death (LCD) have now been described. Cells may exhibit necrotic, apoptotic, or apoptosis-like features depending on the extent of the leakage and the cellular context [16].
Lysosomes and calcium signaling
Lysosome membranes contain several ion channels that establish concentration gradients and maintain the lysosome membrane potential (ΔψLysol). Relative to the cytoplasm, the lysosome has high concentrations of Ca2 + , which is a key regulator of many lysosomal functions. There are at least three main types of Ca2 + channels in mammalian lysosomes: Transient receptor potential mucolipin sub-family (TRPML)/mucolipin 1-3, (Two-Pore) TPC1-2 and P2X4. These respond to a variety of signals including cell stress, phospholipids, nutrients, and ATP depletion. The best-characterized channel is probably TRPML1/mucolipin 1, which regulates several lysosomal processes including lysosomal reformation, exocytosis, motility, and fusion with other organelles. Calcium efflux from TRPML1 also activates the calcium-dependent phosphatase calcineurin, which is important for TFEB nuclear translocation [17].
Luminal essential amino acid (EAA) sensing role
The lysosome can sense both luminal and cytosolic amino acid levels—with crosstalk between these pathways to maintain homeostasis or respond to nutrient-related signaling. Lysosomal luminal arginine is sensed by the sodium-coupled amino-acid transporter SLC38A9. The binding of arginine induces a conformational change in SLC38A9 leading to stimulation of the Rag A/B (Ras-related GTP-binding) GTPase and Ragulator complex localized on the lysosomal surface. This recruits the mechanistic target of rapamycin complex 1 (mTORC1), which is one of two protein kinase complexes incorporating the serine-threonine kinase mTOR. The mTORC1 complex consists of the mTOR kinase itself, Raptor, GβL, and DEPTOR. mTORC1 is then activated by Rheb (Ras homolog enriched in brain) GTPase, on the lysosomal membrane. Simultaneously, SLC38A9 also allows efflux of other amino acids such as leucine into the cytosol. This enables cross talk with other cytosolic amino acid sensors associated with mTORC1 complex (outlined below) [13, 18]. High levels of intra-luminal leucine can also activate mTORC1 by promoting ATP hydrolysis by the V-ATPase. This also stimulates the Ragulator/Rag complex to recruit mTORC1 [19] (Fig. 2).
Lysosomal membranes harbor a variety of proteins that regulate multiple signaling pathways. The transporter protein NPC1 releases free cholesterol from the lumen to maintain lipid homeostasis. The V-ATPase is a proton pump responsible for maintaining luminal pH. mTORC1 links metabolic functions to environmental signaling. The components of the mTORC1 signaling pathway can be regulated by both intra- and extra-luminal amino acid signaling. Arginine (R) in the lysosomal lumen is sensed by the amino acid transporter SLC38A9 which eventually activates mTORC1 with the help of the Rag GTPases and Ragulator complex. In the cytosol, Sestrin proteins serve as leucine (L) sensors and with the help of GATOR proteins, regulate mTORC1 activity. See text for further description. mTORC1 also negatively regulates lysosome biogenesis. Luminal Ca2 + initiates lysosome biogenesis. It is released into the cytoplasm through the lysosomal calcium channel TRPML1 to activate calcineurin, which dephosphorylates TFEB, enabling it to translocate into the nucleus where it facilitates transcription of the CLEAR network genes. This pathway is inhibited by the transcription repressor ZKSCAN3. LAMPs are transmembrane proteins that account for about 80% of all membrane proteins and take part in various functions including, autophagy, lipid transport, and immune response. SYT7 is another calcium-dependent membrane protein that takes part in lysosomal exocytosis. Created with BioRender.com.
The lysosome surface as a signaling hub
As with other organelles, most intra-organelle communication and signaling is orchestrated at the surface. The lysosomal surface is a platform for the assembly of several signaling hubs that link metabolic functions to environmental signaling. A central link in this signaling is the mTORC1 complex. The primary role of active mTORC1 is to drive anabolic/biosynthetic pathways to fuel growth and proliferation and to inhibit catabolic pathways such as autophagy. Several signaling cues converge at mTORC1 including (i) nutrients (ii) growth factors and (iii) energy status.
Cytosolic amino acid signaling
Amino acids promote the translocation of mTORC1 to the lysosomal surface where it can be activated. This process is mediated by the coordinated actions of several complexes including Ragulator and Rag GTPases (A, B, C, and D). The Ragulator complex consists of 5 subunits; p18, p14, MP1, C7orf59, and HBXIP (also referred to as LAMTOR -1, 2, 3, 4, 5 respectively). The p18 subunit (LAMTOR-1) is anchored to the lysosomal membrane. In response to amino acids, Ragulator acts as a guanine nucleotide exchange factor (GEF) for Rag A/B—promoting their GTP bound state and anchoring both the Rag GTPases and mTORC1 to the membrane where it can be activated by the small GTPase Rheb [20, 21].
The presence of specific amino acid residues also influences other GEFs and GTPase-activating proteins (GAPs), upstream of GTP-RagA and GDP-RagC and mTORC1 signaling. A key integrator of amino acid signaling is the GTPase-activating proteins toward Rags (GATOR) complex [22]. GATOR is composed of two sub-complexes: GATOR1 and GATOR2. GATOR1 acts as a GAP (off switch) for RagA/B, releasing mTORC1 from the lysosome, thus acting as a negative regulator. GATOR2 positively regulates mTORC1 by inhibiting the GAP activity of GATOR1. Leucine and arginine residues act through stress-inducible proteins (Sestrins) and cytosolic arginine sensors for mTORC1 subunit 1 (CASTOR1) respectively. Upon amino acid binding, they dissociate from GATOR2, releasing their suppressive effects on GATOR2 and activate mTORC1 [23]. In the absence of amino acids, mTORC1 activity is also regulated by the cyclin-CDK inhibitor, cyclin-dependent kinase inhibitor 1C (CDKN1B) or p27. It binds to LAMTOR1 and prevents mTORC1 activation, which in turn, activates macroautophagy. In this context, p27 serves as a tumor suppressor [24].
Growth factors
Many external signals, such as growth factors converge at a large protein complex referred to as tuberous sclerosis complex (TSC), which negatively regulates the MTORC1 kinase [25]. TSC is a complex of TSC1, TSC2, and TBC1D7 subunits. TSC2 displays GTPase-activating protein (GAP) activity for Rheb, inactivating Rheb through induction of the GDP-bound state. Initially, signaling from receptors such as insulin or IGF (insulin-like growth factor) generates phosphatidylinositol lipids (3,4,5)P3 (PIP3), which stimulates protein kinase B (Akt) phosphorylation and inactivation of TSC2. This inactivation leads to an increase in GTP-loaded Rheb, which fuels activation of mTORC1 [26].
Energy status
In contrast to growth factor signaling, energy depletion activates AMP kinase at the lysosomal surface, which stimulates TSC2 through phosphorylation and inhibits RAPTOR. This leads to inhibition of mTORC1 and promotion of catabolic pathways [25].
mTORC1 and transcriptional regulation of anabolic/catabolic pathways
mTORC1 also plays a major role in the phosphorylation of MiT/TFE factors. Under nutrient-rich conditions, mTORC1 phosphorylates TFEB on Ser 122 and Ser 211 and sequesters it to the cytoplasm. Inhibition of mTORC1 under a stressful condition, such as starvation, disrupts this association allowing nuclear translocation of TFEB and transcription of CLEAR network genes [3]. Another important regulator of TFEB nuclear translocation is the Ca2 + -dependent phosphatase, calcineurin. During starvation, Ca2 + is released through the lysosomal calcium channel, TRPML1/mucolipin-1, activating calcineurin, which dephosphorylates TFEB. A zinc finger family DNA-binding protein, ZKSCAN3, has been shown to be a transcriptional repressor of autophagy by repressing the expression of more than 60 essential genes associated with lysosomal biogenesis and autophagy including Microtubule Associated Protein 1 Light Chain 3 Beta (Map1lc3b) and Wipi2. ZKSCAN3 and TFEB have also been found to be oppositely regulated by starvation [27] (Fig. 2).
Autophagy
Autophagy is a process by which cells transport dysfunctional or excessive cellular components into the lysosomes for degradation. Autophagy can eradicate harmful organelles, pathogens or aggregated material and the recycling of macromolecules from the lysosome enables cells to cope with stresses such as nutrient deficiency [28]. The three main types of autophagic pathways are macroautophagy, microautophagy and chaperone-mediated autophagy.
Macroautophagy is mediated by several protein conjugation reactions, eventually leading to the formation and expansion of a double-membraned autophagosome which carries the cargo to the lysosome to be degraded [29]. The genes involved in the process are known as ATG (AuTophaGy-related) genes, first identified in yeast, followed by studies of orthologues in other species [30]. In mammals, macroautophagy is initiated through activation of two key upstream regulatory complexes: (i) The class III phosphatidylinositol 3-kinase (PI3K-III/ VPS34) complex containing Beclin-1 (a mammalian homolog of yeast Atg6), hVps34, p150 (a mammalian homolog of yeast Vps15), and Atg14-like protein (Atg14L) and (ii) the ULK1 kinase complex (ULK1-Atg13-FIP200-ATG101)—this activates the class III PI3K/ Beclin-1 complex enabling autophagosome nucleation. These complexes are regulated by phosphorylation from the following pathways:
The PI3K/AKT/mTOR signaling pathway: Under abundant supply of nutrients and growth factors, cells activate the PI3K/AKT/mTOR signaling pathway and suppress autophagy, by inhibitory phosphorylation. In contrast, when cells are experiencing stress or nutrient starvation, they respond by inactivating the PI3K/AKT/mTOR pathway, which de-represses and activates autophagy.
The AMPK signaling pathway: AMPK (AMP-activated Protein Kinase) is a critical energy-sensing protein, which detects changes in the ratio of ATP to AMP or ADP. Phosphorylated AMPK phosphorylates TSC2 thereby inhibiting mTORC1 and activating autophagy. AMPK can also induce autophagy by directly phosphorylating and activating ULK-1/2. [31]
CMA is a client-specific, selective autophagic pathway. CMA clients contain a signature motif containing a short stretch of 5 amino acid residues, KFERQ (Lysine, Phenylalanine, Glutamate, Arginine, Glutamine) [32]. This motif is recognized by the chaperone protein HSPA8, also known as HSC70 or HSP73, which is a member of the HSP70 family of proteins [33]. The client protein is received on the lysosomal surface by a single molecule of LAMP-2A. LAMP-2A receptors form multimers that are stabilized by HSP90 on the lysosomal surface and aid translocation of the client. HSPA8 is also present inside the lysosome and helps in the translocation of the substrate by interacting with LAMP-2A [34].
In microautophagy, the dynamics of the lysosomal membrane allows it to wrap itself around cytosolic contents and engulf them into the lumen for degradation. Molecular details of microautophagy have not been elucidated as thoroughly as the other two pathways. Client specificity has been reported in endosomal microautophagy. Similar to CMA, the protein clients harbor a KFERQ-like motif and are delivered to the lysosomes by HSPA8 with the help of the endosomal sorting complex required for transport (ESCRT) machinery [35]. The ESCRT machinery helps to load protein into multivesicular bodies (MVBs), which are a special form of late endosome, that can then fuse with the lysosome [36].
Subcellular localization of lysosomes
The signaling properties of lysosomes are affected by their position within the cell. Lysosomes can move from the center of the cell to the periphery or vice versa with the help of kinesin motors KIF2A and KIF1Bβ as well as the small GTPase ARL8B [37]. The consequences of lysosomal positioning seem to be dependent on the environmental signaling. Localization of lysosomes towards the periphery has been associated with increased interaction with mTORC1 and elevation of mTOR activity, whereas perinuclear localization in starved conditions inhibits mTORC1 and activates autophagy [38]. Hong and colleagues have shown that mechanistically, the FYVE-domain proteins Protrudin and FYVE and coiled-coil domain autophagy adaptor 1 (FYCO1) help to bring mTOR positive lysosomes towards the plasma membrane in a VPS34-dependent process [39]. Perinuclear clustering also delays mTORC2 reactivation in response to serum depletion [40]. In contrast, it has also been reported that under hypoxic conditions, lysosomes localize to the periphery and this actually reduces mTORC1 activity [41]. The tumor suppressor, folliculin is responsible for the association of lysosomes with the perinuclear membrane and restricting their localization in the area [42]. The subcellular localization of lysosomes also determines their luminal pH. Peripheral lysosomes are less acidic due to lowered V-ATPase activity [43]. The movement of lysosomes within the cell is essential for lysosomal exocytosis, which is a process that leads to the secretion of lysosomal content upon lysosome fusion with the plasma membrane. It is a ubiquitous, Ca2 + -regulated mechanism, which plays a role in various physiological processes such as, plasma membrane repair and immunogenic ATP release [44].
Galectins and lysophagy
The elimination of defective lysosomes by autophagy is referred to as ‘lysophagy’. Several β-galactoside-binding proteins (galectins) have been implicated in mediating lysophagy. GAL 3, 8 and 9 are recruited by exposure of luminal glycans following lysosomal damage. Galectin 8 can inhibit mTORC1 and Galectin 9 activates Mitogen-activated protein kinase kinase kinase 7 (TAK1 or MAP3K7) that stimulates AMPK [45]. Members of the galectin family have also been implicated in the control of differentiation and self-renewal of hematopoietic cells [46], and have been associated with prognosis in AML [47].
Lipid homeostasis
Lysosomes are important regulators of lipid homeostasis and play a key role in cholesterol trafficking [48]. Low-density lipoproteins (LDLs) are trafficked to lysosomes via clathrin-mediated endocytosis. The release of free cholesterol from the lysosome is mediated by two transporter proteins, NPC 1 and 2 [49]. Lysosomes also contribute to maintaining cellular lipid balance through a selective autophagy pathway, referred to as lipophagy. Lipids are stored as triglycerides in lipid droplets (LD) which are delivered to lysosomes via lipophagy and degraded to release free fatty acids (FFAs) that can be used as an energy source [50].
Immune responses/inflammation
Lysosomes can participate at several stages in the generation of an immune response. This includes pathogen sensing, phagocytosis, antigen processing and presentation, and inflammation/secretion.
Sentinel cells such as macrophages and dendritic cells (DCs) use toll-like receptors (TLR) to sense pathogens and elicit an immune response. Members of the TLR family, TLR 3 TLR7, TLR 8, and TLR9 have been shown to signal from endolysosomes. In addition, TLR9 responds to mitochondrial DNA, delivered to the lysosome via mitophagy [51, 52]. Foreign particles are internalized by phagosomes which eventually mature and fuse with lysosomes to become phagolysosomes. TFEB enhances phagocytosis in a calcium-dependent manner by transcriptional activation of immune-related genes [53]. Macrophages, DCs, and B cells are APCs that process antigens from degraded pathogens and incorporate them into MHCs on their surfaces. Lysosomal pH is important in this process as too much acidification results in excessive proteolysis of engulfed microbes, thus impairing cross-presentation. On the other hand, increased lysosomal pH can also alter lysosomal properties and impair antigen processing as seen in lupus, for example [54]. Lysosomes also participate in downregulation of inflammation in immune cells. The inflammasome complex, which is responsible for the secretion of pro-inflammatory cytokines such as IL-1β and IL-18, is selectively degraded via autophagy [55, 56]
Lysosomes in malignancy
Cell transformation and cancer progression require an increase in biomass production and adaptation to nutrient stress, processes that are aided by lysosomes. Accordingly, cancer cells have higher lysosomal activity compared to healthy adjacent tissue [57]. Lysosomes contribute to the maintenance of many hallmarks of cancer such as, sustaining proliferative signaling (mTORC1 signaling), metabolism (catabolic reactions, autophagy), and invasion (lysosomal exocytosis) [58]. For example, in sarcomas, downregulation of the protein NEU1 aggravates lysosomal exocytosis, which releases hydrolases and exosomes that facilitate extracellular matrix remodeling and invasion into the adjacent tissue [59]. Lysosomes are also known to sequester chemotherapeutic drugs and thus play an active role in the development of drug resistance [60]. In addition, lysosomes in metastatic cells differ greatly in content, localization, and activity from the lysosomes in normal cells which make them potentially more susceptible to lysosome targeting agents [61]. Since lysosomes are central to many cellular pathways, several ways exist to target them indirectly for cancer therapy (Fig. 3).
Lysosomes are associated with various pathways which makes it possible to target them in different ways. Lysosomotropic agents can disrupt the lysosomal membrane and release cathepsins into the cytosol leading to LCD. Other lysosomotropic agents such as chloroquine (or hydroxychloroquine) can impede lysosomal membrane fusion which is important for turnover in autophagy. Due to the protective role of HSP70 against lysosomotropic agents, HSP70 inhibitors can be used to intensify their effect. During oncogenic activation, lysosomal exocytosis causes release of cathepsins into extracellular space, which promotes extracellular matrix degradation and invasion of malignant cells. This makes protease inhibitors targeting cathepsins also a therapeutic option. Autophagic pathways have context-dependent roles in cancer and thus, there may be several strategies to either block or stimulate the pathways. Upstream pathways such as mTORC1 and 2 can be inhibited to stimulate autophagy in an indirect manner. V-ATPase inhibitors can elevate lysosomal pH, thus impairing the function of many pH-sensitive lysosomal enzymes. Created with BioRender.com.
Direct impairment of lysosomal function can be achieved through several strategies. Most of the therapeutic agents being developed to target lysosomes either raise the luminal pH to inactivate cathepsins thus rendering them unable to mediate invasion, or aid in lysosomal membrane permeabilization and induce LCD. Lysosome targeting agents and the diseases they are being tested against has already been adequately reviewed [62]. In Table 1, we have summarized the mechanism of a few emerging preclinical options which can affect lysosomes, the diseases they have been tested with, and their toxicity profile.
Lysosomes in AML
Hematopoiesis is a complex process that gives rise to cells with strikingly distinct morphology and functions. Such processes require drastic reorganization of cellular homeostasis. Lysosomes play a vital role here as they maintain a balance between anabolic and catabolic pathways. Due to their association with the master regulator of cell growth, mTORC1, and the ability to degrade macromolecules via autophagic pathways, they are in a central position to modulate metabolism, differentiation, proliferation, and cell death. All of these components are essential in the maintenance of functioning hematopoiesis and their dysregulation can be a feature of hematological malignancies.
Lysosome derived structures in myeloid leukemia cells
Auer rods (or Auer bodies) are large structures sometimes observed in AML, acute promyelocytic leukemia (APL), and myelodysplastic syndromes (Fig. 4). Auer rods are azurophilic, can resemble needles, commas, diamonds, rectangles, corkscrews, and can appear bundled. They are not always present—but are a highly specific finding and virtually pathognomonic for a myeloid neoplastic disorder. They are apparently composed of fused lysosomes and are rich in lysosomal enzymes. Their method of formation and functional significance are not known but several processes have been described that cause tubulation of lysosomes including autophagic lysosome reformation (ALR) [63] and activation of antigen-presenting cells. Activation of macrophages and DCs causes dramatic tubulation of endolysosomes—which increases their surface area to volume ratio and is also proposed to assist with the delivery of MHC-II peptides to the cell surface for presentation [64].
In positive AML cases, the percentage of cells with Auer rods and the number of Auer rods per cell can vary greatly. Auer rods are found in AML M1, M2, M3, M4, M5, and M6, but not in AML M0 or M7 of the French–American–British (FAB) classification, or in blast crisis chronic myeloid leukemia, suggesting an association with a more mature myeloid phenotype. While Auer rods clearly have diagnostic value in determining the neoplastic nature of myeloid cells, they do not seem to have a clear prognostic significance [65].
Lysosomes in hematopoietic stem cells (HSC)
HSC are responsible for repopulating blood cells and maintaining blood homeostasis. Therefore, in order to protect themselves from stress, they usually remain quiescent. Liang et al., reported that, quiescent HSCs are defined by their low mitochondrial membrane potential and glycolysis. Enlarged lysosomes are found in quiescent HSCs, which are speculated to remove toxic products and contribute to the maintenance of healthy HSCs [66]. Lysosomes are asymmetrically inherited by the daughter cells during cell division of HSCs and therefore determine their cellular fate by reorganizing the metabolic and translational machinery of the cells [67].
Autophagy in HSCs
Autophagic pathways, in particular, macroautophagy contributes to the maintenance of energy metabolism, which is crucial in HSCs. The transcription factor FOXO3A, drives a gene expression program that induces autophagy to protect HSCs during stress [68]. The specific knockout of Atg7 in the hematopoietic system of mice results in severe clinical symptoms (lethargy, piloerection, and weight loss), leading to their death within 12 weeks. The Atg7−/− LSK (Lin− Sca-1+ Kit+) cells showed accumulation of mitochondria and DNA damage with complete loss of stem cell function [69]. Similar pre-leukemic phenotypes were observed in Atg5 knockout mice [70]. FIP200, a key protein in the autophagosome nucleation process, is necessary for fetal HSC maintenance and its loss leads to a block in erythroid maturation, depletion, and loss of reconstituting capacity of HSCs, and aberrant expansion of myeloid cells [71]. A recent study reported that CMA is crucial for protein quality control and metabolic reprogramming during HSC activation. It also maintains the functionality of aged HSCs [72].
Autophagy in AML
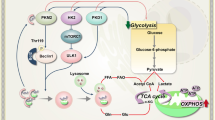
Autophagy dysregulation can lead to and drive AML. An in silico analysis has shown that a deleted chromosomal region often found in AML coincides with the location of autophagy genes and that lowered autophagic flux is advantageous to AML [70]. Heterozygous deletion of Atg5 in Mixed Lineage Leukemia-Eleven Nineteen Leukemia (MLL-ENL) induced murine HSCs led to a higher proliferation rate and a more aggressive leukemia in vivo due to a shift towards glycolytic metabolism [70]. Depletion of ATG7 was found to enhance chemoresistance in mice transplanted with OCI-AML3 cells harboring Atg7 targeting shRNA [73]. In a MLL-AF9 driven model, deletion of Atg5 accelerated the initiation of leukemia but had no effect on therapeutic intervention [74]. Conversely, another study on MLL-ENL leukemic mice showed that homozygous deletion of Atg5 or Atg7 in bone marrow cells reduced the frequency of leukemia-initiating cells (LICs) and decelerated leukemia progression [75]. In FLT3-ITD driven AML, the receptor tyrosine kinase, RET, has been identified as an essential driver of leukemogenesis. Mechanistically, it does so by activating mTORC1, thus, suppressing the ability of autophagy to degrade the FLT3 protein [76]. In contrast, another group found that FLT3-ITD upregulates basal autophagy mediated by the transcription factor, ATF4 and inhibition of autophagy could in fact, facilitate increased survival of mice with FLT3-ITD driven AML [77]. The aforementioned examples aptly demonstrate that, while the relevance of autophagy in AML is undeniable, inconsistencies between results exist possibly because the process has different implications at different levels of leukemic transformation.
Autophagy in myeloid differentiation
Several studies indicate that autophagy plays a key role in myeloid differentiation. Macroautophagy is upregulated during colony-stimulating factor-1 mediated monocyte-macrophage differentiation and is necessary for cell survival as it can direct cells towards differentiation in lieu of apoptosis [78, 79]. Mechanistically, it has been shown that autophagy-mediated lipolysis is necessary for neutrophil differentiation [80].
Granulocytic differentiation can be induced in the AML subtype APL by treatment with all-trans retinoic acid (ATRA). Autophagy can facilitate degradation of the PML-RARA onco-protein in APL, but it is also important for differentiation of AML cells that do not have a fusion protein [81, 82]. ATRA-induced differentiation and autophagy were found to be dependent upon upregulation of TFEB [83]. More recently, it was shown that degradation of fatty acid synthase via ATRA-induced macroautophagy activates lysosomal biogenesis and accelerates granulocytic differentiation [84].
Targeting lysosomes in AML therapy
The lysosome is clearly a multi-functional organelle and there may be many ways to target it for therapeutic gain in leukemia. Moreover, lysosomes in AML cells are bigger in size which could make AML cells more susceptible towards lysosomal disruption than normal cells [85]. Several studies have investigated areas such as: direct structural damage, interference with pH or other luminal homeostasis, blocking associated signaling or interference with key trafficking events (Fig. 5). Similar to solid cancers, lysosomes also sequester chemotherapeutic drugs and confer resistance in AML [86]. Therefore, targeting lysosomes can not only serve as a way to destroy AML cells while sparing normal hematopoietic cells, but may also be a strategy to combat chemotherapy resistance.
A Lysosomes play important roles in HSC maintenance and during cell division their asymmetric inheritance can influence cell fate. B Lysosomes are enlarged in AML cells and these can sequester therapeutic drugs to promote chemoresistance. C The various agents designed to target lysosomes in AML lead to five main consequences: (i) disruption of lysosome membranes, leading to release of luminal enzymes into the cytosol to cause LCD, (ii & iv) hyper- or hypo-acidification of the lysosomal lumen which disrupts lysosome function and can increase susceptibility towards certain drugs, (iii) alteration of luminal lipid profiles leading to lysosome-dependent cell death, (iv) blocking activity of pumps or channels such as the vacuolar-type H+ ATPase to reduce luminal acidity or impair other lysosomal homeostasis, and (v) targeting various signaling proteins on the membrane surface to functionally impair lysosomes. Created with BioRender.com.
Structural damage
In 1994, Craig Rosenfeld demonstrated the antileukemic property of the lysosomotropic compound phenylalanine methyl ester in leukemic specimens derived from patients [87].
A screen of a library of 100 drugs showed that the anti-malarial drug, mefloquine, is effective against AML cell lines as well as leukemia patient samples. A gene set enrichment analysis carried out in yeast identified genes associated with lysosomal functions to be enriched in gene deletion strains that were sensitive to mefloquine, indicating that mefloquine targets lysosomes. Mefloquine disrupts the lysosomal membrane of AML cells allowing the release of cathepsins B and L, which results in cell death [85]. Another in silico screening assay of FDA-approved drugs identified a distinct subset of cationic-amphiphilic antihistamines that selectively killed AML cell lines and primary patient samples. Their toxicity was associated with the simultaneous disruption of both lysosomes and mitochondria which triggered both autophagy and apoptosis [88]. High expression of p53 induced gene 7 (pig7), which encodes for the small integral membrane protein of lysosome/late endosome (SIMPLE) was found to induce lysosomal membrane permeabilization in AML cell lines and render them more susceptible to chemotherapies such as VP16 and daunorubicin [89].
Interference with pH or other aspects of luminal homeostasis
The compound, deoxysappanone B 7,4′-dimethyl ether (Deox B 7,4) was identified from a chemical screen of small molecules with anti-leukemic activity. It is a microtubule inhibitor, which indirectly increases lysosomal V-ATPase activity causing hyper-acidification. This effect leads to lysosomal disruption and the death of AML cells. Interestingly, AML cells were found to be more vulnerable to Deox B 7,4, compared to normal hematopoietic cells [90]. Conversely, blocking lysosomal acidification via the V-ATPase inhibitor Archazolid A also has anti-leukemic effect. Archazolid A reduces the anti-apoptotic protein survivin which leads to cell death [91]. Several small molecules known as cationic amphiphilic drugs (CAD) are already used in the treatment of various disorders and possess lysosomotropic properties [92]. CADs are defined structurally by the presence of both a hydrophobic and hydrophilic domain. Many antihistamines, antidepressents, and even mefloquine are classified as CADs. A recent study has reported that CADs can induce lysosome-dependent cell death in various AML cell lines by altering the lipid profile within the lysosomal lumen. The compounds tested were the following antihistamines: desloratadine, ebastine, loratadine, astemizole, and terfenadine; the antimalarials: chloroquine and mefloquine; and the antidepressants: desipramine, penfluridol, and siramesine, among which siramesine and terfenadine had the strongest effects [93]. The flavonoid polyphenol, quercetin has also been reported to induce lysosome-dependent cell death in leukemia and is effective against multi-drug resistant HL60 cell lines [94].
Lysosomal associated signaling
Several mTORC1 inhibitors, including rapamycin and its analogous (rapalogs) have been developed. Many second-generation inhibitors have also been developed that target both mTORC1 and mTORC2, or mTORC1 and PI3K. Phase I/II studies with rapalogs as single agents in AML have been disappointing, whereas combination studies with chemotherapy have demonstrated some promise [95]. Interestingly, the mTORC1/2 dual inhibitor AZD2014 was found to reduce the pH of lysosomes in AML cells and this enhanced the cytotoxicity of the antibody-drug conjugate Gemtuzumab ozogamicin (GO) [96]. The use of an antibody conjugate such as GO in AML is particularly appealing as most AMLs express CD33. GO requires an acidic environment inside lysosomes for hydrolysis of the linker molecule and cytocidal activity. The mTORC1-associated protein, raptor, represents a potential target in AML, as its ablation blocks leukemia progression. However, it does not affect leukemic stem cell (LSC) renewal [97]. mTORC1 inhibition sensitizes the cells to lysine-specific demethylase 1 inhibitors and triggers differentiation in MLL leukemia [98]. The lysosomal membrane protein LAMP5 acts as an autophagy suppressor and targeting it promotes autophagic degradation of the MLL fusion protein, which extends survival [99]. A novel ATRA derivative, 4-amino-2-trifluoromethyl-phenyl retinate (ATPR) has demonstrated superior anti-cancer efficacy against AML by inducing ferroptosis in AML cells through a mechanism involving macroautophagy [100].
Other novel approaches
An innovative approach to target AML cells involved the development of a biohybrid with a tumor-associated peptide somatostatin and the photosensitizer ruthenium, named RU-SST. The stomatostatin receptor type 2 (SSRT2) is expressed more highly on AML cell lines and leukemic cells of patients with AML compared to HSCs from healthy donors. Therefore, the biohybrid more specifically targets AML cells. Interestingly, RU-SST localizes within the lysosomes, indicating their involvement in the eradication mechanism [101]. Non-thermal plasma (NTP) is a relatively new addition in the field of cancer therapy. It is a form of ionized gas consisting of atoms and molecules in an excited state. NTP has been shown to induce cell death in AML cell lines, HL60, and KG-1 via lysosome inhibition [102].
Lysosomes can be utilized in a rather unique manner by harnessing them as a biologically derived nano-carrier. Encapsulation of cancer drugs within nano-carriers such as liposomes has already been put into practice. Lysosomes, due to their robustness within the biological environment and low immunogenic reaction can serve as a suitable carrier. Yeast-derived lysosomes, engineered to carry the anthracycline drug, daunorubicin, have exhibited efficacy against the AML cell line, HL60 [103].
Conclusion and perspectives
This review highlights the variety of functions associated with lysosomes and emphasizes their central role in maintaining cellular homeostasis. It is hardly surprising that deregulation of lysosomal structure and function can play a key role in malignant diseases such as AML. Tumor-specific lysosomal alterations have been identified, opening the possibility of targeting lysosomes as a way to exert maximum effect on cancer cells while sparing normal ones. To date, the cornerstone of chemotherapy treatment in AML is the combination of cytarabine (Ara-C) with an anthracycline and/or allogeneic stem cell transplantation. However, patients above the age of 65 respond poorly to such a taxing treatment regime. On the other hand, targeted therapies, while less strenuous, are usually not effective as a monotherapy [104]. In the scope of this review, we have highlighted lysosomal functions that may be targeted therapeutically and potentially convey a synergistic effect when combined with other modes of targeted therapies.
One major issue in the field of lysosomal research is that the lysosome has become synonymous with its degradative functions and more attention needs to be drawn towards the lysosome as a signaling organelle. Importantly, targeting the lysosome is independent of the molecular lesions in AML and is not dependent upon the disruption of up-stream receptor-mediated signaling events—for which resistance is inevitable. New targeting or chemo-sensitizing strategies of this type would therefore be extremely valuable. We hope that this review reiterates the importance of studying lysosomes and reinforces the growing interest in exploring its value as a therapeutic target in leukemia.
References
de Duve C. The lysosome turns fifty. Nat Cell Biol. 2005;7:847–9.
Baba M, Takeshige K, Baba N, Ohsumi Y. Ultrastructural analysis of the autophagic process in yeast: detection of autophagosomes and their characterization. J Cell Biol. 1994;124:903–13.
Settembre C, Zoncu R, Medina DL, Vetrini F, Erdin S, Erdin S, et al. A lysosome-to-nucleus signalling mechanism senses and regulates the lysosome via mTOR and TFEB. EMBO J. 2012;31:1095–108.
Abu-Remaileh M, Wyant GA, Kim C, Laqtom NN, Abbasi M, Chan SH, et al. Lysosomal metabolomics reveals V-ATPase- and mTOR-dependent regulation of amino acid efflux from lysosomes. Science. 2017;358:807–13.
Platt FM. Emptying the stores: lysosomal diseases and therapeutic strategies. Nat Rev Drug Discov. 2018;17:133–50.
Ohkuma S, Moriyama Y, Takano T. Identification and characterization of a proton pump on lysosomes by fluorescein-isothiocyanate-dextran fluorescence. Proc Natl Acad Sci USA. 1982;79:2758–62.
Perera RM, Zoncu R. The lysosome as a regulatory hub. Annu Rev Cell Dev Biol. 2016;32:223–53.
Saftig P, Klumperman J. Lysosome biogenesis and lysosomal membrane proteins: trafficking meets function. Nat Rev Mol Cell Biol. 2009;10:623–35.
Settembre C, Di Malta C, Polito VA, Arencibia MG, Vetrini F, Erdin S, et al. TFEB links autophagy to lysosomal biogenesis. Science. 2011;332:1429–33.
Luzio JP, Pryor PR, Bright NA. Lysosomes: fusion and function. Nat Rev Mol Cell Biol. 2007;8:622–32.
Embgenbroich M, Burgdorf S. Current concepts of antigen cross-presentation. Front Immunol. 2018;9:1–10.
Saftig P, Puertollano R. How lysosomes sense, integrate, and cope with stress. Trends Biochem Sci. 2021;46:97–112.
Wyant GA, Abu-Remaileh M, Wolfson RL, Chen WW, Freinkman E, Danai LV, et al. mTORC1 activator SLC38A9 is required to efflux essential amino acids from lysosomes and use protein as a nutrient. Cell. 2017;171:642–54.e12.
Yadati T, Houben T, Bitorina A, Shiri-Sverdlov R. The ins and outs of cathepsins: physiological function and role in disease management. Cells. 2020;9:1–26.
Aits S, Jäättelä M. Lysosomal cell death at a glance. J Cell Sci. 2013;126:1905–12.
Kirkegaard T, Jäättelä M. Lysosomal involvement in cell death and cancer. Vol. 1793, Biochimica et Biophysica Acta - Molecular Cell Research. Elsevier B.V.; 2009. p. 746–54.
Medina DL, Di Paola S, Peluso I, Armani A, De Stefani D, Venditti R, et al. Lysosomal calcium signalling regulates autophagy through calcineurin and TFEB. Nat Cell Biol. 2015;17:288–99.
Rebsamen M, Pochini L, Stasyk T, De Araújo MEG, Galluccio M, Kandasamy RK, et al. SLC38A9 is a component of the lysosomal amino acid sensing machinery that controls mTORC1. Nature. 2015;519:477–81.
Zoncu R, Bar-Peled L, Efeyan A, Wang S, Sancak Y, Sabatini DM. mTORC1 senses lysosomal amino acids through an inside-out mechanism that requires the vacuolar H+-ATPase. Science. 2011;334:678–83.
Sancak Y, Sabatini DM. Rag proteins regulate amino-acid-induced mTORC1 signalling. Biochem Soc Trans. 2009;37:289–90.
Zhang T, Wang R, Wang Z, Wang X, Wang F, Ding J. Structural basis for Ragulator functioning as a scaffold in membrane-anchoring of Rag GTPases and mTORC1. Nat Commun. 2017;8:1–10.
Bar-Peled L, Chantranupong L, Cherniack AD, Chen WW, Ottina KA, Grabiner BC, et al. A tumor suppressor complex with GAP activity for the Rag GTPases that signal amino acid sufficiency to mTORC1. Science. 2013;340:1100–6.
Liu GY, Sabatini DM. mTOR at the nexus of nutrition, growth, ageing and disease. Nat Rev Mol Cell Biol. 2020;21:183–203.
Nowosad A, Jeannot P, Callot C, Creff J, Perchey RT, Joffre C, et al. p27 controls Ragulator and mTOR activity in amino acid-deprived cells to regulate the autophagy-lysosomal pathway and coordinate cell cycle and cell growth. Nat Cell Biol. 2020;22:1076–90.
Inoki K, Li Y, Zhu T, Wu J, Guan KL. TSC2 is phosphorylated and inhibited by Akt and suppresses mTOR signalling. Nat Cell Biol. 2002;4:648–57.
Kim J, Guan KL. mTOR as a central hub of nutrient signalling and cell growth. Nat Cell Biol. 2019;21:63–71.
Chauhan S, Goodwin JG, Chauhan S, Manyam G, Wang J, Kamat AM, et al. ZKSCAN3 is a master transcriptional repressor of autophagy. Mol Cell. 2013;50:16–28.
Yim WW-Y, Mizushima N. Lysosome biology in autophagy. Cell Discov. 2020;6:6.
Mizushima N, Yoshimori T, Ohsumi Y. The role of Atg proteins in autophagosome formation. Annu Rev Cell Dev Biol. 2011;27:107–32.
Ohsumi Y. Historical landmarks of autophagy research. Cell Res. 2014;24:9–23.
Death C, Dikic I. Mechanism and medical implications of mammalian autophagy. Nat Rev Mol Cell Biol. 2018;19:349–64.
Kirchner P, Bourdenx M, Madrigal-Matute J, Tiano S, Diaz A, Bartholdy BA, et al. Proteome-wide analysis of chaperone-mediated autophagy targeting motifs. Simonsen A, editor. PLoS Biol. 2019;17:e3000301.
Stricher F, Macri C, Ruff M, Muller S. HSPA8/HSC70 chaperone protein: Structure, function, and chemical targeting. Autophagy. 2013;9:1937–54.
Kaushik S, Cuervo AM. The coming of age of chaperone-mediated autophagy. Nat Rev Mol Cell Biol. 2018;19:365–81.
Mukherjee A, Patel B, Koga H, Cuervo AM, Jenny A. Selective endosomal microautophagy is starvation-inducible in Drosophila. Autophagy. 2016;12:1984–99.
Mijaljica D, Prescott M, Devenish RJ. Microautophagy in mammalian cells: revisiting a 40-year-old conundrum. Autophagy. 2011;7:673–82.
Pu J, Guardia CM, Keren-Kaplan T, Bonifacino JS. Mechanisms and functions of lysosome positioning. J Cell Sci. 2016;129:4329–39.
Korolchuk VI, Saiki S, Lichtenberg M, Siddiqi FH, Roberts EA, Imarisio S, et al. Lysosomal positioning coordinates cellular nutrient responses. Nat Cell Biol. 2011;13:453–62.
Hong Z, Pedersen NM, Wang L, Torgersen ML, Stenmark H, Raiborg C. PtdIns3P controls mTORC1 signaling through lysosomal positioning. J Cell Biol. 2017;216:4217–33.
Jia R, Bonifacino JS. Lysosome positioning influences mTORC2 and AKT signaling. Mol Cell. 2019;75:26–38. e3.
Walton ZE, Patel CH, Brooks RC, Yu Y, Ibrahim-Hashim A, Riddle M, et al. Acid suspends the Circadian clock in hypoxia through inhibition of mTOR. Cell. 2018;174:72–87.
Starling GP, Yip YY, Sanger A, Morton PE, Eden ER, Dodding MP. Folliculin directs the formation of a Rab34– RILP complex to control the nutrient‐dependent dynamic distribution of lysosomes. EMBO Rep. 2016;17:823–41.
Johnson DE, Ostrowski P, Jaumouillé V, Grinstein S. The position of lysosomes within the cell determines their luminal pH. J Cell Biol. 2016;212:677–92.
Buratta S, Tancini B, Sagini K, Delo F, Chiaradia E, Urbanelli L, et al. Lysosomal exocytosis, exosome release and secretory autophagy: the autophagic- and endo-lysosomal systems go extracellular. Int J Mol Sci. 2020;21:2576.
Jia J, Abudu YP, Claude-Taupin A, Gu Y, Kumar S, Choi SW, et al. Galectins control MTOR and AMPK in response to lysosomal damage to induce autophagy. Autophagy. 2019;15:169–71.
Tazhitdinova R, Timoshenko AV. The emerging role of galectins and O-GlcNAc homeostasis in processes of cellular differentiation. Cells. 2020;9:1–35.
Ruvolo PP. Galectins as regulators of cell survival in the leukemia niche. Adv Biol Regul. 2019;71:41–54.
Thelen AM, Zoncu R. Emerging Roles for the Lysosome in Lipid Metabolism. Trends Cell Biol. 2017;27:833–50.
Infante RE, Wang ML, Radhakrishnan A, Hyock JK, Brown MS, Goldstein JL. NPC2 facilitates bidirectional transfer of cholesterol between NPC1 and lipid bilayers, a step in cholesterol egress from lysosomes. Proc Natl Acad Sci USA. 2008;105:15287–92.
Singh R, Kaushik S, Wang Y, Xiang Y, Novak I, Komatsu M, et al. Autophagy regulates lipid metabolism. Nature. 2009;458:1131–5.
Miyake K, Shibata T, Ohto U, Shimizu T, Saitoh SI, Fukui R, et al. Mechanisms controlling nucleic acid-sensing toll-like receptors. Int Immunol. 2018;30:43–51.
Xu Y, Shen J, Ran Z. Emerging views of mitophagy in immunity and autoimmune diseases. Autophagy. 2020;16:3–17.
Nabar NR, Kehrl JH. The transcription factor EB links cellular stress to the immune response. Yale J Biol Med. 2017;90:301–15.
Wang F, Tasset I, Cuervo AM, Muller S. In vivo remodeling of altered autophagy-lysosomal pathway by a phosphopeptide in lupus. Cells. 2020;9:1–15.
Saitoh T, Fujita N, Jang MH, Uematsu S, Yang BG, Satoh T, et al. Loss of the autophagy protein Atg16L1 enhances endotoxin-induced IL-1β production. Nature. 2008;456:264–8.
Shi CS, Shenderov K, Huang NN, Kabat J, Abu-Asab M, Fitzgerald KA, et al. Activation of autophagy by inflammatory signals limits IL-1β production by targeting ubiquitinated inflammasomes for destruction. Nat Immunol. 2012;13:255–63.
Glunde K, Guggino SE, Solaiyappan M, Pathak AP, Ichikawa Y, Bhujwalla ZM. Extracellular acidification alters lysosomal trafficking in human breast cancer cells. Neoplasia. 2003;5:533–45.
Davidson SM, Vander, Heiden MG. Critical functions of the lysosome in cancer biology. Annu Rev Pharm Toxicol. 2017;57:481–507.
Machado E, White-Gilbertson S, van de Vlekkert D, Janke L, Moshiach S, Campos Y, et al. Regulated lysosomal exocytosis mediates cancer progression. Sci Adv. 2015;1:e1500603.
Zhitomirsky B, Assaraf YG. Lysosomes as mediators of drug resistance in cancer. Drug Resist Updat. 2016;24:23–33.
Morgan MJ, Fitzwalter BE, Owens CR, Powers RK, Sottnik JL, Gamez G, et al. Metastatic cells are preferentially vulnerable to lysosomal inhibition. Proc Natl Acad Sci USA. 2018;115:E8479–88.
Bonam SR, Wang F, Muller S. Lysosomes as a therapeutic target. Nat Rev Drug Discov. 2019;18:923–48.
Saffi GT, Botelho RJ. Lysosome fission: planning for an exit. Trends Cell Biol. 2019;29:635–46.
Perrin P, Jongsma ML, Neefjes J, Berlin I. The labyrinth unfolds: architectural rearrangements of the endolysosomal system in antigen-presenting cells. Curr Opin Immunol. 2019;58:1–8.
Yoshida Y, Oguma S, Ohno H. John Auer and Auer rods; controversies revisited. Leuk Res. 2009;33:614–6.
Liang R, Arif T, Kalmykova S, Kasianov A, Lin M, Menon V, et al. Restraining lysosomal activity preserves hematopoietic stem cell quiescence and potency. Cell Stem Cell. 2020;26:359–76.e7.
Loeffler D, Wehling A, Schneiter F, Zhang Y, Müller-Bötticher N, Hoppe PS, et al. Asymmetric lysosome inheritance predicts activation of haematopoietic stem cells. Nature. 2019;573:426–9.
Warr MR, Binnewies M, Flach J, Reynaud D, Garg T, Malhotra R, et al. FOXO3A directs a protective autophagy program in haematopoietic stem cells. Nature. 2013;494:323–7.
Mortensen M, Soilleux EJ, Djordjevic G, Tripp R, Lutteropp M, Sadighi-Akha E, et al. The autophagy protein Atg7 is essential for hematopoietic stem cell maintenance. J Exp Med. 2011;208:455–67.
Watson A, Riffelmacher T, Stranks A, Williams O, De Boer J, Cain K, et al. Autophagy limits proliferation and glycolytic metabolism in acute myeloid leukemia. Cell Death Discov. 2015;1:1–10.
Liu F, Lee JY, Wei H, Tanabe O, Engel JD, Morrison SJ, et al. FIP200 is required for the cell-autonomous maintenance of fetal hematopoietic stem cells. Blood. 2010;116:4806–14.
Dong S, Wang Q, Kao Y-R, Diaz A, Tasset I, Kaushik S, et al. Chaperone-mediated autophagy sustains haematopoietic stem-cell function. Nature. 2021;591:117–23.
Piya S, Kornblau SM, Ruvolo VR, Mu H, Ruvolo PP, McQueen T, et al. Atg7 suppression enhances chemotherapeutic agent sensitivity and overcomes stroma-mediated chemoresistance in acute myeloid leukemia. Blood. 2016;128:1260–9.
Liu Q, Chen L, Atkinson JM, Claxton DF, Wang HG. Atg5-dependent autophagy contributes to the development of acute myeloid leukemia in an MLL-AF9-driven mouse model. Cell Death Dis. 2016;7:1–12.
Sumitomo Y, Koya J, Nakazaki K, Kataoka K, Tsuruta-Kishino T, Morita K, et al. Cytoprotective autophagy maintains leukemia-initiating cells in murine myeloid leukemia. Blood. 2016;128:1614–24.
Rudat S, Pfaus A, Cheng YY, Holtmann J, Ellegast JM, Bühler C, et al. RET-mediated autophagy suppression as targetable co-dependence in acute myeloid leukemia. Leukemia. 2018;2018:1.
Heydt Q, Larrue C, Saland E, Bertoli S, Sarry JE, Besson A, et al. Oncogenic FLT3-ITD supports autophagy via ATF4 in acute myeloid leukemia. Oncogene. 2018;37:787–97.
Jacquel A, Obba S, Boyer L, Dufies M, Robert G, Gounon P, et al. Autophagy is required for CSF-1–induced macrophagic differentiation and acquisition of phagocytic functions. Blood. 2012;119:4527–31.
Zhang Y, Morgan MJ, Chen K, Choksi S, Liu Z. Induction of autophagy is essential for monocyte-macrophage differentiation. Blood. 2012;119:2895–905.
Riffelmacher T, Clarke A, Richter FC, Stranks A, Pandey S, Danielli S, et al. Autophagy-dependent generation of free fatty acids is critical for normal neutrophil differentiation. Immunity. 2017;47:466–80.e5.
Wang Z, Cao L, Kang R, Yang M, Liu L, Zhao Y, et al. Autophagy regulates myeloid cell differentiation by p62/SQSTM1-mediated degradation of PML-RARα oncoprotein. Autophagy. 2011;7:401–11.
Orfali N, O’Donovan TR, Nyhan MJ, Britschgi A, Tschan MP, Cahill MR, et al. Induction of autophagy is a key component of all-trans-retinoic acid-induced differentiation in leukemia cells and a potential target for pharmacologic modulation. Exp Hematol. 2015;43:781–93.e2.
Orfali N, O’Donovan TR, Cahill MR, Benjamin D, Nanus DM, McKenna SL, et al. All-trans retinoic acid (ATRA)-induced TFEB expression is required for myeloid differentiation in acute promyelocytic leukemia (APL). Eur J Haematol. 2020;104:236–50.
Humbert M, Seiler K, Mosimann S, Rentsch V, Sharma K, Pandey AV, et al. Reducing FASN expression sensitizes acute myeloid leukemia cells to differentiation therapy. Cell Death Differ. 2021;28:2465–81.
Sukhai MA, Prabha S, Hurren R, Rutledge AC, Lee AY, Sriskanthadevan S, et al. Lysosomal disruption preferentially targets acute myeloid leukemia cells and progenitors. J Clin Invest. 2013;123:315–28.
Yamagishi T, Sahni S, Sharp DM, Arvind A, Jansson PJ, Richardson DR. P-glycoprotein mediates drug resistance via a novel mechanism involving lysosomal sequestration. J Biol Chem. 2013;288:31761–71.
Rosenfeld CS. Antileukemic activity of phenylalanine methyl ester (PME): A lysosomotropic peptide methyl ester. Stem Cells. 1994;12:198–204.
Cornet-Masana JM, Banús-Mulet A, Carbó JM, Torrente MÁ, Guijarro F, Cuesta-Casanovas L, et al. Dual lysosomal-mitochondrial targeting by antihistamines to eradicate leukaemic cells. EBioMedicine. 2019;47:221–34.
Liu J, Peng L, Niu T, Wu Y, Li J, Wang F, et al. PIG7 promotes leukemia cell chemosensitivity via lysosomal membrane permeabilization. Oncotarget. 2016;7:4841–59.
Bernard D, Gebbia M, Prabha S, Gronda M, MacLean N, Wang X, et al. Select microtubule inhibitors increase lysosome acidity and promote lysosomal disruption in acute myeloid leukemia (AML) cells. Apoptosis. 2015;20:948–59.
Zhang S, Schneider LS, Vick B, Grunert M, Jeremias I, Menche D, et al. Anti-leukemic effects of the V-ATPase inhibitor Archazolid A. Oncotarget. 2015;6:43508–28.
Nielsen IØ, Groth-Pedersen L, Dicroce-Giacobini J, Jonassen ASH, Mortensen M, Bilgin M, et al. Cationic amphiphilic drugs induce elevation in lysoglycerophospholipid levels and cell death in leukemia cells. Metabolomics. 2020;16:91.
Nielsen IØ, Groth-Pedersen L, Dicroce-Giacobini J, Jonassen ASH, Mortensen M, Bilgin M, et al. Cationic amphiphilic drugs induce elevation in lysoglycerophospholipid levels and cell death in leukemia cells. Metabolomics. 2020;16:1–13.
Maruszewska A, Tarasiuk J. Quercetin triggers induction of apoptotic and lysosomal death of sensitive and multidrug resistant leukaemia HL60 cells. Nutr Cancer. 2020;0:1–18.
Ghosh J, Kapur R. Role of mTORC1-S6K1 signaling pathway in regulation of hematopoietic stem cell and acute myeloid leukemia. Exp Hematol. 2017;50:13–21.
Mizutani Y, Inase A, Maimaitili Y, Miyata Y, Kitao A, Matsumoto H, et al. An mTORC1/2 dual inhibitor, AZD2014, acts as a lysosomal function activator and enhances gemtuzumab ozogamicin-induced apoptosis in primary human leukemia cells. Int J Hematol. 2019;110:490–9.
Hoshii T, Tadokoro Y, Naka K, Ooshio T, Muraguchi T, Sugiyama N, et al. mTORC1 is essential for leukemia propagation but not stem cell self-renewal. J Clin Invest. 2012;122:2114–29.
Deb G, Wingelhofer B, Amaral FMR, Maiques-Diaz A, Chadwick JA, Spencer GJ, et al. Pre-clinical activity of combined LSD1 and mTORC1 inhibition in MLL-translocated acute myeloid leukaemia. Leukemia. 2020;34:1266–77.
Zeng Z-C, Han C, Chen Z-H, Huang W, Luo X-Q, Chen Y-Q, et al. Activation of the lysosome-associated membrane protein LAMP5 by DOT1L serves as a bodyguard for MLL fusion oncoproteins to evade degradation in leukemia. Clin Cancer Res. 2019;:clincanres.1474.2018.
Du Y, Bao J, Zhang MJ, Li LL, Xu XL, Chen H, et al. Targeting ferroptosis contributes to ATPR-induced AML differentiation via ROS-autophagy-lysosomal pathway. Gene. 2020;755:1–7.
Vegi NM, Chakrabortty S, Zegota MM, Kuan SL, Stumper A, Rawat VPS, et al. Somatostatin receptor mediated targeting of acute myeloid leukemia by photodynamic metal complexes for light induced apoptosis. Sci Rep. 2020;10:371.
Kim S-Y, Kim HJ, Kim HJ, Kim C-H. Non-thermal plasma induces antileukemic effect through mTOR ubiquitination. Cells. 2020;9:595.
Choi W, Heo MY, Kim SY, Wee JH, Kim YH, Min J. Encapsulation of daunorubicin into Saccharomyces cerevisiae-derived lysosome as drug delivery vehicles for acute myeloid leukemia (AML) treatment. J Biotechnol. 2020;308:118–23.
Molica M, Breccia M, Foa R, Jabbour E, Kadia TM. Maintenance therapy in AML: the past, the present and the future. Am J Hematol. 2019;94:1254–65.
Solitro AR, MacKeigan JP. Leaving the lysosome behind: novel developments in autophagy inhibition. Future Med Chem. 2016;8:73–86.
Della Porta A, Bornstein K, Coye A, Montrief T, Long B, Parris MA. Acute chloroquine and hydroxychloroquine toxicity: A review for emergency clinicians. Am J Emerg Med. 2020;38:2209–17.
Chugh R, Sangwan V, Patil SP, Dudeja V, Dawra RK, Banerjee S, et al. A preclinical evaluation of minnelide as a therapeutic agent against pancreatic cancer. Sci Transl Med. 2012;4:1–10.
Owa C, Messina ME, Halaby R. Triptolide induces lysosomal-mediated programmed cell death in MCF-7 breast cancer cells. Int J Women’s Health. 2013;5:557–69.
Zhang K, Ma Y, Guo Y, Sun T, Wu J, Pangeni RP, et al. Cetuximab-triptolide conjugate suppresses the growth of EGFR-overexpressing lung cancers through targeting RNA polymerase II. Mol Ther - Oncolytics. 2020;18:304–16.
Giri B, Gupta VK, Yaffe B, Modi S, Roy P, Sethi V, et al. Pre-clinical evaluation of Minnelide as a therapy for acute myeloid leukemia. J Transl Med. 2019;17:163.
Xi C, Peng S, Wu Z, Zhou Q, Zhou J. Toxicity of triptolide and the molecular mechanisms involved. Biomed Pharmacother. 2017;90:531–41.
Borkowska M, Siek M, Kolygina DV, Sobolev YI, Lach S, Kumar S, et al. Targeted crystallization of mixed-charge nanoparticles in lysosomes induces selective death of cancer cells. Nat Nanotechnol. 2020;15:331–41.
McGuire C, Cotter K, Stransky L, Forgac M. Regulation of V-ATPase assembly and function of V-ATPases in tumor cell invasiveness. Biochim Biophys Acta - Bioenerg. 2016;1857:1213–8.
Garcia-Rodriguez J, Mendiratta S, White MA, Xie XS, De Brabander JK. Synthesis and structure-activity studies of the V-ATPase inhibitor saliphenylhalamide (SaliPhe) and simplified analogs. Bioorg Med Chem Lett. 2015;25:4393–8.
Yambire KF, Rostosky C, Watanabe T, Pacheu-Grau D, Torres-Odio S, Sanchez-Guerrero A, et al. Impaired lysosomal acidification triggers iron deficiency, necrotic cell death and inflammation in vivo. bioRxiv. 2019;5:1–36.
Söderholm S, Anastasina M, Islam MM, Tynell J, Poranen MM, Bamford DH, et al. Immuno-modulating properties of saliphenylhalamide, SNS-032, obatoclax, and gemcitabine. Antivir Res. 2016;126:69–80.
Massey AJ, Williamson DS, Browne H, Murray JB, Dokurno P, Shaw T, et al. A novel, small molecule inhibitor of Hsc70/Hsp70 potentiates Hsp90 inhibitor induced apoptosis in HCT116 colon carcinoma cells. Cancer Chemother Pharm. 2010;66:535–45.
Kirkegaard T, Roth AG, Petersen NHT, Mahalka AK, Olsen OD, Moilanen I, et al. Hsp70 stabilizes lysosomes and reverts Niemann-Pick disease-associated lysosomal pathology. Nature. 2010;463:549–53.
Ikonomov OC, Sbrissa D, Shisheva A. Small molecule PIKfyve inhibitors as cancer therapeutics: translational promises and limitations. Toxicol Appl Pharm. 2019;383:114771.
Sharma G, Guardia CM, Roy A, Vassilev A, Saric A, Griner LN, et al. A family of PIKFYVE inhibitors with therapeutic potential against autophagy-dependent cancer cells disrupt multiple events in lysosome homeostasis. Autophagy. 2019;15:1694–718.
Acknowledgements
This study was supported by grants from the Swiss National Science Foundation (31003A_173219 to MPT), the Bernese Cancer League, “Stiftung für klinisch-experimentelle Tumorforschung”, and the Werner and Hedy Berger-Janser Foundation for Cancer Research (to MH). SLM is supported by Breakthrough Cancer Research, Ireland. SLM, SM, MPT, and MH also acknowledge the support of the TRANSAUTOPHAGY COST Action (CA15138). The Muller laboratory was funded by the French Centre National de la Recherche Scientifique, Région Grand-Est, and the University of Strasbourg Institute for Advanced Study (USIAS) and the Interdisciplinary Thematic Institute 2021-2028 program of the University of Strasbourg, CNRS, and Inserm (ANR-10-IDEX-0002 and ANR-20-SFRI-0012) in the frame of the Strasbourg drug discovery and development Institute (IMS). SM is supported by the Club francophone de l’autophagie (CFATG), the European Regional Development Fund of the European Union in the framework of the INTERREG V Upper Rhine program. The authors would like to express gratitude to Myriam Legros at the Center of Laboratory Medicine, University Hospital Bern, Switzerland for providing the image of auer rods. All schematic representations were created with BioRender.com.
Funding
Open Access funding provided by Universität Bern.
Author information
Authors and Affiliations
Contributions
SR conceptualized and drafted the article and prepared all figures. SLM, SM, MPT, and MH conceptualized and revised the article. MPT and MH gave the final approval for the publication.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interest.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Rafiq, S., McKenna, S.L., Muller, S. et al. Lysosomes in acute myeloid leukemia: potential therapeutic targets?. Leukemia 35, 2759–2770 (2021). https://doi.org/10.1038/s41375-021-01388-x
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41375-021-01388-x
This article is cited by
-
The dual role of autophagy in acute myeloid leukemia
Journal of Hematology & Oncology (2022)
-
The PIP4K2 inhibitor THZ-P1-2 exhibits antileukemia activity by disruption of mitochondrial homeostasis and autophagy
Blood Cancer Journal (2022)