Abstract
Gliomas are the most common primary intracranial tumor, accounting for more than 70% of brain malignancies. Studies indicate that highly upregulated in liver cancer (HULC), a long noncoding RNA (lncRNA), functions as an oncogene in gliomas. However, the underlying mechanism of HULC in gliomas remains under-studied and was subsequently investigated in the current study. Brain tissues were clinically collected from 50 patients with glioblastoma (GBM) and 35 patients with acute craniocerebral injury, followed by immunohistochemical detection of the expression patterns of Forkhead box M1 (FOXM1), anterior gradient 2 (AGR2), and hypoxia-inducible factor-1α (HIF-1α). After flow cytometry-based sorting of the CD133+ glioma stem cells (GSCs) from the U251 cell line, the obtained cells were subjected to lentivirus infection. Afterwards, the proliferation, stemness, and apoptosis of GSCs were evaluated using sphere formation, immunofluorescence, and flow cytometry assays, respectively. In addition, the interactions among HULC, FOXM1, AGR2, and HIF-1α were identified using RNA immunoprecipitation (RIP), RNA pull-down, Chromatin immunoprecipitation (ChIP), IP, and dual luciferase reporter assays. Last, the specific effects were validated in vivo. HULC was upregulated in GBM tissues and GSCs, which may promote the progression of glioma. On the other hand, silencing of HULC reduced the stemness, inhibited the proliferation, and promoted the apoptosis and differentiation of GSCs. In addition, HULC further stabilized FOXM1 expression in GSCs through ubiquitination, while FOXM1 activated AGR2 transcription to promote HIF-1α expression. Moreover, HULC promoted the glycolysis and stemness of GSCs through its regulation of the FOXM1/AGR2/HIF-1α axis, consequently exacerbating the occurrence and development of glioma. The findings obtained in our study indicate that HULC stabilizes the FOXM1 protein by ubiquitination to upregulate the expression of AGR2 and HIF-1α, which further promote the glycolysis of and maintain the stemness of GSCs, to enhance the tumorigenicity of GSCs, highlighting a novel therapeutic target for glioma.
Similar content being viewed by others
Introduction
Gliomas, the most widespread primary brain tumors in adults, account for over 70% of brain malignancies1. Gliomas usually are derived from glial progenitor cells and glial cells, which have been historically diagnosed and classified into ependymoma, oligodendroglioma, glioblastoma (GBM), and astrocytoma2,3. According to the latest World Health Organization (WHO) classification criteria, GBM is regarded as the most prevalent and malignant type of glioma (WHO grade IV)4. GBM is known to contain self-renewing and tumorigenic glioma stem cells (GSCs), which participate in the initiation and development of tumors and may lead to therapeutic resistance. GSCs are not self-autonomous units that function within tumor microenvironment, and play critical roles in microenvironment remodeling and maintenance5. Thus, a prudent approach would be to improve our understanding of the molecular mechanism underlying GSCs in glioma in order to identify novel therapies to inhibit dynamics of GSCs in glioma treatment.
Long non-coding RNAs (lncRNAs), which are functional RNA molecules without protein-coding functions, are also known to contribute to the development of glioma6. The lncRNA highly upregulated in liver cancer (HULC) is upregulated in tumors and regarded as a therapeutic target for numerous human malignancies, with studies even indicating its potential role in carcinogenesis7,8. Moreover, a previous study documented high expression levels of HULC in glioma tissues, and further highlighted that upregulated HULC enhances the proliferation and colony formation capability of glioma cells9. Moreover, HULC also exhibits pro-angiogenesis activity in human gliomas, while its silencing inhibits angiogenesis in glioma10. In addition, accumulating evidence has suggested an interaction between HULC and forkhead box M1 (FOXM1), and highlighted its function in cancer progression and development11,12. Very coincidentally, downregulation of FOXM1 by a small interfering RNA (siRNA) was further indicated to induce cell cycle arrest, apoptosis, and the epithelial-mesenchymal transition of glioma cells13. Meanwhile, our peers in the research field further suggest that hypoxia-inducible factor-1 (HIF-1) may regulate the expression of anterior gradient 2 (AGR2) in GBM, thereby affecting the growth and angiogenesis of GBM cells14. Based on these data, we hypothesized in the current study that HULC may participate in the regulation of glycolysis and stemness properties of GSCs in glioma through the FOXM1/AGR2/HIF-1α regulatory axis, and thus we performed a series of experiments to validate our hypothesis with the aim of discovering a novel therapeutic strategy for GBM.
Materials and methods
Ethical approval
Human specimen collection and experimentation in the current study were approved by the ethics committee of the First Affiliated Hospital of Nanchang University, and conformed to the guidelines of the Declaration of Helsinki. Signed informed consent forms were obtained from all participants or their relatives prior to specimen collection. Animal experiments were performed in accordance with the Guide for the Care and Use of Laboratory Animals published by the US National Institutes of Health. Extensive efforts were made to minimize the number and suffering of the experimental animals.
Microarray analysis
First, the Gene Expression Omnibus (GEO) database (https://www.ncbi.nlm.nih.gov/geo/) was employed to obtain the GSE12657 dataset related to glioma, which included four sample types: GBM samples, control samples, oligodendroglioma samples, and fibrous astrocytoma samples. Next, 5 control samples and 7 GBM samples were included in the subsequent differential expression analysis, but data from the other two samples were excluded. The differential expression analysis was then performed to screen significantly differentially expressed genes using the R language “limma” package, with a false discovery rate (FDR) < 0.05 and a | log fold change (FC) | > 1.5.
Clinical sample collection
Tumor tissue samples were collected from 50 patients pathologically diagnosed with GBM from June 2017 to June 2019 at the Neurosurgery Department of the First Affiliated Hospital of Nanchang University, comprising 28 males and 22 females, with a mean calculated age of 54.74 ± 16.32 years (20–76 years). None of the included patients received chemotherapy or radiotherapy prior to surgery. According to magnetic resonance imaging (MRI) data collected from the head, the specific location of GBM was as follows: 19 cases of frontal tumors, 7 cases of parietal tumors, 9 cases of occipital tumors, and 15 cases of temporal tumors. Twenty-nine cases of isocitrate dehydrogenase 1 (IDH1)-wild type (Wt) and 21 cases of IDH1-mutant (Mut) tumors were identified. Additionally, 33 tumors possessed a telomerase reverse transcriptase (TERT) promoter mutation and 17 possessed p53 mutations (TP53). The GBM diagnosis and classification were conducted by a pathological examination using the WHO classification system. Furthermore, 35 brain tissue samples (at least 1 cm away from hemorrhage, contusions or extensive edema) were collected from patients with acute craniocerebral injury as a control, including 22 males and 13 females, with a mean calculated age of 49.06 ± 8.46 years (29–60 years). The clinical characteristics of enrolled patients with GBM are listed in Supplementary Table 1.
Cell culture
The 293 T cells (CRL-3216; American Type Culture Collection, ATCC, Manassas, VA, USA), normal brain glial cell line (HEB) (Shanghai Meilian Biotechnology Co., Ltd., Shanghai, China), and glioma cell lines U251 (TCHu 58; Cell Bank of Chinese Academy, Shanghai, China), LN18 (CM-H291; Shanghai GaiNing Biotechnology Co., Ltd, Shanghai, China), T98G (CRL-1690; ATCC), and A172 (TCHu171; Cell Bank of Chinese Academy) were cultured in Dulbecco’s Modified Eagle’s Medium (DMEM) (10569044, Gibco, Carlsbad, CA, USA) containing 10% fetal bovine serum (FBS; 10099141, Gibco) at 37 °C with 5% CO2.
GSC sorting
The U251 cell line was cultured in serum-free DMEM-F12 (A4192001, Gibco) supplemented with 10 ng/mL basic fibroblast growth factor (FGF; 100-18b, Peprotech, Rocky Hill, NJ, USA), 20 ng/mL epidermal growth factor (EGF; AF-100-15, Peprotech), and 2% B27 (17504044, Gibco) at 37 °C with 5% CO2. Next, the CD133-positive cells were isolated from the suspended U251 cells with immunomagnetic beads using a CD133+ cell sorting kit (Miltenyi Biotech, Bergisch Gladbach, Germany). Briefly, the U251 cells were first separated into a single-cell suspension by adding sorting solution (200 μL/108 cells), which were then coincubated with the CD133 antibody bead complex (100 μL/105 cells) at 4 °C for 30 min and rinsed with the sorting solution (1 mL/108 cells) again. After centrifugation and removal of the supernatant, the cells were collected and resuspended in the sorting solution (500 μL/108 cells). Subsequently, the cells were eluted with 2 mL of sorting solution to separate the CD133+ and CD133− cells, followed by the addition of 50 μL of CD133/2 (293C3)-PE antibody (130-113-748, Miltenyi) or 50 μL of IgG2b-PE antibody (IC0041P, R&D Systems, Minneapolis, MN, USA) as the isotype control. Later, a flow cytometer (Becton, Dickinson and Company, Sparks, MD, USA) was employed for cell sorting, and the sorted cells were cultured in serum-free DMEM-F12 containing 10 ng/mL FGF, 20 ng/mL EGF, and 2% B27, with FGF and EGF added every other day15.
Cell treatment
For cell infection, the core plasmids containing target gene silencing sequences (PLKO.1) and helper plasmids (RRE, REV, and VSVG) were used to package the short hairpin RNA lentivirus (sh-), while the core plasmids (Fugw-GFP and Plx304) containing target gene cDNA sequences and the helper plasmids (RRE, REV, and VSVG) were used to package the overexpression lentivirus (oe-). The lentiviruses were purchased from Sangon Biological Engineering Technology & Services Co., Ltd. (Shanghai, China), and the primer sequences and plasmids were also constructed by Sangon. All silencing sequences are listed in Supplementary Table 2. Meanwhile, the glucose transporter 1 expression (GLUT1) inhibitor (WZB117) (HY-19331, MedChemExpress, Monmouth Junction, NJ, USA) was dissolved in dimethyl sulfoxide (DMSO) (D2650, Sigma Aldrich, St. Louis, MO, USA), and used to treat cells for 48 h at a concentration of 10 mmol/L. Next, the U251 cells were treated with (1) sh-HULC-1, sh-HULC-2, sh-FOXM1-1, sh-FOXM1-2, or the negative control (NC) (sh-NC); (2) oe-FOXM1, oe-AGR2, oe-HIF-1α, oe-HULC-low (lentivirus expressing a small amount of oe-HULC, multiplicity of infection [MOI] = 2), oe-HULC-high (lentivirus expressing a large amount of oe-HULC, MOI = 10) or oe-NC; or (3) sh-NC + oe-NC, sh-FOXM1 + oe-NC, sh-FOXM1 + oe-AGR2, sh-FOXM1 + oe-HIF-1α, sh-FOXM1 + oe-AGR2 + sh-HIF-1α, sh-NC + oe-NC + DMSO, sh-HULC + oe-NC + DMSO, sh-HULC + oe-HIF-1α + DMSO, or sh-HULC + oe-HIF-1α + WZB117. All the aforementioned experiments were repeated three times. After 48 h of culture, the cells were collected to detect the transfection efficacy prior to use in subsequent experiments.
Reverse transcription-quantitative polymerase chain reaction (RT-qPCR)
Trizol reagent (16096020, Thermo Fisher Scientific, Waltham, MA, USA) was used to extract the total RNA content from tissues and cells. The obtained RNA was reverse-transcribed into complementary DNAs (cDNAs) using the First Strand cDNA Synthesis kit (D7168L, Institute of Biotechnology, Shanghai, China). RT-qPCR was subsequently performed using a RT-qPCR kit (Q511-02, Vazyme Biotech, Nanjing, China) with a Bio-Rad real-time qPCR instrument (Bio-Rad, Hercules, CA, USA). Primers for HULC, FOXM1, AGR2, and HIF-1α were designed and synthesized by Sangon (Supplementary Table 3). With glyceraldehyde-3-phosphate dehydrogenase (GAPDH) serving as the internal reference, the relative transcription expression of target genes was calculated using the 2−ΔΔCT method.
Western blot analysis
Total protein was extracted from tissues and cells using radioimmunoprecipitation assay (RIPA) lysis buffer containing phenylmethylsulfonyl fluoride (PMSF; P0013B, Beyotime, Shanghai, China), while the proteins from cell nucleus and cytoplasm were extracted using Nuclear and Cytoplasmic Protein Extraction kits (P0028, Beyotime, Shanghai, China). A bicinchoninic acid (BCA) assay kit (P0011, Beyotime, Shanghai, China) was used to detect the total protein concentration. Next, the protein samples were dissolved in 8‒12% sodium dodecyl sulfate (SDS) sample buffer and subjected to SDS polyacrylamide gel electrophoresis. Afterward, the proteins were transferred to a polyvinylidene fluoride membrane (1620177, Bio-Rad) using the wet transfer method, followed by blocking with 5% skim milk or 5% bovine serum albumin (BSA) at room temperature for 1 h. The PVDF membrane was subsequently incubated with diluted rabbit antibodies against GAPDH (5174, dilution ratio of 1:5000, Cell Signaling Technology, Beverly, MA, USA), FOXM1 (5436, dilution ratio of 1:1000, Cell Signaling Technology), AGR2 (NBP2-27393, dilution ratio of 1:1000, Novus Biologicals Littleton, CO, USA), HIF-1α (20960-1-AP, dilution ratio of 1:1000, Proteintech, Chicago, IL, USA), GLUT1 (ab652, dilution ratio of 1:1000, Abcam Inc., Cambridge, UK), and GFAP (ab7260, dilution ratio of 1:1000, Abcam) overnight at 4 °C. Next, the membrane was incubated with a horseradish peroxidase-labeled goat anti-rabbit against immunoglobulin G (IgG) secondary antibody (ab6721, dilution ratio of 1:2000, Abcam) at room temperature for 1 h. Enhanced chemiluminescence detection kits (705062, Bio-Rad) were employed to visualize the samples with the ImageQuant LAS4000C instrument (GE Healthcare, Madison, WI, USA). The relative protein expression was reported as the ratio between the gray value of the target protein band to that of the β-actin protein band.
Sphere forming assay of GSCs
The GSC spheres were dissociated into single cells using Accutase (40506ES60, Yeasen Biotechnology Co., Ltd., Shanghai, China) for 10 min. Next, the cells were cultured in serum-free DMEM-F12 supplemented with 10 ng/mL FGF, 20 ng/mL EGF, and 2% B27 and resuspended into a single-cell suspension. The cells growing under nonadherent conditions were seeded in 96-well plates (at a density of 500‒1000 cells/well), followed by the renewal of half of the medium every other day. After 9 days, the cells were fixed with 4% formalin, and the spheres larger than 50 μm were photographed and counted.
Glucose uptake and lactate production assays
Following 16 h of culture in glucose-free DMEM, the cells were further incubated for an additional 24 h in high-glucose DMEM under normoxic conditions. After the removal of culture medium, a fluorescence-based glucose assay kit (K666, BioVision, Milpitas, CA, USA) was used to determine the intracellular glucose levels, and a D-lactate assay kit (colorimetric) (ab83429, Abcam) was used to measure the lactate levels.
Immunofluorescence staining
The cells were seeded in a confocal cell dish. After 24 h, the cells were fixed with 4% paraformaldehyde for 15 min and rinsed with 3% poly(butylene succinate-co-terephthalate) (PBST) three times (5 min/time). After blocking with 5% BSA in 3% PBST for 1 h at room temperature, the cells were then incubated at room temperature for 1 h with the following primary antibodies: mouse anti-CD133 (14-1331, dilution ratio of 1:300, Invitrogen, Carlsbad, CA, USA) and rabbit anti-Nestin (AB5922, dilution ratio of 1:300, Millipore, Bedford, MA, USA). Subsequently, the cells were incubated with a red fluorescent protein (Cy3)-labeled goat anti-rat secondary antibody (ab6953, dilution ratio of 1:500, Abcam) and fluorescein isothiocyanate (FITC)-labeled goat anti-rabbit secondary antibody (ab67173, dilution ratio of 1:500, Abcam) at room temperature for 1 h. Finally, after the nuclei were stained with 4′6-diamidino-2-phenylindole (DAPI), the cells were observed under a fluorescence microscope (BX63, Olympus Optical Co. Ltd., Tokyo, Japan).
Immunohistochemistry
The paraffin-embedded glioma tissues were sliced into sections, and incubated at 60 °C for 20 min, dewaxed with xylene and hydrated with a gradient of ethanol solutions. The sections were subsequently treated with 3% hydrogen peroxide (H2O2) for 10 min at room temperature to inactivate endogenous peroxidases. After an incubation with normal goat serum (Sangon), the sections were incubated with the following antibodies overnight at 4 °C: rabbit anti-FOXM1 (5436, dilution ratio of 1:500, Cell Signaling Technology), rabbit anti-AGR2 (NBP2-27393, dilution ratio of 1:500, Novus Biologicals), rabbit anti-HIF-1α (20960-1-AP, dilution ratio of 1:500, Proteintech), rabbit anti-Ki67 (ab15580, dilution ratio of 1:500, Abcam). Thereafter, sections were incubated with a secondary goat anti-rabbit IgG antibody (ab6721, dilution ratio of 1:500, Abcam), followed by colorimetric detection using a 3,3′-diaminobenzidine (DAB) detection kit (Sigma) for 6 min. Afterward, the sections were stained with hematoxylin for 30 s and dehydrated in an ascending series of ethanol solutions, followed by microscopic observation (BX63, Olympus).
Dual-luciferase reporter assay
The fragment of the AGR2 promoter binding to FOXM1 (AGR2-Wt) and its mutated fragment (AGR2-Mut) were subcloned into the pGL3-Promoter (E1761, Promega, Madison, WI, USA) using restriction enzymes and T4 DNA ligases (M0204S, New England Biolabs, Ipswich, MA, USA). Next, the 293 T cells were transfected with sh-NC or sh-FOXM1, along with AGR2-Wt or AGR2-Mut, while the Renilla luciferase reporter plasmid was used to normalize the transfection efficiency (relative luciferase activity). After a 48-h incubation in the presence of 5% CO2 at 37 °C and saturation humidity, a dual-luciferase assay was performed using a Dual-Luciferase® Reporter Assay System kit (E1910, Promega) and a GloMax® 20/20 Luminometer (E5311, Promega). All vectors were constructed by Sangon.
Chromatin immunoprecipitation (ChIP)
According to the manufacturers’ instructions provided with the ChIP kit (ab117138, Abcam), the cells were fixed with 1% formaldehyde and incubated with glycine for 10 min at room temperature for DNA-protein crosslinking. Afterward, the cells were lysed and randomly disrupted by ultrasonification to produce 200‒300 bp chromatin fragments. Next, the fragments were respectively incubated with rabbit anti-IgG (ab172730, dilution ratio of 1:100, Abcam) as the NC and rabbit anti-FOXM1 (13147-1-AP, dilution ratio of 1:100, Proteintech). Protein A magnetic beads were then used to precipitate the endogenous DNA protein complex. Next, the non-specific complex was washed, followed by de-crosslinking and DNA fragment purification. Finally, the AGR2 promoter was analyzed using RT-qPCR. The promoter primers were F: 5ʹ-ATTTGGCACTCTGCCCATCG-3ʹ and R: 5ʹ-GGGCATACCTCAATCCCACC-3ʹ.
RNA pull-down assay
A Pierce RNA 3ʹ End Desthiobiotinylation Kit (20163, Thermo Fisher Scientific) was used to label the HULC RNA, and a Pierce Magnetic RNA–Protein Pull-Down Kit (20164, Thermo Fisher Scientific) was employed for the RNA pull-down assay. Next, the cell lysate was incubated with the labeled HULC RNA and a buffer containing M-280 streptavidin magnetic beads (11205D, Thermo Fisher Scientific). Finally, the beads were eluted for Western blot analysis.
RNA immunoprecipitation (RIP) assay
A Magna RIP RNA-Binding Protein Immunoprecipitation Kit (17-701, Millipore) was utilized to detect the binding between HULC and the FOXM1 protein. Upon reaching 80–90% confluence, the cells in the 6-well plates were lysed in RIPA buffer (P0013B, Beyotime) on ice for 5 min, followed by centrifugation at 14,000 × g at 4 °C. The supernatant was then randomly separated into two parts, with one part used as the input. Meanwhile, the other part of supernatant was incubated with 50 μL of magnetic beads resuspended in 100 μL of RIP wash buffer, which was then conjugated with 5 μg of the FOXM1 antibody (13147-1-AP, dilution ratio of 1:100, Proteintech) or IgG (ab200699, dilution ratio of 1:100, Abcam) as the NC. Next, the bead-antibody complex was washed and resuspended in 900 μL of RIP wash buffer and incubated overnight with 100 μL of the cell extraction solution. Afterward, the precipitated RNA was collected for RT-qPCR analysis.
Ubiquitination assay
The cells were incubated with 100 μM MG132 (HY-13259, MedChemExpress) for 6 h to detect endogenous FOXM1. Next, the differently treated cells were lysed in 1% SDS RIPA buffer, followed by ultrasonification. Subsequently, the cell extract was immunoprecipitated with the FOXM1 antibody (sc-376471, dilution ratio of 1:100, Santa Cruz Biotechnology) overnight at 4 °C, and then the Protein G magnetic beads were added for another 8-h incubation at 4 °C. Following three washes in IP buffer, the ubiquitination of FOXM1 was detected by Western blot analysis using a rabbit anti-Ubiquitin antibody (ab7780, dilution ratio of 1:1000, Abcam).
Flow cytometry
Forty-eight hours after cell transfection, Annexin V/FITC and propidium iodide (PI) staining kits were used for the cell apoptosis determination (C1062L, Beyotime). Briefly, the cells were resuspended in 200 µL of binding buffer and incubated with 10 µL of PI staining solution and 5 µL of Annexin V-FITC solution at room temperature for 15 min in the dark. Afterward, flow cytometry was immediately performed (Becton), and the apoptotic rate was calculated.
Terminal deoxynucleotidyl transferase-mediated dUTP-biotin nick end labeling (TUNEL) staining
TUNEL staining was performed with using a TUNEL apoptosis assay kit (C1098; Beyotime). Briefly, samples were fixed with 4% paraformaldehyde for 40 min. After PBS washes, the samples were permeabilized for 2 min on ice using immunostaining wash buffer, followed by endogenous peroxidase inactivation by an incubation in methanol containing 0.3% H2O2 at room temperature for 20 min. Subsequently, the samples were incubated with 2 μL of the TdT enzyme and 48 μL of Biotin-dUTP at 37 °C for 60 min. Afterward, the samples were incubated with the streptavidin-horseradish peroxidase (HRP) conjugate for 30 min, followed by an incubation with the DAB solution for 10 min and microscopic observation.
In vivo animal experiments
Seventy male BALB/c nude mice (aged 5 weeks) purchased from the Animal Experimental Center of Nanchang University (Jiangxi, China) were maintained under specific pathogen-free (SPF) conditions. The mice were randomly classified into 7 groups (GSCs, oe-NC, oe-HULC-low, oe-HULC-high, sh-NC + oe-NC, sh-HULC + oe-NC, and sh-HULC + oe-HIF-1α), with 10 mice in each group, for subcutaneous tumorigenesis experiment. Briefly, stably transfected GSCs (1 × 106 cells in 200 μL) were subcutaneously injected into the mice. Next, the tumor volume was calculated every week using the following formula: volume (mm3) = π/6 × length × (width)2. After 4 weeks, the mice were intraperitoneally injected with 9% pentobarbital sodium (P3761; Sigma) and then euthanatized to harvest tumors. After tumor sections were prepared, TUNEL staining was subsequently performed to detect cell apoptosis and Ki67 staining was conducted to detect cell proliferation. All tumor sections were subjected to microscopic observation (BX63, Olympus).
Statistical analyses
Statistical analyses were performed using the SPSS 21.0 statistical software (IBM Corp. Armonk, NY, USA). The count data were analyzed using the Chi-square test. Measurement data are presented as the means ± standard deviations. Comparisons between the two groups were conducted using unpaired Student’s t test. Differences among multiple groups were compared using one-way analysis of variance (ANOVA), while differences observed at different time points between multiple groups were compared using two-way ANOVA or repeated measures ANOVA followed by Tukey’s post hoc test. A value of p < 0.05 was regarded statistically significant.
Results
HULC is expressed at high levels in glioma tissues and GSCs
As reported in a previous study, HULC may participate in the occurrence and development of glioma9. RT-qPCR was performed to detect the expression patterns of HULC in glioma tissues and further elucidate the role of HULC in glioma. HULC was expressed at higher levels in the tumor tissues from patients with GBM compared to the controls (Fig. 1A). In addition, compared to the HEB cell line, all glioma cell lines (U251, T98G, A172, and LN18) exhibited increased levels of HULC in vitro (Fig. 1B).
A Expression of HULC in clinical tissue samples determined using RT-qPCR (n = 50 in the GBM group, n = 35 in the control group); *p < 0.05 compared with the control group. B Expression of HULC in the glioma cell lines determined using RT-qPCR; *p < 0.05 compared with the HEB group. C Sorting of CD133 + GSCs using flow cytometry. D Morphological observation of the spheroids that formed from cultured GSCs in vitro. E Expression of CD133 and Nestin in the GSC spheres determined using immunofluorescence staining. F Expression of the GSC marker genes (OCT4, SOX2, and Nanog) determined using RT-qPCR; *p < 0.05 compared with the U251 group. G Expression of HULC in GSCs determined using RT-qPCR; *p < 0.05 compared with the U251 group. All experiments were repeated three times.
Furthermore, flow cytometry was employed to sort the CD133+ GSCs (Fig. 1C), while the GSCs cultured in vitro formed a significantly greater number of spheroids (Fig. 1D). As depicted in Fig. 1E, immunofluorescence staining revealed increased expression levels of the CD133 and Nestin proteins in the GSC spheres. RT-qPCR results further indicated that the expression levels of the GSC marker genes OCT4, SOX2, and Nanog were all increased in the GSCs (Fig. 1F). In addition, the expression levels of HULC were upregulated in the GSCs (Fig. 1G). Together, these findings revealed that HULC was expressed at high levels in glioma tissues and GSCs.
HULC silencing inhibits the stemness of GSCs and promotes apoptosis and differentiation
The expression of HULC in GSCs was knocked down to further explore the specific role of HULC in maintaining the stemness and differentiation of GSCs in subsequent experiments. Based on the results of RT-qPCR detection of the efficiency of two silencing sequences, the expression levels of HULC were obviously lower in the GSCs following the transfection of sh-HULC-1 and sh-HULC-2 compared to those in sh-NC group, which exhibited significantly increased levels compared with cells transfected with the sh-HULC1 sequence (Fig. 2A). Thus, the sh-HULC-1 silencing sequence was employed in subsequent experiments. Sphere formation assay results revealed a remarkably reduced number of GSC spheres following sh-HULC treatment (Fig. 2B). Meanwhile, the proliferation of GSCs was also decreased as a result of HULC silencing (Fig. 2C). In addition, as displayed in Fig. 2D, flow cytometry showed an increasing trend in the percentage of apoptotic GSCs after HULC silencing. The results of immunofluorescence staining showed that sh-HULC transfection reduced the expression of CD133 and Nestin in GSCs (Fig. 2E), while the expression of OCT4, SOX2, and Nanog was also decreased by HULC silencing, as detected using RT-qPCR (Fig. 2F). Moreover, Western blot analysis showed significantly higher expression levels of GFAP in the GSCs following HULC silencing (Fig. 2G). Furthermore, the microscopic observation of cell morphology showed that HULC silencing promoted the differentiation of GSCs (Fig. 2H). Altogether, these findings highlighted that HULC silencing inhibits the stemness and proliferation of GSCs while enhancing apoptosis and differentiation.
A Silencing efficiency of two silencing sequences on HULC expression determined using RT-qPCR. B GSC sphere formation after HULC silencing. C Proliferation of GSCs after HULC silencing. D Apoptosis rate of GSCs after HULC silencing determined using flow cytometry. E Expression of CD133 and Nestin in the GSCs after HULC silencing was determined using immunofluorescence staining. F Expression of the GSC marker genes (OCT4, SOX2, and Nanog) after HULC silencing determined using RT-qPCR. G Expression of the GFAP protein in GSCs after HULC silencing determined using Western blot analysis. H Microscopic views of GSC morphology after HULC silencing. *p < 0.05 compared with the sh-NC group. All experiments were repeated three times.
HULC stabilizes the FOXM1 protein through ubiquitination
Next, we aimed to explore the relationship between HULC and FOXM1 in glioma. The results of immunohistochemical staining (Fig. 3A) indicated markedly higher FOXM1 expression levels in the tumor tissues from patients with GBM relative to those in the control tissues. Pearson’s correlation coefficient suggested an inverse correlation between HULC and FOXM1 expression in GBM clinical samples (Fig. 3A). Furthermore, the results of the Western blot analysis (Fig. 3B) showed markedly higher FOXM1 expression levels in the GSCs and glioma cell lines. As illustrated in Fig. 3C, FOXM1 was detected in the HULC pull-down complex in RNA pull-down assay, while the results of RIP assay showed that the amount of HULC pulled down by FOXM1 was higher than that pulled down by IgG (Fig. 3D). In addition, HULC exerted no remarkable effects on the expression of the FOXM1 mRNA, as detected using RT-qPCR (Fig. 3E). Moreover, consistent with the in vitro experiments, the expression of the FOXM1 mRNA did not change significantly in GBM clinical samples compared to that in control samples (Fig. 3E). Furthermore, HULC silencing significantly downregulated the levels of the FOXM1 protein, as determined using Western blot analysis (Fig. 3F). Meanwhile, no significant differences were observed in the expression of the FOXM1 protein following treatment with MG132 (Fig. 3F). As shown in Fig. 3G, the protein synthesis inhibitor CHX exerted a marked inhibitory effect on HULC-mediated FOXM1 expression. Subsequently, an IP assay was performed, and HULC silencing induced a significant increase in the level of ubiquitination in the GSCs (Fig. 3H). Overall, HULC stabilizes FOXM1 expression in GSCs through ubiquitination.
A Expression of FOXM1 in clinical tissue samples determined using immunohistochemistry and correlation between HULC and FOXM1 expression in GBM clinical samples analyzed by calculating Pearson’s correlation coefficient (n = 50 in the GBM group, n = 35 in the control group); *p < 0.05 compared with the control group. B Expression of the FOXM1 protein in GSCs determined using Western blot analysis; *p < 0.05 compared with the HEB group and #p < 0.05 compared with the U251 group. C Interaction between HULC and FOXM1 analyzed using the RNA-pull down assay. D Interaction between HULC and FOXM1 analyzed using the RIP assay; *p < 0.05 compared with the anti-IgG group. E Expression of the FOXM1 mRNA determined using RT-qPCR in GSCs and GBM clinical samples (n = 50 in the GBM group, n = 35 in the control group). F Expression of the FOXM1 protein determined using Western blot analysis; *p < 0.05 compared with the sh-NC + DMSO group. G Expression of the FOXM1 protein in GSCs treated with CHX as determined using Western blot analysis. H IP assay showing the level of FOXM1 ubiquitination. All experiments were repeated three times.
FOXM1 upregulates the expression of HIF-1α through the activation of AGR2
An analysis of the glioma-related dataset GSE12657 further indicated that the expression of HIF-1α (HIF1A) was prominently upregulated in GBM samples (Fig. 4A). Subsequently, immunohistochemical staining was performed, and patients with GBM presented with significantly increased expression of AGR2 and HIF-1α (Fig. 4B). In addition, based on the Western blot analysis, levels of the AGR2 and HIF-1α proteins were increased in the U251 cell line and GSCs compared to the HEB cell line (Fig. 4C). Furthermore, RT-qPCR was applied to detect the efficiency of two FOXM1 silencing sequences. As shown in Fig. 4D, both the sh-FOXM1-1 and sh-FOXM1-2 sequences resulted in significant reductions in FOXM1 expression, while the sh-FOXM1-1 (sh-FOXM1) sequence was used for subsequent experiments due to its more pronounced silencing effect.
A Expression of HIF-1α in the normal and tumor samples from the GSE12657 dataset. B Immunohistochemical staining showing the expression of AGR2 and HIF-1α in clinical tissue samples (n = 50 in the GBM group, n = 35 in the control group); *p < 0.05 compared with the control group. C Western blots showing AGR2 and HIF-1α levels in GSCs; *p < 0.05 compared with the HEB group and #p < 0.05 compared with the U251 group. D Expression of the FOXM1 mRNA determined using RT-qPCR; *p < 0.05 compared with the sh-NC group. E Enrichment of FOXM1 at the AGR2 promoter region determined using the ChIP assay; *p < 0.05 compared with the sh-NC group. F Binding of FOXM1 to the AGR2 promoter determined using dual-luciferase gene reporter assay; *p < 0.05 compared with the sh-NC group. G Expression of the AGR2 mRNA after FOXM1 silencing as determined using RT-qPCR; *p < 0.05 compared with the sh-NC group. H Expression of the AGR2 mRNA after AGR2 overexpression determined using RT-qPCR; *p < 0.05 compared with the oe-NC group; (I) Expression of the FOXM1, AGR2, and HIF-1α mRNAs after FOXM1 silencing and AGR2 overexpression, as determined using RT-qPCR; *p < 0.05 compared with the sh-NC + oe-NC group and #p < 0.05 compared with the sh-FOXM1 + oe-NC group. All experiments were repeated three times.
Furthermore, aiming to validate whether FOXM1 transcriptionally activates AGR2 expression, we initially conducted a ChIP assay, which indicated that the enrichment of FOXM1 in the AGR2 promoter region was significantly reduced following sh-FOXM1 treatment (Fig. 4E). In addition, as suggested by the dual-luciferase gene reporter assay, AGR2-Wt and sh-FOXM1 cotransfection resulted in reduced a fluorescence signal for AGR2, while no significant changes were observed following cotransfection with AGR2-Mut and sh-FOXM1 (Fig. 4F). Next, RT-qPCR was further performed, and the AGR2 mRNA expression levels were decreased after FOXM1 knockdown (Fig. 4G), indicating that FOXM1 targets AGR2 and transcriptionally activates its expression. As manifested in the RT-qPCR results presented in Fig. 4H, oe-AGR2 plasmids significantly upregulated the expression of AGR2 in the cells. Furthermore, the expression of FOXM1, AGR2, and HIF-1α was decreased in cells following sh-FOXM1 + oe-NC treatment. Compared to the sh-FOXM1 + oe-NC treatment, treatment with sh-FOXM1 + oe-AGR2 upregulated the expression of AGR2 and HIF-1α, but did not alter FOXM1 expression (Fig. 4I). Collectively, these data revealed the activation of the AGR2/HIF-1α axis by FOXM1.
FOXM1 facilitates glycolysis in GSCs by activating the AGR2/HIF-1α axis
After discovering that the FOXM1-activated AGR2/HIF-1α axis may participate in glioma development, we focused on determining its effects on glycolysis in GSCs. As detected using RT-qPCR, GSCs treated with oe-HIF-1α presented significantly increased expression levels of HIF-1α compared with GSCs treated with oe-NC (Fig. 5A). In addition, compared to the sh-NC treatment, both sh-HIF-1α-1 and sh-HIF-1α-2 resulted in downregulated expression of HIF-1α, where the sh-HIF-1α-1 treatment resulted in more pronounced downregulation (Fig. 5B). Therefore, oe-HIF-1α and sh-HIF-1α-1 (sh-HIF-1α) were used in subsequent experiments. Meanwhile, the results of RT-qPCR revealed that, compared with sh-NC + oe-NC treatment, the expressions of FOXM1, AGR2, and HIF-1α were all decreased following sh-FOXM1 + oe-NC treatment. Relative to GSCs following transfection of sh-FOXM1 + oe-NC, GSCs transfected with sh-FOXM1 + oe-AGR2 presented with increased expressions of AGR2, while those transfected with sh-FOXM1 + oe-HIF-1α exhibited increased expressions of HIF-1α. In addition, relative to sh-FOXM1 + oe-AGR2 treatment, the expression of HIF-1α was found to be decreased as a result of sh-FOXM1 + oe-AGR2 + sh-HIF-1α treatment (Fig. 5C). Next, we further detected the glycolysis of GSCs. As indicated by the Western blot analysis, GLUT1 expression was significantly decreased in the GSCs following sh-FOXM1 transfection but was increased upon additional transfection with oe-AGR2 or oe-HIF-1α. Compared with the transfection of sh-FOXM1 + oe-AGR2, GLUT1 expression exhibited a decreasing trend following transfection with sh-FOXM1 + oe-AGR2 + sh-HIF-1α (Fig. 5D). Next, we evaluated the glucose uptake and lactate production of GSCs transfected with different constructs. The results are illustrated in Fig. 5E and revealed that FOXM1 silencing inhibited the glucose uptake and lactate production of GSCs, while these processes were enhanced by AGR2 or HIF-1α overexpression. Compared with the GSCs transfected with sh-FOXM1 + oe-AGR2, glucose uptake and lactate production were decreased in the GSCs transfected with sh-FOXM1 + oe-AGR2 + sh-HIF-1α (Fig. 5E). Collectively, these findings suggested that FOXM1 promotes glucose uptake and lactate production in GSCs by activating the AGR2/HIF-1α axis.
A The expression of HIF-1α in the GSCs transfected with oe-HIF-1α was determined using RT-qPCR; *p < 0.05 compared with the oe-NC group. B Efficiency of two silencing sequences at silencing HIF-1α expression determined using RT-qPCR; *p < 0.05 compared with the sh-NC group. C Expression of FOXM1, AGR2, and HIF-1α in the GSCs transfected with different constructs, as determined using RT-qPCR. D Expression of the GLUT1 protein in the GSCs transfected with different constructs, as determined using Western blot analysis. E Detection of glucose uptake and lactate production in the GSCs transfected with different constructs; *p < 0.05 compared with the sh-NC + oe-NC group, #p < 0.05 compared with. the sh-FOXM1 + oe-NC group, and ^p < 0.05 compared with the sh-FOXM1 + oe-AGR2 group; All experiments were repeated three times.
HULC regulates glycolysis and the stemness of GSCs through the FOXM1/AGR2/HIF-1α axis
In addition, we investigated whether HULC affects glycolysis and the stemness of GSCs through the FOXM1/AGR2/HIF-1α axis. As illustrated in Fig. 6A, the RT-qPCR results showed significantly decreased expression of HULC, FOXM1, AGR2 and HIF-1α in the GSCs after sh-HULC + oe-NC + DMSO treatment compared with the sh-NC + oe-NC + DMSO treatment. Compared to the GSCs transfected with sh-HULC + oe-NC + DMSO, HIF-1α expression was increased in cells transfected with sh-HULC + oe-HIF-1α + DMSO, while no significant differences in the expression levels of HULC, FOXM1 and AGR2 were observed. Meanwhile, no significant differences were observed in the expression of HULC, FOXM1, AGR2 and HIF-1α between the GSCs treated with sh-HULC + oe-HIF-1α + DMSO treatment and those treated with sh-HULC + oe-HIF-1α + WZB117 (Fig. 6A). In addition, a Western blot analysis was performed, and treatment with sh-HULC + oe-NC + DMSO reduced levels of the GLUT1 protein in the GSCs compared with the sh-NC + oe-NC + DMSO treatment. Compared with the GSCs treated with sh-HULC + oe-NC + DMSO, GLUT1 protein levels were increased in cells treated with sh-HULC + oe-HIF-1α + DMSO; meanwhile, lower levels of the GLUT1 protein were detected in GSCs treated with sh-HULC + oe-HIF-1α + WZB117 compared to those in the sh-HULC + oe-HIF-1α + DMSO treatment group (Fig. 6B). In addition, glucose uptake and lactate production in GSCs exposed to different treatments were evaluated, and the experimental data are presented in Fig. 6C. Compared with the sh-HULC + oe-NC + DMSO treatment, sh-HULC + oe-NC + DMSO treatment resulted in decreased glucose uptake and lactate production. Compared with the GSCs treated with sh-HULC + oe-NC + DMSO, glucose uptake and lactate production were increased following treatment with sh-HULC + oe-HIF-1α + DMSO. However, compared to the sh-HULC + oe-HIF-1α + DMSO treatment, the GSCs presented significantly reduced glucose uptake and lactate production after the sh-HULC + oe-HIF-1α + WZB117 treatment.
A Expression of HULC, FOXM1, AGR2, and HIF-1α in the GSCs transfected with different constructs, as determined using RT-qPCR. B Expression of the GLUT1 protein in the GSCs transfected with different constructs, as determined using Western blot analysis. C Detection of glucose uptake and lactate production in the GSCs transfected with different constructs. D Number of spheres formed by GSCs transfected with different constructs. E Proliferation of the GSCs transfected with different constructs. F Expression of CD133 and Nestin in the GSCs transfected with different constructs, as determined using immunofluorescence staining. G Expression of the GSC marker genes (OCT4, SOX2, and Nanog) in the GSCs transfected with different constructs, as determined using RT-qPCR. H Expression of the GFAP protein in the GSCs transfected with different constructs, as determined using Western blot analysis. I Apoptosis of the GSCs transfected with different constructs, as determined using flow cytometry; *p < 0.05 compared with the sh-NC + oe-NC + DMSO group, #p < 0.05 compared with the sh-HULC + oe-NC + DMSO group, and ^p < 0.05 compared with the sh-HULC + oe-HIF-1α + DMSO group. All experiments were repeated three times.
Furthermore, the maintenance of GSC stemness and the differentiation of GSCs were evaluated. Compared with the sh-HULC + oe-NC + DMSO treatment, the sh-HULC + oe-NC + DMSO treatment significantly reduced the sphere number (Fig. 6D) and suppressed cell proliferation (Fig. 6E). In addition, GSCs treated with sh-HULC + oe-NC + DMSO exhibited decreased levels of the CD133 and Nestin proteins (Fig. 6F), reduced expression of the OCT4, SOX2 and Nanog mRNAs (Fig. 6G), and increased expression of the GFAP protein (Fig. 6H) and cell apoptosis (Fig. 6I). In GSCs, sh-HULC + oe-HIF-1α + DMSO treatment led to an increase in the sphere number (Fig. 6D), reduced cell proliferation (Fig. 6E), increased levels of the CD133 and Nestin proteins (Fig. 6F), elevated mRNA expression of OCT4, SOX2, and Nanog (Fig. 6G), decreased expression of the GFAP protein (Fig. 6H) and reduced cell apoptosis (Fig. 6I) compared to the sh-HULC + oe-NC + DMSO treatment. In addition, compared with GSCs treated with sh-HULC + oe-HIF-1α + DMSO, GSCs treated with sh-HULC + oe-NC + DMSO showed a significantly reduced sphere number (Fig. 6D), decreased cell proliferation (Fig. 6E), decrease levels of the CD133 and Nestin proteins (Fig. 6F), reduced expression of the OCT4, SOX2 and Nanog mRNAs (Fig. 6G), increased expression of the GFAP protein (Fig. 6H), and the induction of cell apoptosis (Fig. 6I). Furthermore, we overexpressed HULC in HEB cells (Supplementary Fig. 1A) and observed increased levels of FOXM1, AGR2, HIF-1α and GLUT1 increased in HEB cells overexpressing HULC (Supplementary Fig. 1B). Moreover, increasing trends were observed in cell proliferation (Supplementary Fig. 1C), the expression of CD133, Nestin (Supplementary Fig. 1E), OCT, SOX2 and NANOG (Supplementary Fig. 1F), and sphere formation ability (Supplementary Fig. 1G), while cell apoptosis (Supplementary Fig. 1D) was reduced following HULC overexpression. In addition, glucose uptake and lactate production were increased following HULC overexpression (Supplementary Fig. 1H), indicating increased glycolysis. Based on these findings, HULC regulates glycolysis in GSCs to inhibit GSC proliferation and promote apoptosis and differentiation through the FOXM1/AGR2/HIF-1α axis.
HULC promotes glioma development through the FOXM1/AGR2/HIF-1α axis in vivo
We detected the effect of HULC/FOXM1/AGR2/HIF-1α regulatory axis on tumor formation by conducting subcutaneous tumor formation experiments to further explore the role of HULC/FOXM1/AGR2/HIF-1α axis in the development of glioma. The Western blot analysis showed that compared with the oe-NC treatment, the oe-HULC-low treatment increased the expression of HULC, which was further significantly increased after the oe-HULC-high treatment (Fig. 7A). Mice treated with oe-HULC-low and oe-HULC-high exhibited an increased tumor volume and weight, changes that were more significant after oe-HULC-high treatment (Fig. 7B). As revealed by TUNEL and Ki67 staining, both of oe-HULC-low and oe-HULC-high treatments led to a significantly reduced cell apoptosis rate and increased cell proliferation rate in tumors, especially tumors from mice receiving the oe-HULC-high treatment (Fig. 7C, D).
A Expression of HULC in the brain tissues from oe-HULC-low- or oe-HULC-high-treated mice determined using RT-qPCR. B Tumor volumes and weights of oe-HULC-low- or oe-HULC-high-treated mice. C TUNEL-positive cells in the brain tissues from oe-HULC-low- or oe-HULC-high-treated mice. D Immunohistochemical staining showing Ki67-positive cells in the brain tissues from oe-HULC-low- or oe-HULC-high-treated mice. E Expression of HULC in each group of mice determined using RT-qPCR. F Expression of the FOXM1, AGR2, and HIF-1α proteins in each group of mice, as determined using Western blot analysis. G Representative images of the tumors in nude mice, with the measurements of the tumor volume and tumor weight. H Cell apoptosis determined using TUNEL staining. I Immunohistochemical staining of Ki67-positive cells. *p < 0.05 compared with the sh-NC + oe-NC group or the oe-NC group and #p < 0.05 compared with the sh-HULC + oe-NC group or the oe-HULC-low group; n = 10 mice in each group. All experiments were repeated three times.
Last, we aimed to identify the role of HULC in the development of glioma in vivo. RT-qPCR and Western blot analyses showed reduced expression of the HULC, FOXM1, AGR2, and HIF-1α mRNAs and proteins in response to the sh-HULC + oe-NC treatment, while the sh-HULC + oe-HIF-1α treatment exerted no significant effects on the expression of HULC, FOXM1, and AGR2, but upregulated HIF-1α expression compared to the sh-HULC + oe-NC treatment (Fig. 7E, F). In addition, the tumor volume and weight in the nude mice treated with sh-HULC + oe-NC were decreased relative to those treated with sh-NC + oe-NC, but were increased after sh-HULC + oe-HIF-1α treatment (Fig. 7G). TUNEL staining results showed a significantly increased apoptosis rate in nude mice treated with sh-HULC + oe-NC compared to nude mice treated with sh-NC + oe-NC, which was inhibited by the sh-HULC + oe-HIF-1α treatment (Fig. 7H). As suggested in Fig. 7I, Ki67 detection indicated a decrease in the cell proliferation rate after sh-HULC + oe-NC treatment compared with sh-NC + oe-NC treatment, which was promoted by sh-HULC + oe-HIF-1α treatment.
Collectively, these findings suggested that HULC promotes glioma development by regulating the FOXM1/AGR2/HIF-1α axis in vivo.
Discussion
Glioma, a malignancy of the brain arising from glial cells, is one of the most commonly occurring intracranial tumors16. The upregulation of HULC promotes glioma development9. The current study aimed to elucidate the role of HULC in glioma and found that HULC contributes to the development of glioma through the FOXM1/AGR2/HIF-1α axis.
First, findings obtained in our study indicated that HULC was expressed at high levels in the brain tissues from patients with GBM and GSCs, which suggested an effect of HULC on promoting glioma development. Moreover, as an lncRNA, HULC was recently identified as a key regulator of the progression of various malignancies, such as hepatocellular carcinoma, gastric cancer, and prostate cancer12,17,18. Consistent with our findings, a previous study documented the aberrant upregulation of HULC in gliomas, where its enforced expression promoted the occurrence and progression of glioma9. Meanwhile, analyses of the obtained GBM samples in our study revealed that a large proportion IDH1-Mut cases presented low expression of HULC, whereas the majority of IDH1-Wt cases exhibited high expression of HULC. The aforementioned finding is particularly important, as the existing literature indicates that the prognosis of patients with IDH1-Wt GBM is significantly lower than that of patients with IDH1-Mut GBM19. Notably, another study recently suggested that HULC represents a prognostic marker of glioma9, while HULC was also reported to promote GBM cell proliferation, invasion, and migration by regulating RRAS expression20. Here, the current study revealed that HULC promotes the stemness of GSCs through the FOXM1/AGR2/HIF-1α axis and induces the occurrence and development of glioma. Therefore, we propose that HULC expression is potentially useful as a prognostic marker of GBM, and patients with high HULC expression may have a poor prognosis. To some extent, this finding is consistent with the use of IDH1-Mut as a prognostic marker of GBM. In addition, HULC silencing suppressed the stemness of GSCs, which further inhibited cell proliferation and promoted apoptosis and differentiation in our study. On the other hand, HULC overexpression was reported to induce cell proliferation, migration, invasion, while inhibiting the apoptosis of epithelial ovarian carcinoma cells21. Furthermore, various studies have highlighted the oncogenic role of HULC in glioma. For example, Zhu et al. illustrated the proangiogenic activity of HULC in glioma, as evidenced by the inhibited angiogenesis, restrained glioma cell proliferation and invasion after HULC silencing10. Overall, the aforementioned findings support our results of the role of HULC in promoting glioma development and the inhibitory effect of HULC silencing on promoting tumor cell stemness.
Furthermore, mechanistic experiments in our study indicated that HULC stabilized FOXM1 through ubiquitination and promoted the expression of AGR2/HIF-1α in glioma. Elaborating on the interaction between HULC and FOXM1, a previous study found that HULC suppressed autophagy and reduced the CDDP resistance of drug-resistant gastric cancer cells by regulating FOXM112. In fact, an increasing number of studies have documented the involvement of FOXM1 in glioma13,22,23. According to Zhang et al., nuclear translocation of FOXM1 is vital for FOXM1-mediated β-catenin nuclear accumulation in glioma tumor cells, which further controls glioma formation and development24. Moreover, FOXM1 activates AGR2, which promotes the development of the tumors from lung adenomas to invasive mucinous adenocarcinomas25. Similarly, Hong and colleagues showed that AGR2 expression is regulated by HIF-1, which is implicated in the growth and angiogenesis of GBM14. In addition, glucose metabolism induced by AGR2 may activate the MUC1/HIF-1α pathway to promote the development of endometrial carcinoma26. In our study, FOXM1 enhanced the glycolysis of GSCs by activating the AGR2/HIF-1α axis. This result is consistent with a previous study showing that FOXM1 regulates glycolysis in hepatocellular carcinoma cells through the transactivation of GLUT1 expression27. Together, these findings highlight the involvement of the FOXM1-regulated AGR2/HIF-1α axis in HULC-mediated changes in the glycolysis and stemness of GSCs.
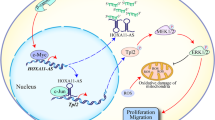
In conclusion, findings obtained in the current study indicated that HULC stabilizes the expression of FOXM1 by ubiquitination to activate the AGR2/HIF-1α axis, thereby regulating glycolysis and thus promoting the stemness of GSCs, which ultimately accelerates the development of glioma (Fig. 8). Our discoveries might provide insights into novel treatment strategies for glioma. However, further investigations are warranted to determine how HULC regulates the ubiquitination of FOXM1, as the types of E3 ubiquitinase and deubiquitinase required and the involved miRNAs are uncharacterized. In addition, we have only conducted experiments using xenograft tumor models in nude mice, and a large number of experiments are needed to verify the feasibility of translating this work into the clinic for better treatment of GBM in the near future.
Data availability
The datasets generated and/or analyzed during the current study are available from the corresponding author upon reasonable request.
References
Ostrom, Q. T. et al. CBTRUS statistical report: primary brain and other central nervous system tumors diagnosed in the United States in 2009–2013. Neuro. Oncol. 18, v1–v75 (2016).
Rasmussen, B. K. et al. Epidemiology of glioma: clinical characteristics, symptoms, and predictors of glioma patients grade I-IV in the the Danish Neuro-Oncology Registry. J. Neurooncol. 135, 571–579 (2017).
Gladson, C. L., Prayson, R. A. & Liu, W. M. The pathobiology of glioma tumors. Annu. Rev. Pathol. 5, 33–50 (2010).
Chen, R., Smith-Cohn, M., Cohen, A. L. & Colman, H. Glioma subclassifications and their clinical significance. Neurotherapeutics 14, 284–297 (2017).
Lathia, J. D., Mack, S. C., Mulkearns-Hubert, E. E., Valentim, C. L. & Rich, J. N. Cancer stem cells in glioblastoma. Genes Dev. 29, 1203–1217 (2015).
Shi, J. et al. Long non-coding RNA in glioma: signaling pathways. Oncotarget 8, 27582–27592 (2017).
Ghafouri-Fard, S., Esmaeili, M., Taheri, M. & Samsami, M. Highly upregulated in liver cancer (HULC): An update on its role in carcinogenesis. J. Cell Physiol. 235, 9071–9079 (2020).
Klec, C., Gutschner, T., Panzitt, K. & Pichler, M. Involvement of long non-coding RNA HULC (highly up-regulated in liver cancer) in pathogenesis and implications for therapeutic intervention. Expert Opin. Ther. Targets 23, 177–186 (2019).
Yan, H. et al. High expression of long noncoding RNA HULC is a poor predictor of prognosis and regulates cell proliferation in glioma. Onco. Targets Ther. 10, 113–120 (2017).
Zhu, Y. et al. HULC long noncoding RNA silencing suppresses angiogenesis by regulating ESM-1 via the PI3K/Akt/mTOR signaling pathway in human gliomas. Oncotarget 7, 14429–14440 (2016).
He, J. et al. Liver X receptor inhibits the growth of hepatocellular carcinoma cells via regulating HULC/miR-134-5p/FOXM1 axis. Cell Signal 74, 109720 (2020).
Xin, L. et al. METase/lncRNA HULC/FoxM1 reduced cisplatin resistance in gastric cancer by suppressing autophagy. J. Cancer Res. Clin. Oncol. 145, 2507–2517 (2019).
Zhang, X., Lv, Q. L., Huang, Y. T., Zhang, L. H. & Zhou, H. H. Akt/FoxM1 signaling pathway-mediated upregulation of MYBL2 promotes progression of human glioma. J. Exp. Clin. Cancer Res. 36, 105 (2017).
Hong, X. Y., Wang, J. & Li, Z. AGR2 expression is regulated by HIF-1 and contributes to growth and angiogenesis of glioblastoma. Cell Biochem Biophys. 67, 1487–1495 (2013).
Wang, Z. F., Liao, F., Wu, H. & Dai, J. Glioma stem cells-derived exosomal miR-26a promotes angiogenesis of microvessel endothelial cells in glioma. J. Exp. Clin. Cancer Res. 38, 201 (2019).
Ostrom, Q. T., Gittleman, H., Stetson, L., Virk, S. M. & Barnholtz-Sloan, J. S. Epidemiology of gliomas. Cancer Treat Res. 163, 1–14 (2015).
Xin, X. et al. Long noncoding RNA HULC accelerates liver cancer by inhibiting PTEN via autophagy cooperation to miR15a. Mol. Cancer 17, 94 (2018).
Chen, C., Wang, K., Wang, Q. & Wang, X. LncRNA HULC mediates radioresistance via autophagy in prostate cancer cells. Braz J. Med. Biol. Res. 51, e7080 (2018).
Louis, D. N. et al. The 2016 World Health Organization Classification of Tumors of the Central Nervous System: a summary. Acta Neuropathol. 131, 803–820 (2016).
Hu, Y. et al. Quantitative proteomics analysis indicates that upregulation of lncRNA HULC promotes pathogenesis of glioblastoma cells. Onco. Targets Ther. 13, 5927–5938 (2020).
Chen, S. et al. The lncRNA HULC functions as an oncogene by targeting ATG7 and ITGB1 in epithelial ovarian carcinoma. Cell Death Dis. 8, e3118 (2017).
Zhang, S. et al. m(6)A demethylase ALKBH5 maintains tumorigenicity of glioblastoma stem-like cells by sustaining FOXM1 expression and cell proliferation program. Cancer Cell 31, 591–606 e596 (2017).
Tang, J. H. et al. Bortezomib inhibits growth and sensitizes glioma to temozolomide (TMZ) via down-regulating the FOXM1-Survivin axis. Cancer Commun. 39, 81 (2019).
Zhang, N. et al. FoxM1 promotes beta-catenin nuclear localization and controls Wnt target-gene expression and glioma tumorigenesis. Cancer Cell 20, 427–442 (2011).
Milewski, D. et al. FOXM1 activates AGR2 and causes progression of lung adenomas into invasive mucinous adenocarcinomas. PLoS Genet. 13, e1007097 (2017).
Gong, W., Ekmu, B., Wang, X., Lu, Y. & Wan, L. AGR2-induced glucose metabolism facilitated the progression of endometrial carcinoma via enhancing the MUC1/HIF-1alpha pathway. Hum Cell 33, 790–800 (2020).
Shang, R., Pu, M., Li, Y. & Wang, D. FOXM1 regulates glycolysis in hepatocellular carcinoma by transactivating glucose transporter 1 expression. Oncol. Rep. 37, 2261–2269 (2017).
Funding
The study was supported by National Natural Science Foundation of China No.82060560 and Natural Science Foundation of Jiangxi Province 20192BAB205067.
Author information
Authors and Affiliations
Contributions
Y.-P.L. and Y.L. designed the study. Y.-P.L., L.-M.X., and L.-K.C. collated the data, carried out data analyses and produced the initial draft of the manuscript. S.-Y.Z., E.-M.Z., and C.-H.X. contributed to drafting the manuscript. All authors have read and approved the final submitted manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
About this article
Cite this article
Li, YP., Liu, Y., Xiao, LM. et al. Induction of cancer cell stemness in glioma through glycolysis and the long noncoding RNA HULC-activated FOXM1/AGR2/HIF-1α axis. Lab Invest 102, 691–701 (2022). https://doi.org/10.1038/s41374-021-00664-9
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41374-021-00664-9
This article is cited by
-
Forkhead box transcription factors (FOXOs and FOXM1) in glioma: from molecular mechanisms to therapeutics
Cancer Cell International (2023)
-
The anterior gradient homologue 2 (AGR2) co-localises with the glucose-regulated protein 78 (GRP78) in cancer stem cells, and is critical for the survival and drug resistance of recurrent glioblastoma: in situ and in vitro analyses
Cancer Cell International (2022)